Abstract
Acute lymphoblastic leukemia (ALL) is the most common type of childhood cancer, presenting with approximately 5,000 new cases each year in the United States. An interesting enzyme implicated in this disease is terminal deoxynucleotidyl transferase (TdT), a specialized DNA polymerase involved in V(D)J recombination. TdT is an excellent biomarker for ALL as it is overexpressed in ~90% of ALL patients, and these higher levels correlate with a poor prognosis. These collective features make TdT an attractive target to design new selective anti-cancer agents against ALL. In this report, we evaluate the anti-leukemia activities of two non-natural nucleotides designated 5-nitroindolyl-2′-deoxynucleoside triphosphate (5-NITP) and 3-ethynyl-5-nitroindolyl-2′-deoxynucleoside triphosphate (3-Eth-5-NITP). Using purified TdT, we demonstrate that both non-natural nucleotides are efficiently utilized as TdT substrates. However, 3-Eth-5-NITP is poorly elongated, and this observation validates its activity as a chain-terminator for blunt-end DNA synthesis. Cell based experiments validate that the corresponding non-natural nucleoside produces robust cytostatic and cytotoxic effects against leukemia cells that overexpress TdT. The strategic placement of the ethynyl moiety allows the incorporated nucleoside triphosphate to be selectively tagged with an azide-containing fluorophore via “click” chemistry. This reaction allows the extent of nucleotide incorporation to be quantified such that the anti-cancer effects of the corresponding non-natural nucleoside can be self-assessed. The applications of this novel nucleoside are discussed, focusing on its use as a “theranostic” agent that can improve the accuracy of dosing regimens and accelerate clinical decisions regarding therapeutic intervention.
Keywords: DNA polymerization, terminal deoxynucleotidyl transferase, non-natural nucleotides, click chemistry, chemotherapeutic agent
DNA replication is the process by which genetic information is duplicated to produce two identical copies of an organism’s genome (1). This complex biological process typically involves a conglomerate of different proteins working in an orderly fashion (2,3). However, the most important enzyme in DNA replication is the DNA polymerase that adds mononucleotides into a growing primer using a nucleic acid template as a guide for directing each incorporation event. All DNA polymerases characterized to date share a common mechanism for DNA chain synthesis in which a nucleotide is covalently linked to the end of a preexisting DNA chain serving as a primer. Most DNA polymerases perform this reaction using a DNA or RNA template to direct each incorporation event. This template-dependent polymerization reaction results in a DNA chain that is complementary to the template strand and sequenced according to proper Watson-Crick nucleotide base pairing rules. There is, however, one unique polymerase denoted as terminal deoxynucleotidyl transferase (TdT) that performs DNA synthesis without the use of a templating strand (4,5). The biological function of TdT is to add random dNTPs at double-strand DNA breaks that form during V(D)J recombination (6,7). The ability of TdT to randomly incorporate dNTPs promotes immunological diversity during an immune response (8). Indeed, TdT activity increases antigen receptor diversity by facilitating the generation of ~1014 different immunoglobulins and ~1018 unique T cell antigen receptors that are required for the neutralization of potential antigens (9,10).
TdT is also noteworthy for its role in cancer as it is overexpressed in acute lymphocytic leukemia (ALL) and acute myelocytic leukemia (AML) (11–13). While the frequency of TdT overexpression in AML is less than that observed in ALL (~20% vs ~90%, respectively), the levels of this polymerase are still higher than those found in other blood cancers such as chronic lymphocytic leukemia (CLL) or chronic myelocytic leukemia (CML) (13). Furthermore, higher levels of TdT activity correlate with a poor prognosis as lower remission rates are observed in patients with TdT-positive leukemia (36%) compared to patients with normal TdT levels (61%) (14). Finally, ALL is the most common form of childhood cancer in the United States, presenting with approximately 5,000 new cases each year (15). If left untreated, ALL typically causes death within a few months (16).
These collective features have prompted efforts to develop selective inhibitors of TdT as anti-cancer agents against TdT-positive leukemias. One noteworthy example is the nucleoside analog cordycepin (3′-deoxyadenosine) (Figure 1A) that lacks a 3′-OH and thus terminates DNA synthesis after its incorporation into DNA. This analog produces cytotoxic effects against TdT-positive leukemia, especially when combined with the adenosine deaminase inhibitor, deoxycoformycin (17,18). Unfortunately, cordycepin is not a truly selective TdT inhibitor as it is also utilized by template-dependent DNA polymerases involved in chromosomal DNA synthesis (19). The ability of these polymerases to incorporate but not elongate cordycepin terminates DNA synthesis in both cancerous and healthy cells. This non-selective killing can cause adverse side effects such as immunosuppression, fatigue, nausea, vomiting, and alopecia (20).
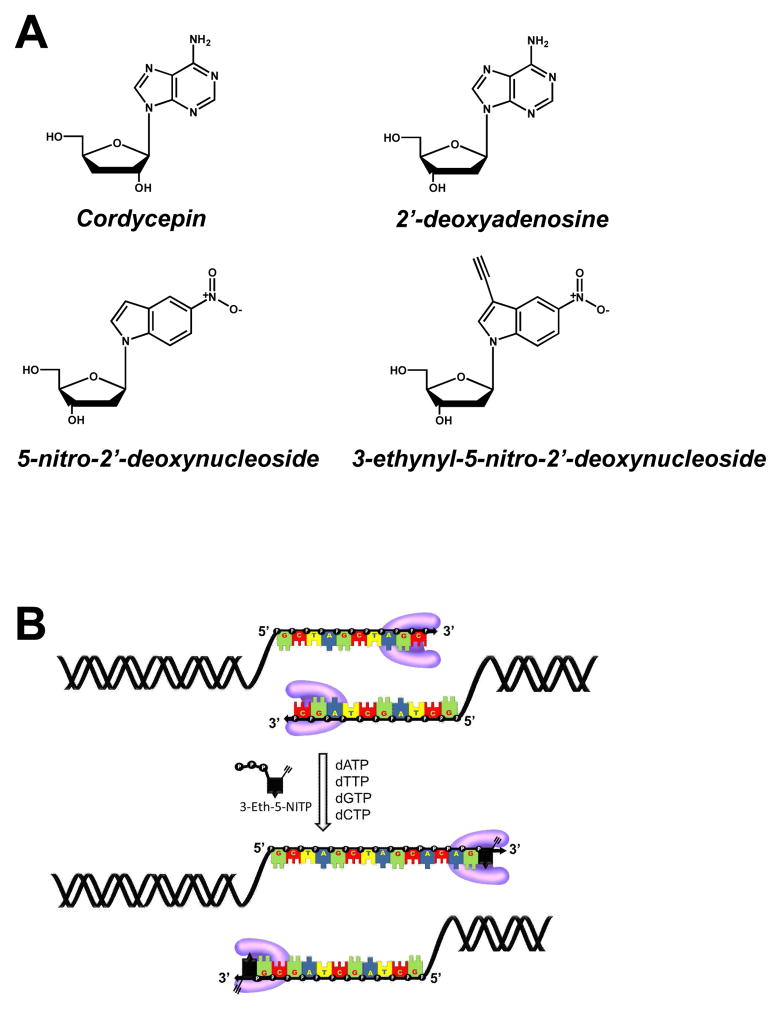
Figure 1.
(A) Chemical structures for cordycepin (3′-deoxyadenosine), 2′-deoxyadenosine, 5-nitroindolyl-2′-deoxyribose, 3-ethynyl-5-nitroindolyl-2′-deoxyribose. (B) Strategy for using “clickable” nucleotides to monitor template-independent DNA synthesis. TdT is designated in pink.
Template-independent synthesis catalyzed by TdT is mechanistically similar to the replication of non-instructional DNA lesions such as abasic sites. We have studied the molecular mechanism for how abasic sites are replicated using non-natural nucleotides that are incorporated with variable efficiencies (21–25). One exceptional analog is 5-nitroindolyl-2′-deoxynucleoside triphosphate (5-NITP) (Figure 1A) as it displays an incredibly high catalytic efficiency of 106 M−1sec−1 for insertion opposite this lesion (21). This value is ~1,000-fold higher than dATP, the preferred natural nucleotide substrate (26). Although 5-NI can pair equally with all four natural nucleobases (27, 28), we demonstrated that the non-natural nucleotide, 5-NITP, is inserted poorly opposite undamaged DNA with low catalytic efficiencies of ~103 M−1sec−1 (21). Finally, we recently described a synthetic protocol to convert 5-NITP into a chemical probe that can visualize and quantify the replication of non-instructional abasic sites under in vitro conditions (29). In this case, placement of an ethynyl moiety at the 3-position of the non-natural nucleotide allows the analog to be tagged with a fluorogenic probe via click chemistry once it is incorporated opposite the DNA lesion (Figure 1B). In this report, we describe the ability of 3-ethynyl-5-nitroindolyl-2′-deoxynucleoside triphosphate (3-Eth-5-NITP) to function as an efficient and potent chain-terminating nucleotide for TdT. In addition, we demonstrate that the corresponding nucleoside, 3-ethynyl-5-nitroindolyl-2′-deoxynucleoside (3-Eth-5-NIdR), functions as a novel theranostic anti-cancer agent against leukemia cells that overexpress TdT. The unique activities of this non-natural nucleoside against ALL highlights the selective inhibition of TdT activity. Potential clinical applications of this novel nucleoside analog are discussed.
RESULTS AND DISCUSSION
Utilization of Non-Natural Nucleotides by TdT
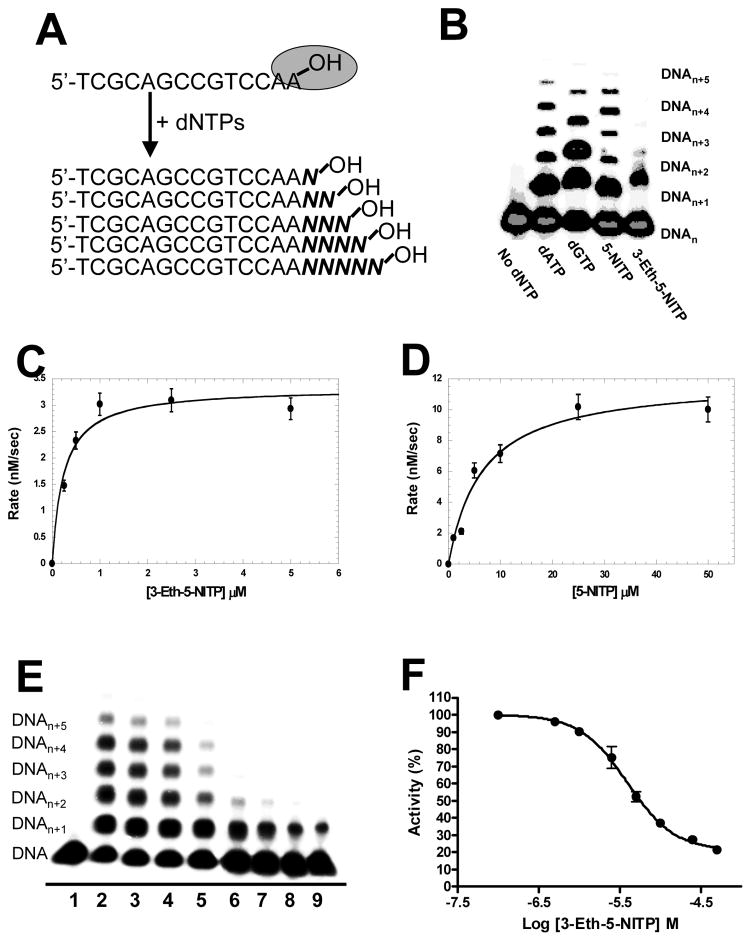
The biological function of TdT is to expand immunological diversity by randomly incorporating dNTPs into single-stranded DNA during V(D)J recombination (8). In vitro studies with purified TdT have demonstrated that while the polymerase utilizes all four natural dNTPs, there is a bias for incorporating dGTP and dCTP versus dATP and dTTP (30). TdT also utilizes nucleotide analogs including 2′,3′-dideoxynucleotides (31), dinucleoside 5′,5′-tetraphosphates (32), and intrinsically fluorescent nucleotide analogs (33). In this report, we tested if TdT also incorporates indolyl-2′-deoxyribose-5′-triphosphates that bear non-natural moieties such as ethynyl and nitro groups at the 3- and 5-position, respectively, of the indole base. Polymerization reactions were performed as outlined in Figure 2A in which 100 μM of natural (dATP and dGTP) or non-natural (5-NITP or 3-Eth-5-NITP) nucleotides were added to a solution containing 6 units of TdT pre-incubated with 1.5 μM single-stranded DNA substrate (14-mer). Aliquots of the reaction were quenched with EDTA at variable time points, and the polymerization reactions were then subjected to denaturing polyacrylamide gel electrophoresis to separate extended primers from unreacted substrate. Representative data provided in Figure 2B shows that both 5-NITP and 3-Eth-5-NITP are utilized by TdT as efficiently as the natural purines, dATP and dGTP as judged by the elongation of 14-mer DNA substrate to longer products.
Figure 2.
Non-Natural Nucleotides are Efficient Substrates for Terminal Deoxynucleotidyl Transferase. (A) DNA substrate and assay used to monitor nucleotide incorporation catalyzed by TdT. (B) Denaturing gel electrophoresis image for the incorporation and extension of dATP, dGTP, 5-NITP, and 3-Eth-5-NITP by TdT. Data points were obtained by quenching an aliquot of the reaction in EDTA (Δt = 2 minutes). (C) Michaelis-Menten plot for the incorporation of 3-Eth-5-NITP by TdT. Rates of incorporation (closed circle) were plotted against 3-Eth-5-NITP concentration. A fit of the data to the Michaelis-Menten equation yielded a Vmax of 3.2 ± 0.1 nM sec−1 and a Km of 0.19 ± 0.04 VM. (D) Michaelis-Menten plot for the incorporation of 5-NITP by TdT. Rates of incorporation (closed circle) were plotted against 5-NITP concentration. A fit of the data to the Michaelis-Menten equation yielded a Vmax of 11.5 ± 0.4 nM/sec and a Km of 4.6 ± 0.6 VM. (E) Representative gel image illustrating the dose-dependence of the chain-termination capabilities of 3-Eth-5-NITP. The assay was performed using 6 units TdT preincubated with 1.5 VM 32P-radio-labeled 14-mer DNA substrate in 100 mM cacodylate buffer (pH 6.8) containing 1 mM CoCl2, and 0.1 mM DTT. The reaction was initiated by the addition of 10 μM dNTPs in the absence and presence of 3-Eth-5-NITP. Lane 1 = 14-mer DNA alone. Lane 2 = 10 VM dNTPs. Lane 3 = 10 VM dNTPs + 0.5 VM 3-Eth-5-NITP. Lane 4 = 10 VM dNTPs + 1 VM 3-Eth-5-NITP. Lane 5 = 10 VM dNTPs + 2.5 VM 3-Eth-5-NITP. Lane 6 = 10 VM dNTPs + 5 VM 3-Eth-5-NITP. Lane 7 = 10 VM dNTPs + 10 VM 3-Eth-5-NITP. Lane 8 = 10 VM dNTPs + 25 VM 3-Eth-5-NITP. Lane 9 = 10 VM dNTPs + 50 VM 3-Eth-5-NITP. Data points were obtained by quenching an aliquot of the reaction in EDTA (Δt = 2 minutes). (F) Dose-response curve used to calculate an IC50 value of 3.9 ± 1.0 VM. The IC50 value of 3.9 +/− 1.0 μM represents an average from three independent experiments performed as described in the text.
The utilization of 5-NITP and 3-Eth-5-NITP by TdT was further quantified by measuring the kinetic parameters, Km, Vmax, and Vmax/Km. Time courses in product formation were generated using pseudo-first order reaction conditions in which 6 units of TdT were incubated with 1.5 μM DNA substrate and mixed with variable concentrations of non-natural nucleotide (0.05–50 μM). Each time course was fit to an equation for a straight line to define the rate of nucleotide incorporation (data not shown). The resulting plot of rate versus 3-Eth-5-NITP concentration is hyperbolic (Figure 2C), and a fit of the data to the Michaelis-Menten equation yields a Vmax of 3.2 +/− 0.1 nM/sec, a Km of 0.19 +/− 0.04 μM, and a Vmax/Km of 0.0168 +/− 0.0004 s−1. Identical experiments performed with 5-NITP yield a Vmax of 11.5 +/− 0.4 nM/sec, a Km of 4.6 +/− 0.6 μM, and a Vmax/Km of 0.0025 +/− 0.0003 s−1 (Figure 2D). Kinetic analyses performed using dATP and dGTP as natural nucleotide substrates are provided as Supplemental Figure 1.
Table 1 summarizes the kinetic parameters for the utilization of natural and non-natural nucleotides by TdT. The catalytic efficiencies for 5-NITP and dATP are essentially identical, thus confirming reports that hydrogen-bonding interactions are not required by TdT for efficient DNA synthesis (34–36). More importantly, the catalytic efficiency for 3-Eth-5-NITP is 7-fold higher than 5-NITP. This enhancement is caused by a 24-fold decrease in the Km for 3-Eth-5-NITP that offsets a 3.5-fold decrease in Vmax. This result is noteworthy as similar effects on Km were observed for incorporating 3-Eth-5-NITP opposite an abasic site (29). These results highlight the universal requirement of nucleobase hydrophobicity and pi-electron interactions during template-independent DNA synthesis (25). However, one important distinction is that TdT extends beyond 5-NITP with an overall efficiency that rivals that of the natural purines, dATP and dGTP (Figure 2B). In contrast, 3-Eth-5-NITP is poorly extended (Figure 2B), indicating that it behaves as a universal chain terminator of DNA synthesis. Indeed, extension beyond 3-Eth-5-NITP is not observed, even after longer reaction times of up to 20 minutes (Supplemental Figure 2).
Table 1.
Summary of kinetic parameters for the incorporation of natural and non-natural nucleotides by terminal deoxynucleotidyl transferase.
| dXTP | Km [μM] | Vmax (nM sec−1) | Vmax/Km (sec−1) |
|---|---|---|---|
| dATP | 1.5 +/− 0.4 | 3.3 +/− 0.3 | 0.0022 +/− 0.0005 |
| dGTP | 8.0 +/− 0.9 | 29.2 +/− 1.1 | 0.0037 +/− 0.0005 |
| 5-NITP | 4.6 +/− 0.6 | 11.5 +/− 0.4 | 0.0025 +/− 0.0003 |
| 3-Eth-5-NITP | 0.19 +/− 0.04 | 3.2 +/− 0.1 | 0.0168 +/− 0.0004 |
The ability of 3-Eth-5-NITP to terminate DNA synthesis catalyzed by TdT was further evaluated through a series of competition experiments. TdT was pre-incubated with single-stranded DNA substrate and then mixed with a solution containing 10 μM dNTPs in the absence and presence of variable concentrations of 3-Eth-5-NITP (0.5–50 μM). Reactions were terminated after 3 minutes by adding 200 mM EDTA, and the replication products were separated via denaturing gel electrophoresis. Representative data provided in Figure 2E shows that in the absence of 3-Eth-5-NITP, TdT randomly incorporates dNTPs to generate replication products with lengths ranging from DNAn+1 to DNAn+5. As the concentration of 3-Eth-5-NITP is increased, there is a concomitant decrease in the amount of replication products greater than DNAn+1. Note that under these conditions, products corresponding to DNAn+1 accumulate since 3-Eth-5-NITP is a non-extendable nucleotide substrate for TdT (vide supra). As such, the percent TdT activity as a function of 3-Eth-5-NITP concentration was plotted (Figure 2F), and the resulting sigmoidal curve was fit to Eqn 3 to yield an IC50 value of 3.9 +/− 1.0 μM for 3-Eth-5-NITP. A true Ki value for 3-Eth-5-NITP of 0.51 +/− 0.13 μM was obtained using the Cheng-Prusoff equation (Eqn 4) to normalize this IC50 value for the concentration of dNTPs and their corresponding Km values (37). The calculated Ki value of 0.51 μM for 3-Eth-5-NITP is in good agreement with the Km value of 0.19 μM measured through initial velocity studies (Table 1). Collectively, these data indicate that 3-Eth-5-NITP is a bona fide chain-terminating substrate for TdT, even in the presence of physiological concentrations of dNTPs.
Defining the Potency of Non-Natural Nucleosides Against ALL Cell Lines
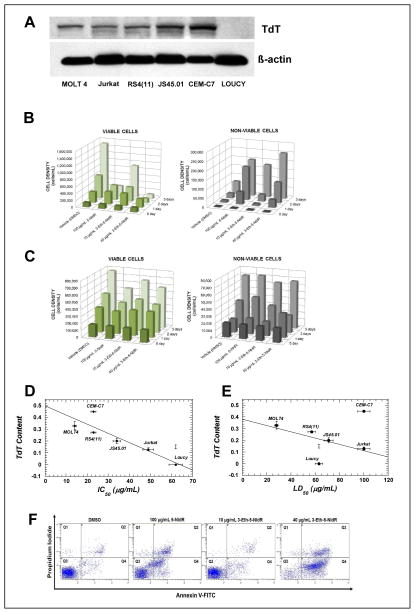
The facile utilization and chain termination capabilities of 3-Eth-5-NITP against TdT suggests that it may have therapeutic potential against ALL. This hypothesis was tested by measuring the cytostatic and/or cytotoxic effects of the corresponding nucleoside, 3-Eth-5-NIdR, and 5-nitroindolyl-2′-deoxynucleoside (5-NIdR) against various ALL cell lines including MOLT4, CEM-C7, Jurkat, RS4(11), JS45.01, and Loucy. Western blot analyses was first performed to determine TdT levels in each ALL cell line. As illustrated in Figure 3A, each cell line contains variable levels of TdT, ranging from none (Loucy) to low (Jurkat, RS4(11)) to high (MOLT4, JS45.01, and CEM-C7). We next tested the efficacy of 5-NIdR and 3-Eth-5-NIdR by treating exponentially growing cells with variable concentrations (0.1–100 μg mL−1) of each non-natural nucleoside for time periods of up to 3 days. Initial experiments used the MOLT4 cell line since this ALL cell line displays resistance to various chemotherapeutic agents due to higher levels of TdT (38). Figure 3B provides representative time courses for the number of viable (left panel) and non-viable (right panel) MOLT4 cells in the absence and presence of 100 μg mL−1 5-NIdR or 3-Eth-5-NIdR (10 and 40 μg mL−1). Treatment with 100 μg mL−1 of 5-NIdR produces robust cytostatic and cytotoxic effects as early as two days post-treatment. In this case, the number of viable cells is reduced ~5-fold while there is a 2.5-fold increase in the number of non-viable cells. Analyses of the time courses in cell growth as a function of nucleoside concentration yield an IC50 of 36.4 +/− 5.8 μg/mL and an LD50 value of ~100 μg mL−1 for 5-NIdR (Supplemental Figure 3). More striking effects are observed with 3-Eth-5-NIdR as strong cytostatic and cytotoxic effects are observed at a low concentration of 10 μg mL−1 (Figure 3B). These effects are also dose-dependent as treatment with 40 μg mL−1 shows a significant reduction in the number of viable cells coupled with a substantial increase in the number of non-viable cells (Figure 3B). Quantitative analyses of the data yield an IC50 of 14.1 +/− 2.4 μg mL−1 and an LD50 value 27.7 +/− 1.7 μg mL−1 value for 3-Eth-5-NIdR (Supplemental Figure 3).
Figure 3.
Correlating the Anti-Cancer Effects of Non-Natural Nucleosides with Cellular Levels of TdT. (A) Western blot analyses examining TdT content in ALL cell lines including MOLT4, Jurkat, RS4(11), JS45.01, CEM-C7, and Loucy. TdT content was normalized against cellular levels of β-actin in each resepective cell line. (B) Time courses in the number of viable (left) and non-viable (right) MOLT4 cells in the absence and presence of non-natural nucleosides. (C) Time courses in the number of viable (left) and non-viable (right) Loucy cells in the absence and presence of non-natural nucleosides. (D) Plot correlating IC50 values of 3-Eth-5-NIdR against various ALL cell lines versus the cellular content of TdT in each cell line. (E) Plot correlating LD50 values of 3-Eth-5-NIdR against various ALL cell lines versus the respective cellular content of TdT in each ALL cell line. (F) Dual parameter flow cytometry of MOLT4 cells treated with (i) DMSO, (ii) 100 μg mL−1 5-NIdR, (iii) 10 μg mL−1 3-Eth-5-NIdR, and (iv) 40 μg mL−1 3-Eth-5-NIdR. Q1 represents cells that are necrotic. Q2 represents late-apoptotic cells. Q3 represents viable cells. Q4 represents early-apoptotic cells.
To interrogate if the cytostatic and cytotoxic effects of 5-NIdR and 3-Eth-5-NIdR are dependent upon the cellular level of TdT, we next measured their effects against the TdT-negative Loucy cell line (vide supra). Data provided in Figure 3C shows that both non-natural nucleosides are significantly less potent against this TdT-negative leukemia cell line compared to the TdT-positive cell line, MOLT4. In particular, treatment with 100 μg mL−1 of 5-NIdR produces only a weak cytostatic effect as the number of viable cells is reduced by <25%. In addition, treatment with 5-NIdR does not cause a substantial increase in the number of non-viable cells. 3-Eth-5-NIdR is also ineffective as significant cytostatic or cytotoxic effects are not observed at a concentration of 40 μg mL−1.
Identical cell-based experiments were performed with the other ALL cell lines, and the corresponding IC50 and LD50 values for 5-NIdR and 3-Eth-5-NIdR against each cell line are summarized in Table 2. Inspection of these data indicates that 3-Eth-5-NIdR is more potent than the parental nucleoside, 5-NIdR. The increased potency of 3-Eth-5-NIdR likely reflects the higher catalytic efficiency for the corresponding nucleoside triphosphate to act as a chain-terminating substrate for TdT. Consistent with this mechanism, the data also show that 3-Eth-5-NIdR displays higher potency against ALL cells that overexpress TdT compared to cells with lower TdT levels. This is best illustrated in Figure 3D which shows a linear relationship between the IC50 values of 3-Eth-5-NIdR in various ALL cell lines as a function of cellular TdT content in each respective cell line. In general, there is an excellent correlation (R2 = 0.695, p< 0.0001) between the anti-cancer effects of the non-natural nucleoside with TdT level. Similar analyses were performed by plotting the LD50 values of 3-Eth-5-NIdR as a function of TdT content (Figure 3E). These data also highlight a correlative effect between the cell killing effects of the non-natural nucleoside with the cellular content of the TdT, the proposed molecular target for the corresponding chain-terminating nucleotide substrate.
Table 2.
Summary of IC50 and LD50 of the non-natural nucleosides against acute lymphoblastic leukemia cell lines.
| Nucleoside | Cell Line | IC50 (μg mL−1) | LD50 (μg mL−1) |
|---|---|---|---|
| 5-NIdR | MOLT4 | 36 +/− 6 | ~100 |
| 3-Eth-5-NIdR | MOLT4 | 14 +/− 2 | 28 +/− 4 |
| 5-NIdR | Loucy | ≫100 | ≫100 |
| 3-Eth-5-NIdR | Loucy | 62 +/− 5 | 63 +/− 3 |
| 5-NIdR | CEM-C7 | ~100 | ≫100 |
| 3-Eth-5-NIdR | CEM-C7 | 23 +/− 2 | ~100 |
| 5-NIdR | Jurkat | ~100 | ≫100 |
| 3-Eth-5-NIdR | Jurkat | 49 +/− 1 | ~100 |
| 5-NIdR | RS4(11) | >100 | ≫100 |
| 3-Eth-5-NIdR | RS4(11) | 23 +/− 2 | 57 +/− 2 |
| 5-NIdR | J45.01 | ~100 | ≫100 |
| 3-Eth-5-NIdR | J45.01 | 34 +/− 6 | 71 +/− 6 |
Defining the Mechanism of Cell Death Induced by Non-Natural Nucleosides
The mechanism by which these non-natural nucleosides induce cell death was interrogated using dual parameter FACS analyses measuring propidium iodide uptake and Alexa Fluor488 annexin V conjugate staining. This allows live cells (unstained with either fluorophore) to be distinguished from cells that are early apoptotic (annexin V staining only), late apoptotic (propidium iodide and annexin V staining), and necrotic (propidium iodide staining only) (39). Representative data provided in Figure 3F shows that MOLT4 cells treated with 5-NIdR have higher levels of early and late stage apoptosis compared to cells treated with DMSO. In this case, treatment with 100 μg mL−1 5-NIdR causes a 5-fold increase in early and late stage apoptosis, respectively. Data summarized in Table 3 shows that the “clickable” nucleoside, 3-Eth-5-NIdR, is significantly more potent as treatment with 10 μg mL−1 causes equivalent levels of apoptosis while treatment with 40 μg mL−1 results in 13- and 7-fold increases in early stage and late stage apoptosis, respectively. Furthermore, cells treated with either 5-NIdR or 3-Eth-5-NIdR show no significant PI uptake, thus indicating that neither non-natural nucleoside causes necrotic cell death. The induction of apoptosis was also independently confirmed by using agarose gel electrophoresis to observe DNA cleavage sites between nucleosomes that occur in chromatin at ~200-base pair intervals (Supplemental Figure 4) (40).
Table 3.
Summary of viable, apoptotic, and necrotic MOLT4 cells after treatment with non-natural nucleosides.
| Condition | Viable Cells (%) | Early Apoptotic Cells (%) | Late Apoptotic Cells (%) | Necrotic Cells (%) |
|---|---|---|---|---|
| DMSO | 89.9 +/− 0.9 | 5.4 +/− 0.1 | 3.5 +/− 1.1 | 1.2 +/− 0.3 |
| 100 μg mL−1 5-NIdR |
53.1 +/− 6.7 | 26.5 +/− 3.7 | 18.9 +/− 3.4 | 1.5 +/− 0.5 |
| 10 μg mL−1 3-Eth-5-NIdR |
83.4 +/− 2.7 | 10.5 +/− 1.5 | 5.4 +/− 1.0 | 0.7 +/− 0.2 |
| 40 μg mL−1 3-Eth-5-NIdR |
6.3 +/− 3.7 | 68.8 +/− 0.9 | 23.7 +/− 4.9 | 1.2 +/− 0.2 |
Diagnostic Activities of Non-Natural Nucleosides
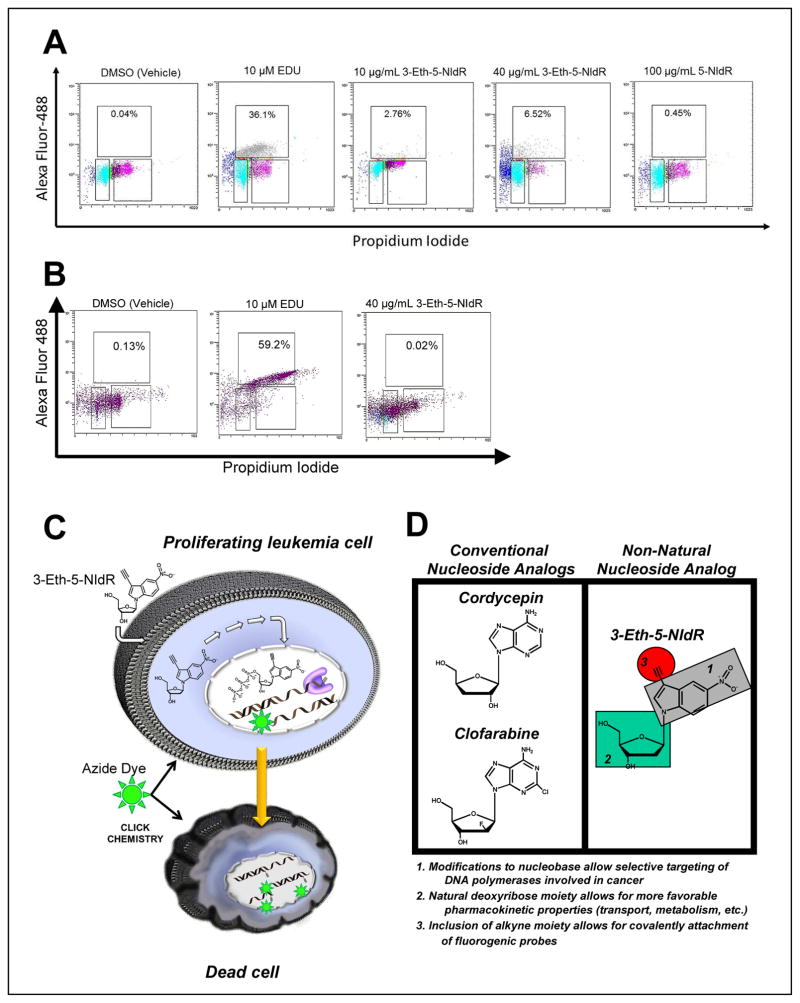
Cell-based experiments were performed to demonstrate that the “clickable” nucleoside, 3-Eth-5-NIdR, functions as a spectroscopic probe to measure the cellular activity of TdT. MOLT4 cells were treated with DMSO, EdU (a “clickable” thymidine analog), 5-NIdR, and 3-Eth-5-NIdR for 2 days and then washed with PBS to remove any nucleoside and/or nucleotide not incorporated into DNA. After fixation and permeabilization, the cells were treated with AlexaFluor488-azide and Cu(I) catalyst to commence clicking of any incorporated non-natural nucleotide. Dual parameter flow cytometry applying PI staining coupled with detection of the AlexaFluor488 fluorophore was used to detect the “clicked” non-natural nucleotide in cellular DNA. As illustrated in Figure 4A, cells treated with DMSO have a low level (<0.5%) of AlexaFluor488 stained DNA while cells treated with 10 μM EdU show significantly higher levels (36.1%). This high amount of “clicked” DNA reflects transport of EdU into the cell, conversion to the corresponding nucleoside triphosphate, and incorporation into the DNA of replicating cells. MOLT4 cells treated with 10 μg mL−1 3-Eth-5-NIdR also show appreciable levels of AlexaFluor488 stained DNA (2.8%) after two days of treatment. Furthermore, the amount of “clicked” DNA is dose-dependent as treatment with 40 μg mL−1 Eth-5-NIdR yields a 2.5-fold increase in AlexaFluor488 staining (6.5%).
Figure 4.
Diagnostic Applications of Non-Natural Nucleotides. (A) Dual parameter flow cytometry of MOLT 4 cells treated with (i) DMSO, (ii) 10 μM EdU, (iii) 10 μg mL−1 3-Eth-5-NIdR, (iv) 40 μg mL−1 3-Eth-5-NIdR and (v) 100 μg mL−1 5-NIdR. (B) Model for the ability of the non-natural nucleoside, 3-Eth-5-NIdR, to function as a theranostic agent against TdT-positive leukemia. (D) Structural comparison between a conventional nucleoside analogs (cordycepin and clofarabine) with the non-natural nucleoside, 3-Eth-5-NIdR.
Identical experiments were performed using the Loucy cell line that lacks TdT and thus is less sensitive to the cytotoxic effects of the non-natural nucleoside (vide infra). Data provided in Figure 4B show that Loucy cells treated with DMSO have low levels of AlexaFluor488 staining (0.13%) while cells treated with 10 μM EdU have significantly higher levels (59.2%). Treatment with 40 μg mL−1 of 3-Eth-5-NIdR produces low levels of “clicked” DNA (0.02%). These levels are significantly lower than those measured in MOLT4 cells under identical conditions and again correlate well with the lack of measurable TdT activity in the Loucy cell line.
Collectively, these data are consistent with the mechanism outlined in Figure 4C in which the non-natural nucleoside enters the cell via the activity nucleoside transporters, undergo catabolism to the corresponding nucleoside triphosphate, and is then utilized by TdT. Once incorporated into DNA, the non-natural nucleotide inhibits cellular DNA replication and induces apoptosis. Finally, it should be emphasized that low levels (<0.5%) of AlexaFluor488 staining are detected in cells treated with 100 μg mL−1 5-NIdR. While this result could reflect a lack of stable incorporation of the non-natural nucleotide into DNA, it is more likely that the lack of the ethynyl moiety precludes covalent attachment of the azide-containing dye.
Biological and Clinical Applications of Non-Natural Nucleosides
Acute lymphocytic leukemia (ALL) is the most common form of childhood cancer (15). As with all cancers, a fundamental feature of ALL is its hyperproliferative nature that is defined by uncontrollable and pro-mutagenic DNA replication. Nucleoside analogs are effective anti-cancer agents against leukemia as they produce anti-proliferative and/or cytotoxic effects by inhibiting DNA replication (41). Despite their widespread utility, however, most nucleoside analogs possess very narrow therapeutic windows that can create significant clinical problems. Indeed, the ability of conventional nucleoside analogs to non-selectively kill healthy cells causes severe side effects such as anemia, leukopenia, thrombocytopenia, and diarrhea/nausea that adversely influence a patient’s response to chemotherapy (20). This problem is exacerbated since it is nearly impossible to rapidly and accurately assess the efficacy of conventional nucleoside analogs. Patient responses to chemotherapy are typically gauged by qualitative criteria ranging from the absence of overt disease symptoms to quantifying the ratio of normal versus cancerous blood cells (42). These clinical endpoints can take weeks or even months to accurately define. As such, the inability to assess drug action on shorter time scales (hours or days) significantly hinders physicians from making informed decisions regarding optimal dosing regimens. Our studies here demonstrate a pragmatic solution to overcome these complications by using 3-Eth-5-NIdR as a theranostic agent to treat leukemia. The theranostic potential of 3-Eth-5-NIdR is evident as it produces robust and potent therapeutic effects against TdT-positive leukemia cells while also possessing diagnostic properties to monitor its self-efficacy. The sections below elaborate on the properties of this unique theranostic agent, specifically comparing and contrasting its novel characteristics with nucleoside analogs currently used to treat leukemia.
Although conventional nucleoside analogs such as fludarabine and gemcitabine influence DNA synthesis on multiple levels (43), their primary effect is to directly inhibit DNA synthesis via incorporation and chain-termination by the corresponding nucleoside triphosphate (44). In these cases, a template-dependent polymerase inserts the analog into DNA as efficiently as its natural counterpart. However, the absence of a usable 3′-OH group prematurely terminates DNA synthesis by creating a nucleic acid substrate that is refractory to elongation. The inability to complete DNA synthesis in a timely fashion stalls replication fork progression and activates apoptosis to cause cell death. The in vitro studies described here clearly show that 3-Eth-5-NITP is a potent chain-terminator for template-independent synthesis catalyzed by TdT. Indeed, the measured Km value of 200 nM for 3-Eth-5-NITP is ~10-fold lower than that for natural purine nucleotides such as dATP. In addition, 3-Eth-5-NITP is highly selective for template-independent DNA synthesis catalyzed by TdT since it is poorly incorporated opposite normal templating bases (29). These activities alone represent important features required for a potential therapeutic agent. However, 3-Eth-5-NIdR also possesses several other important pharmacological and pharmacokinetic features that make it distinct from conventional nucleoside analogs. Figure 4D highlights three important distinctions between 3-Eth-5-NIdR and conventional nucleosides. First, 3-Eth-5-NIdR contains a unique nucleobase devoid of classical hydrogen-bonding functional groups. Secondly, the presence of an ethynyl moiety allows for facile covalent attachment of fluorogenic molecules. Finally, 3-Eth-5-NidR contains a natural deoxyribose moiety rather than a modified sugar. All three features significantly impact the biological function of 3-Eth-5-NIdR. As discussed earlier, the presence of the non-natural nitro moiety allows for TdT, a specialized DNA polymerase involved in replicating DSBs, to efficiently incorporate 5-NITP and 3-Eth-5-NITP. This feature promotes selectivity to inhibit template-independent DNA synthesis. Indeed, the increased utilization of these analogs by TdT provides a reasonable mechanism to explain their higher potencies against ALL cells with elevated TdT levels compared to those with lower levels of TdT.
While conventional nucleoside analogs function as effective therapeutic agents against leukemia, none possess diagnostic capabilities to monitor their self-effectiveness. The work provided here demonstrates proof-of-concept for the use of 3-Eth-5NIdR as a valuable “theranostic” agent against leukemia. The presence of an ethynyl moiety was used to directly and selectively attach a fluorogenic probe to the non-natural nucleotide after it was incorporated into DNA. This reaction not only validates the mechanism of action of the nucleoside at the cellular level, but it also confirms its diagnostic potential by equating the amount of nucleotide incorporation into genomic DNA with the therapeutic activity of the corresponding non-natural nucleoside. This technology represents an important clinical feature that can be used to directly quantify the location and concentration of the therapeutic nucleotide in patient samples. Indeed, theranostic agents such as 3-Eth-5-NIdR can allow physicians to adjust the dose of nucleoside rapidly and accurately to optimize its therapeutic effectiveness. These features could alleviate possible adverse side effects associated with chemotherapy capability and thus revolutionize patient care by achieving optimal levels of a drug to kill cancer cells without harming normal cells.
The final point to emphasize is that the inclusion of a natural deoxyribose moiety may also provide improved pharmacokinetic features compared to conventional nucleoside analogs. For example, natural nucleosides and their anti-cancer counterparts enter cells via the activity of various equilibrative and concentrative nucleoside transporters (45). The major recognition element for efficient transport is the presence of a natural (deoxy)sugar moiety (46). Transport activity is highly sensitive to the correct sugar conformation as nucleoside analogs such as AZT (zidovudine) and ddC (zalcitabine) that lack a 3′-hydroxyl group show poor cellular uptake (47, 48). As such, the presence of an unmodified deoxyribose group on 5-NIdR and 3-Eth-5-NIdR may facilitate their cellular transport and provide an advantage over conventional nucleoside analogs such as fludarabine and gemcitabine that use modified sugar moieties to inhibit DNA synthesis. The presence of a correct deoxyribose moiety on 5-NIdR and 3-Eth-5-NIdR may also facilitate their conversion to the triphosphate form that is required for incorporation into nucleic acid (49). However, one concern regarding the potential use of 5-NIdR as a therapeutic agent in humans or in animal models is the presence of the nitro pharmacophore. It is well established that certain nitroaromatic compounds can undergo nitroreductive bioactivation reactions which can generate potentially reactive nitroanion radicals, nitroso intermediates, and N-hydroxy derivatives that can react with biological nucleophiles (DNA, RNA, and/or protein) (50). Indeed, therapeutic agents such as flutamide and nitrofurantoin that contain nitroaromatic moieties can produce idiosyncratic liver injury. However, the presence of a nitro moiety on a drug does not unequivocally indicate a high risk for producing toxic effects. For example, several biologically active nitroheterocyclic compounds such as 2- and 5-nitroimidazoles and 5-nitrofurans function as therapeutic agents against protozoan and microbial infections (51). Regardless, these discussions make it clear that metabolic and toxicological studies are needed to evaluate the safety of 5-NIdR as a potential theranostic agent.
METHODS
Materials
Magnesium acetate, MgCl2 and Trizma base were from Sigma. [γ-32P]-ATP was purchased from Perkin Elmer Life. Urea, acrylamide, and bis-acrylamide were from National Diagnostics. All oligonucleotides were synthesized by Operon Technologies. Antibodies directed against TdT were from Santa Cruz Biotechnology. The primary antibody for β-actin was obtained from Cell Signaling Technology, Inc. Horseradish peroxidase (HRP)-conjugated anti-mouse antibody was from Jackson ImmunoResearch Laboratories, Inc. All other materials were obtained from commercial sources and were of the highest quality available. Terminal deoxynucleotidyl transferase was purchased from Fisher Scientific. MOLT4, CEM-C7, Jurkat, Loucy, JS45.01, and RS4(11) cell lines were purchased from ATCC. Phosphate-buffered saline (PBS), antibiotic and anti-fungal agents, amphotericin, L-glutamine (200 mM), RIPA buffer, Novex 4–20% Tris-Glycine gel, 0.2-μm nitrocellulose membranes, apoptosis assay kit containing AlexaFluor488-labeled Annexin V, and “click” reagents containing EdU, AlexaFluor488 azide, saponin-based permeabilization solution, and copper sulfate that are used for cell-culture studies are from Invitrogen.
Polymerization Assays
All enzymatic assays were performed as previously described (26). Briefly, Vmax and Km values were determined using pseudo-first order reaction conditions in which 6 units TdT was preincubated with single-stranded DNA substrate (1.5 μM) in an assay buffer and mixed with variable concentrations of the nucleotide analogue (0.05–50 VM). Reactions were quenched with 200 mM EDTA at variable times (5 – 600 s) and analyzed using denaturing gel electrophoresis. Time courses in product formation were fitted using equation 1:
| (1) |
y is the amount of product, m is the rate of the reaction, t is time, and b is the Y-intercept. Km and kcat values were determined by fits of the data points to the Michaelis-Menten equation (equation 2):
| (2) |
where kcat is the maximal turnover number of the polymerase, Km is the Michaelis constant for dNTP, and [dNTP] is the concentration of nucleotide substrate.
The IC50 value for 3-Eth-5-NITP was obtained using a non-linear regression curve fit of the data to Equation 3:
| (3) |
where y is the fraction of TdT activity, IC50 is the concentration of 3-Eth-5-NITP to inhibit 50% TdT activity, and Inhibitor is the concentration of 3-Eth-5-NITP tested. The Cheng-Prusoff equation (Eq 4) was used to define a true Ki value for 3-Eth-5-NITP by normalizing the measured IC50 value for the concentration of dNTPs and their corresponding Km values used in the experiments.
| (4) |
where Ki is the true inhibition constant for 3-Eth-5-NITP, IC50 is the concentration of 3-Eth-5-NITP that inhibits 50% TdT activity, [dNTP] is the concentration of nucleotide substrate and Km dNTP is the Michaelis constant for dNTP.
Cell Culture Procedures
All cells were cultured in a humidified atmosphere of 5% CO2 at 37° C. MOLT4, Jurkat, J45.01, and RS4(11) cells were maintained in ATCC-formulated RPMI-1640 media supplemented with 10% fetal bovine serum (FBS), 5% L-glutamine, and 2.5% penicillin/streptomycin. Loucy and CEM-C7 cells were maintained in Cellgro® formulated RPMI-1640 supplemented with 10% heat-inactivated FBS, 5% L-glutamine, and 2.5 % penicillin/streptomycin antibiotic. Cells were routinely propagated and used for experiments in logarithmic phase.
Western Blot Analysis of TdT Present in ALL Cell Lines
Cell lysates from each ALL cell line were prepared in RIPA buffer (Thermo Fisher Scientific) containing a complete protease inhibitor cocktail tablet. The lysates (each with 50 μg of total protein content) were loaded on a Novex 4–20% Tris-Glycine gel, transferred into 0.2 μm nitrocellulose membrane and blocked with 1X Blocking buffer (Sigma-Aldrich) overnight at 4°C. The membrane was probed for TdT with primary antibody diluted in blocking solution (1:200) for 1 h at room temperature and then with a secondary donkey anti-goat IgG-HRP (1:5000 dilution) under identical conditions as the primary antibody. Immunoblots were developed by using enhanced chemiluminescence (Thermo Scientific), and the intensities of TdT and β-actin bands were quantified using the ImageJ program (http://imagej.nih.gov/ij/). TdT in each cell line was normalized by the corresponding β-actin content defined by Western blot analysis under identical conditions (TdT content = the intensity of TdT band divided by the intensity of β-actin band). Data represents an average of three independent determinations.
Cell Proliferation Assays
Cells were seeded at a population density of ~200,000 cells mL−1 and treated with variable concentrations of non-natural nucleoside (0.1–100 μg mL−1) for up to 72 hours. The final DMSO concentration in all experiments was 0.1%. Cell viability was assessed via trypan blue staining and counting the number of viable (clear) versus non-viable (blue) cells under a microscopy using a hemocytometer. The IC50 values for both non-natural nucleosides were obtained using a non-linear regression curve fit of the data to Equation 3. LD50 values for the non-natural nucleosides were calculated using identical approaches.
Apoptosis Measurements
Cells were treated with DMSO (vehicle), 5-NIdR (100 μg mL−1), and 3-Eth-5-NIdR (10 and 40 μg mL−1) as described above. Cells were harvested by centrifugation and washed in phosphate-buffered saline and resuspended in 100 μL in binding buffer containing AnnexinV-Alexa Fluor® 488 conjugate. Cells were treated with propidium iodide (PI) and incubated at room temperature for 15 minutes followed by flow cytometry analysis. This enables live cells (unstained with either Alexa Fluor® 488 or PI) to be discriminated from early apoptotic (stained with Alexa Fluor® 488), late apoptotic cells (stained with Alexa Fluor® 488 and PI) and necrotic cells (stained with PI).
In situ “clicking” reactions were performed using cells harvested after 2 days of treatment with DMSO, 3-Eth-5-NIdR (10, 40, or 100 μg mL−1), 5-NIdR (100 μg mL−1) or EdU (10 μM). All cells were fixed with cold methanol. Under minimal lighting, cells were treated with 0.3 mL of saponin-based permeabilization and wash buffer for 45 minutes at 37° C. Click reactions were initiated by adding the click-iT™ reaction cocktail according to the instructions of the manufacturer (Invitrogen) followed by incubation at 37° C for 90 minutes. Cells were then washed two times with saponin-based permeabilization and wash buffer. Cell pellets were dislodged using 0.5 mL solution of 10 μg mL−1 PI and RNAase A in saponin-based permeabilization and wash buffer, and then incubated for at least 15 minutes prior to flow cytometry analysis.
Supplementary Material
Acknowledgments
This research was supported by funds from the National Institute of Health (CA118408 to AJB). EAM was supported by the National Cancer Institute Training Programs in Cancer Pharmacology (CA148052). We would like to thank the Cytometry & Imaging Microscopy Core Facility of the Case Comprehensive Cancer Center supported by the NCI (P30 CA43703).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kornberg A. Ten commandments: lessons from the enzymology of DNA replication. J Bacteriol. 2000;182:3613–3618. doi: 10.1128/jb.182.13.3613-3618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollum FJ. Thermal conversion of nonpriming deoxyribonucleic acid to primer. J Biol Chem. 1959;234:2733–2734. [PubMed] [Google Scholar]
- 5.Bollum FJ. Chemically Defined Templates and Initiators for Deoxypolynucleotide Synthesis. Science. 1964;144:560. doi: 10.1126/science.144.3618.560-b. [DOI] [PubMed] [Google Scholar]
- 6.Baltimore D. Is terminal deoxynucleotidyl transferase a somatic mutagen in lymphocytes? Nature. 1974;248:409–411. doi: 10.1038/248409a0. [DOI] [PubMed] [Google Scholar]
- 7.Benedict CL, Gilfillan S, Thai TH, Kearney JF. Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev. 2000;175:150–157. [PubMed] [Google Scholar]
- 8.Kunkel TA, Gopinathan KP, Dube DK, Snow ET, Loeb LA. Rearrangements of DNA mediated by terminal transferase. Proc Natl Acad Sci USA. 1986;83:1867–1871. doi: 10.1073/pnas.83.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadofsky MJ. The RAG proteins in V(D)J recombination: more than just a nuclease. Nucleic Acids Res. 2001;29:1399–1409. doi: 10.1093/nar/29.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janeaway CA. Immunobiology: The immune System in Health and Disease. 4. Current Biology Publications; 1999. [Google Scholar]
- 11.Hoffbrand AV, Ganeshaguru K, Janossy G, Greaves MF, Catovsky D, Woodruff RK. Terminal deoxynucleotidyl-transferase levels and membrane phenotypes in diagnosis of acute leukaemia. Lancet. 1977;2:520–523. doi: 10.1016/s0140-6736(77)90662-6. [DOI] [PubMed] [Google Scholar]
- 12.Kung PC, Long JC, McCaffrey RP, Ratliff RL, Harrison TA, Baltimore D. Terminal deoxynucleotidyl transferase in the diagnosis of leukemia and malignant lymphoma. Am J Med. 1978;64:788–794. doi: 10.1016/0002-9343(78)90518-1. [DOI] [PubMed] [Google Scholar]
- 13.Stass SA, McGraw TP, Folds JD, Odle B, Bollum FJ. Terminal transferase in acute lymphoblast leukemia in remission. Am J Clin Pathol. 1981;75:838–840. doi: 10.1093/ajcp/75.6.838. [DOI] [PubMed] [Google Scholar]
- 14.Venditti A, Del Poeta G, Buccisano F, Tamburini A, Aronica G, Bruno A, Cox-Froncillo MC, Maffei L, Simone MD, Papa G, Amadori S. Biological pattern of AML-M0 versus AML-M1: response. Blood. 1997;89:345–346. [PubMed] [Google Scholar]
- 15.Hutter JJ. Childhood leukemia. Pediatr Rev. 2010;31:234–241. doi: 10.1542/pir.31-6-234. [DOI] [PubMed] [Google Scholar]
- 16.Hunger SP, Raetz EA, Loh ML, Mullighan CG. Improving outcomes for high-risk ALL: translating new discoveries into clinical care. Pediatr Blood Cancer. 2011;56:984–993. doi: 10.1002/pbc.22996. [DOI] [PubMed] [Google Scholar]
- 17.McCaffrey R, Bell R, Lillquist A, Wright G, Baril E, Minowada J. Selective killing of leukemia cells by inhibition of TdT. Haematol Blood Transfus. 1983;28:24–27. doi: 10.1007/978-3-642-68761-7_4. [DOI] [PubMed] [Google Scholar]
- 18.Spigelman Z, Duff R, Beardsley GP, Broder S, Cooney D, Landau NR, Mitsuya H, Ullman B, McCaffrey R. 2′,3′-Dideoxyadenosine is selectively toxic for TdT-positive cells. Blood. 1988;71:1601–1608. [PubMed] [Google Scholar]
- 19.Plunkett W, Gandhi V. Purine and pyrimidine nucleoside analogs. Cancer Chemother Biol Response Modif. 2001;19:21–45. [PubMed] [Google Scholar]
- 20.Lyman GH. Impact of chemotherapy dose intensity on cancer patient outcomes. J Natl Compr Canc Netw. 2009;7:99–108. doi: 10.6004/jnccn.2009.0009. [DOI] [PubMed] [Google Scholar]
- 21.Reineks EZ, Berdis AJ. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 22.Vineyard D, Zhang X, Donnelly A, Lee I, Berdis AJ. Optimization of non-natural nucleotides for selective incorporation opposite damaged DNA. Org Biomol Chem. 2007;5:3623–3630. doi: 10.1039/b712480e. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Lee I, Berdis AJ. Evaluating the contributions of desolvation and base-stacking during translesion DNA synthesis. Org Biomol Chem. 2004;2:1703–1711. doi: 10.1039/b401732c. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Lee I, Zhou X, Berdis AJ. Hydrophobicity, Shape, and p-Electron Contributions during Translesion DNA Synthesis. JACS. 2005;128:143–149. doi: 10.1021/ja0546830. [DOI] [PubMed] [Google Scholar]
- 25.Motea EA, Lee I, Berdis AJ. Quantifying the energetic contributions of desolvation and pi-electron density during translesion DNA synthesis. Nucleic Acids Res. 2011;39:1623–1637. doi: 10.1093/nar/gkq925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berdis AJ. Dynamics of translesion DNA synthesis catalyzed by the bacteriophage T4 exonuclease-deficient DNA polymerase. Biochemistry. 2001;40:7180–7191. doi: 10.1021/bi0101594. [DOI] [PubMed] [Google Scholar]
- 27.Loakes D, Brown DM, Linde S, Hill F. 3-Nitropyrrole and 5-nitroindole as universal bases in primers for DNA sequencing and PCR. Nucleic Acids Res. 1995;23:2361–2366. doi: 10.1093/nar/23.13.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loakes D, Hill F, Brown DM, Salisbury SA. Stability and structure of DNA oligonucleotides containing non-specific base analogues. J Mol Biol. 1997;270:426–435. doi: 10.1006/jmbi.1997.1129. [DOI] [PubMed] [Google Scholar]
- 29.Motea EA, Lee I, Berdis AJ. Development of a ‘clickable’ non-natural nucleotide to visualize the replication of non-instructional DNA lesions. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr980. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deibel MR, Jr, Coleman MS. Biochemical properties of purified human terminal deoxynucleotidyltransferase. J Biol Chem. 1980;255:4206–4212. [PubMed] [Google Scholar]
- 31.Ono K. Inhibitory effects of various 2′,3′-dideoxynucleoside 5′-triphosphates on the utilization of 2′-deoxynucleoside 5′-triphosphates by terminal deoxynucleotidyltransferase from calf thymus. Biochim Biophys Acta. 1990;1049:15–20. doi: 10.1016/0167-4781(90)90078-g. [DOI] [PubMed] [Google Scholar]
- 32.Krayevsky AA, Victorova LS, Arzumanov AA, Jasko MV. Terminal deoxynucleotidyl transferase. catalysis of DNA (oligodeoxynucleotide) phosphorylation. Pharmacol Ther. 2000;85:165–173. doi: 10.1016/s0163-7258(99)00070-4. [DOI] [PubMed] [Google Scholar]
- 33.Jarchow-Choy SK, Krueger AT, Liu H, Gao J, Kool ET. Fluorescent xDNA nucleotides as efficient substrates for a template-independent polymerase. Nucleic Acids Res. 2011;39:1586–1594. doi: 10.1093/nar/gkq853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jasko MV, Krayevsky AA. Human DNA polymerases and retroviral reverse transcriptases: selectivity in respect to dNTPs modified at triphosphate residues. Nucleosides Nucleotides. 1999;18:1031–1032. doi: 10.1080/15257779908041641. [DOI] [PubMed] [Google Scholar]
- 35.Arzumanov AA, Victorova LS, Jasko MV. Synthesis of non-nucleoside triphosphate analogues, a new type of substrates for terminal deoxynucleotidyl transferase. Nucleosides Nucleotides Nucleic Acids. 2000;19:1787–1793. doi: 10.1080/15257770008045460. [DOI] [PubMed] [Google Scholar]
- 36.Berdis AJ, McCutcheon D. The use of non-natural nucleotides to probe template-independent DNA synthesis. Chembiochem. 2007;8:1399–1408. doi: 10.1002/cbic.200700096. [DOI] [PubMed] [Google Scholar]
- 37.Gray HE, Luttge WG. Linearization of two ligand-one binding site scatchard plot and the “IC50” competitive inhibition plot: application to the simplified graphical determination of equilibrium constants. Life Sci. 1988;42:231–237. doi: 10.1016/0024-3205(88)90631-5. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava BI, Minowada J. Terminal transferase immunofluorescence, enzyme markers and immunological profile of human leukemia-lymphoma cell lines representing different levels of differentiation. Leuk Res. 1983;7:331–338. doi: 10.1016/0145-2126(83)90097-8. [DOI] [PubMed] [Google Scholar]
- 39.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 40.Stewart BW. Mechanisms of apoptosis: integration of genetic, biochemical, and cellular indicators. J Natl Cancer Inst. 1994;86:1286–1296. doi: 10.1093/jnci/86.17.1286. [DOI] [PubMed] [Google Scholar]
- 41.Parker WB. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer. Chem Rev. 2009;109:2880–2893. doi: 10.1021/cr900028p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffman MA, Janson D, Rose E, Rai KR. Treatment of hairy-cell leukemia with cladribine: response, toxicity, and long-term follow-up. J Clin Oncol. 1997;15:1138–1142. doi: 10.1200/JCO.1997.15.3.1138. [DOI] [PubMed] [Google Scholar]
- 43.Peters GJ, van der Wilt CL, van Moorsel CJ, Kroep JR, Bergman AM, Ackland SP. Basis for effective combination cancer chemotherapy with antimetabolites. Pharmacol Ther. 2000;87:227–253. doi: 10.1016/s0163-7258(00)00086-3. [DOI] [PubMed] [Google Scholar]
- 44.Huang P, Chubb S, Plunkett W. Termination of DNA synthesis by 9-beta-D-arabinofuranosyl-2-fluoroadenine. A mechanism for cytotoxicity. Journal of Biological Chemistry. 1990;265:16617–16625. [PubMed] [Google Scholar]
- 45.Griffith DA, Jarvis SM. Nucleoside and nucleobase transport systems of mammalian cells. Biochim Biophys Acta. 1996;1286:153–181. doi: 10.1016/s0304-4157(96)00008-1. [DOI] [PubMed] [Google Scholar]
- 46.Molina-Arcas M, Casado FJ, Pastor-Anglada M. Nucleoside transporter proteins. Curr Vasc Pharmacol. 2009;7:426–434. doi: 10.2174/157016109789043892. [DOI] [PubMed] [Google Scholar]
- 47.Purcet S, Minuesa G, Molina-Arcas M, Erkizia I, Casado FJ, Clotet B, Martinez-Picado J, Pastor-Anglada M. 3′-Azido-2′,3′-dideoxythymidine (zidovudine) uptake mechanisms in T lymphocytes. Antivir Ther. 2006;11:803–811. [PubMed] [Google Scholar]
- 48.Gibbs JE, Thomas SA. The distribution of the anti-HIV drug, 2′3′-dideoxycytidine (ddC), across the blood-brain and blood-cerebrospinal fluid barriers and the influence of organic anion transport inhibitors. J Neurochem. 2002;80:392–404. doi: 10.1046/j.0022-3042.2001.00711.x. [DOI] [PubMed] [Google Scholar]
- 49.Avramis VI, Plunkett W. Metabolism and therapeutic efficacy of 9-beta-D-arabinofuranosyl-2-fluoroadenine against murine leukemia P388. Cancer Res. 1982;42:2587–2591. [PubMed] [Google Scholar]
- 50.Boelsterli UA, Ho HK, Zhou S, Leow KY. Bioactivation and hepatotoxicity of nitroaromatic drugs. Curr Drug Metab. 2006;7:715–727. doi: 10.2174/138920006778520606. [DOI] [PubMed] [Google Scholar]
- 51.Raether W, Hänel H. Nitroheterocyclic drugs with broad spectrum activity. Parasitol Res. 2003;90(Supp 1):S19–39. doi: 10.1007/s00436-002-0754-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.