Abstract
Blood Oxygen Level Dependent (BOLD) functional magnetic resonance imaging (fMRI) changes in blood oxygenation, which is affected by physiological processes, including cardiac pulsation, breathing, and low frequency oscillations (LFO). It is challenging to identify spatial and temporal effects of these processes on the BOLD signal because the low sampling rate of BOLD leads to aliasing of higher frequency physiological signal components. In this study, we used concurrent functional near infrared spectroscopy (fNIRS) and fMRI on 6 subjects during a resting state scan. To reduce aliasing, the BOLD fMRI acquisition was repeatedly performed on a set of sequentially acquired slice stacks to lower the TR to 0.5 sec while retaining high spatial resolution. Regressor interpolation at progressive time delays (RIPTiDe) method was used, in which physiological signal acquired by fNIRS (without aliasing) and its temporal shifts were used as regressors in the fMRI analysis to determine the magnitude and timing of the effects of various physiological processes on the BOLD signal. The details of the timing of the passage of the cardiac pulsation wave and of the cerebral blood itself were mapped. The result suggests that the cardiac signal affects the voxels near large blood vessels (arteries and veins) most strongly, while LFO mostly affected the drainage veins. We hypothesize this could be the result of differences in the cerebral blood path lengths, and differences in the dynamics of the propagation of the signals. Together these results validate and extend a novel imaging technique to dynamically track the pulse-wave and bulk blood flow with concurrent fMRI and fNIRS.
Keywords: fMRI, Near-infrared spectroscopy, Cerebral vasculature, Low frequency oscillation, Physiological noise, BOLD signal
1. Introduction
Blood Oxygen Level Dependent (BOLD) functional magnetic resonance imaging (fMRI) is the most common imaging technique utilized in the study of brain function both in response to tasks and in task-free “resting state” scans. BOLD measures changes in blood oxygenation, which can be caused by neuronal activation through neurovascular coupling. However, there are also prominent variations in cerebral hemodynamics, which arise from physiological factors affecting blood oxygenation, flow or volume that are not influenced by neuronal activations. Moreover, the temporal resolution of fMRI is relatively low, which is about 2–3 seconds at magnetic field strengths of 1.5–3T. While this is sufficient for the hemodynamic effects of neuronal responses, which alter regional blood flow and volume with a slow “hemodynamic response function” (Siero et al., 2011; Tian et al., 2010), it is insufficient to measure the faster global physiological hemodynamics arising from cardiac, respiratory, and possibly other fast blood-related variations. This means the signals from some physiological processes are aliased, which makes them unidentifiable in the detected BOLD signal, and therefore difficult to remove. These processes include respiration (0.2–0.3 Hz) and cardiac pulsation (~1Hz). Many studies have been done to confirm the existence of these signals in the brain (Chang et al., 2009; Chang and Glover, 2009b; Shmueli et al., 2007; Triantafyllou et al., 2005) and efforts have been made to remove them (Birn et al., 2006; Chang and Glover, 2009a; Glover et al., 2000; Verstynen and Deshpande, 2011).
Functional near infrared spectroscopy (fNIRS) is a promising complementary method to fMRI for the study of brain function (Gibson et al., 2005; Hillman, 2007; Hoshi, 2007; Obrig and Villringer, 2003). Like fMRI, it is sensitive to the hemodynamic changes caused by neuronal and physiological processes. Unlike fMRI, its temporal resolution is generally high enough (12.5 Hz in our study) to sample most physiological processes without aliasing, but with low spatial resolution (~1cm) and short depth of penetration (~2 cm). A new combined method for analyzing concurrent fNIRS/fMRI data capitalizes on the strengths of both types of data (Tong et al., 2011b). The method uses the fNIRS signals to generate regressors to analyze BOLD data (based on the fact that both fMRI and fNIRS are sensitive to blood oxygenation and volume changes). The high temporal resolution offered by fNIRS allows the full sampling of the physiological processes in the fNIRS data, as well as their spectral separation. These isolated physiological signals can then be used to probe the high spatial resolution fMRI data in order to understand how physiological processes unrelated to neuronal activation contribute to the BOLD signal over both time and space.
The method was first tested in a previous study (Tong et al., 2011b), in which the main goal was to study the influences of different physiological processes, such as cardiac pulsation, respiration, and low frequency oscillation (LFO) on BOLD fMRI. The method was useful to separate the physiological signals and map brain voxels affected by the cardiac and the LFO signals. In addition, the method allowed dynamic tracking of the cardiac signal through the brain. However, there were three shortcomings in the last study which this study seeks to resolve: 1) since the TR used in the last study was 1.5s, the cardiac signals in BOLD were still heavily aliased, complicating efforts to unambiguously detect voxels with cardiac driven BOLD signal change; 2) for this reason, the dynamic map generated by time-shifting the fNIRS regressor (at the cardiac frequency) may have missed important details in the passage of the wave; 3) due to the fact that the fNIRS probe detects little to no signal at the respiration frequency, the result of using fNIRS regressors in the respiration band was not informative. In this study, we adopted two improvements. Firstly, we used a segmented whole brain fMRI acquisition, which allowed us to lower the TR to 0.5s while maintaining the same spatial resolution (3.5×3.5×3.5mm). Secondly, a respiration belt was used to simultaneously acquire respiration data that was used as regressor for respiration (instead of using respiration data collected with fNIRS). The main goals were: 1) to confirm and validate the new method of mapping the cerebral vasculature and dynamically assessing the passage of the cardiac wave through the brain, 2) to understand how the LFO and cardiac signals affect the BOLD signal differently in space and time, and 3) to understand how respiration directly affects the BOLD fMRI signal.
2. Material and methods
2.1 Protocol
Concurrent fNIRS and fMRI resting state studies were conducted on six healthy volunteers during resting state acquisitions (4M, 2F, average age 28±4.69). The protocol was approved by the Institutional Review Board of McLean Hospital.
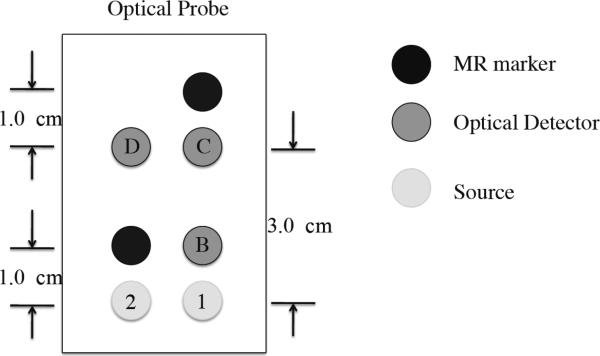
An MRI compatible fNIRS optical probe (as shown in Figure 1), with three collection (detector) and two illumination (source) fibers, with source-detector distances of 1 or 3 cm, was placed over the right prefrontal area of each participant (roughly between Fp1 and F7 in the 10–20 system) and held in place by an elastic band around the head. The position of the probe was chosen due to its easy accessibility (no hair) and relatively short distance from the scalp to the cortex (11–13 mm on average) in this area (Okamoto et al., 2004). The sampling rate of fNIRS data acquisition was 12.5 Hz. fNIRS data was recorded continuously through all the fMRI acquisitions as well as for several minutes before and after the fMRI acquisition.
Figure 1.
Diagram of the fNIRS probe with 2 optical sources and 3 optical detectors.
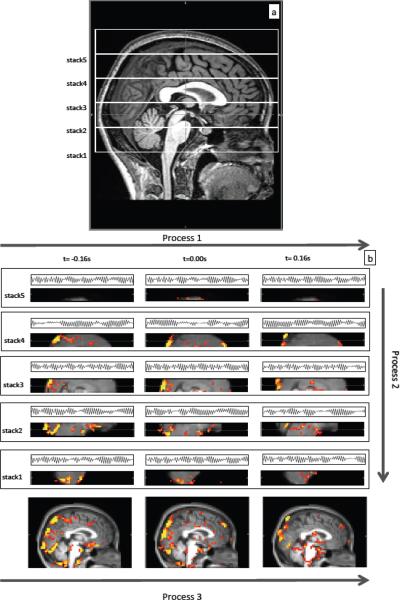
All MRI data were acquired on a Siemens TIM Trio 3T scanner (Siemens Medical Systems, Malvern, PA) using a 12-channel phased array head matrix coil. To reduce the aliasing of the cardiac signal (~1Hz) on the BOLD data, the TR has to be smaller than 0.5s to sample a cardiac frequency of 1Hz (or bigger) based on the Nyquist theorem (five out of six of our subjects had heart rate greater than 1Hz). However, reducing the TR lowers the number of slices that can be obtained, reducing either the spatial resolution or coverage of the fMRI data. To work around this trade-off, we employed a segmented fMRI acquisition, which allowed us to lower the TR to 0.5s while maintaining the same spatial resolution (3.5×3.5×3.5mm). We serially imaged multiple, adjacent subvolumes (segments) of brain. Each segment consisted of a stack of consecutive slices. Segments were acquired from the bottom of the cerebellum to top of the head, and each acquisition was a full resting state scan lasting about 6 minutes and 30 seconds with the following parameters: 240 time points, TR/TE=500/30 ms, flip angle 48 degrees, matrix = 64 × 64 on a 224 × 224 mm FOV, 9 3.5 mm slices with no gap parallel to the AC-PC line extending down from the top of the brain with a brief pause (~10s) between each consecutive stack scan. The total number of segments needed depended on the size of the brain; for this group of subjects, 5 or 6 segments were needed to cover the whole brain. Figure 2(a) illustrates the procedure on one subject. For the depicted subject, five segmental scans were needed for full brain coverage. Physiological waveforms (pulse oximetry, and respiratory timing and depth) were recorded using the scanner's built-in wireless fingertip pulse oximeter and respiratory belt at a rate of 50Hz throughout the entire set of acquisitions. This segmented acquisition is possible because the quantity of interest is the correlation between the fNIRS and fMRI data, which is consistent between task free acquisitions, rather than the actual time courses, which vary. Under these circumstances, data from multiple resting state acquisitions can be combined seamlessly.
Figure 2.
Diagram showing the fMRI data acquisition by segments in Figure 2(a) and combined fNIRS/fMRI data analyses in Figure 2(b) on one subject. Figure 2(a) shows five (segments) of resting state fMRI scans, which were needed for this subject to cover the whole head. Figure 2(b) depicts three processes, which were used in the data analyses. In process 1, the downsampled temporal trace of Δ[HbO] from fNIRS (simultaneously recorded) and its temporal shifts (± 0.16s) were used as regressors in analyzing fMRI data of corresponding sections. In process 2, the z-statistics maps from different sections, but from the same time shift were stitched together to form the z-statistics map of the whole brain at a particular time shift. In process 3, the z-statistics maps (whole brain) were registered on the anatomical scan and the results were connected in time to obtain the movie of the blood flow.
2.2 Combined data analysis
Regular preprocessing steps in FSL (Smith et al., 2004), including motion correction, slice timing correction and spatial smoothing were applied on fMRI data. Then the data analysis procedure (RIPTiDe), similar to the one we presented in the previous work (Tong et al., 2011b) with some important modifications, was used. As in the previous analysis, bandpass filtration was used to spectrally isolate signals of different physiological processes from simultaneously recorded data of oxy-hemoglobin concentration change (Δ[HbO]) from fNIRS, which had high temporal resolution (12.5Hz). The Δ[HbO] is used due to its high sensitivity to global physiological fluctuations, such as cardiac pulsations. For each spectral band, a set of time-shifted versions of the signal was generated. These signals were then downsampled to create a set of regressors at the sample rate of the fMRI data (2 Hz). A set of GLM analyses of the fMRI data was performed to determine the relationship between time shift and fit strength in each voxel. The results of this group of analyses were combined temporally to assess the spatial variation in this relationship as it evolved over time, and allowing visualization of the dynamic wave of correlation passing through the brain.
Several modifications in the data analysis procedure were introduced in this study. Firstly and most importantly, due to the fact that the fMRI data was acquired in sequential segments of slices, combined data analyses was carried out in three steps: first, each segment was processed individually, then the segments were merged, and lastly the results were registered onto the anatomical brain and concatenated over time. This procedure is shown for the cardiac signal in Figure 2(b). In Step 1, for each set of slices, or segment, Δ[HbO] from the fNIRS signal in the cardiac band is successively time shifted and used as a regressor in the GLM based fMRI analysis. For each segment shown in Figure 2(b), only two additional time shifts (±0.16s) of Δ[HbO] and their corresponding results are shown for illustration purposes. In the actual procedure, 200 analyses (with 200 shifts ranging from −8s to +8s at step of 0.08s) were used. This 16 second range was chosen in order to fully cover the passage time needed for the LFO wave (Tong and Frederick, 2010). After completing 200 analyses for each section, the results obtained for each individual time shift in each segment were concatenated in space to form a whole brain image, resulting in 200 whole brain images, one for each time-shift. This procedure is shown as Step 2 in Figure 2(b). The final step (Step 3) involves registering the results for each time shift onto the subject's own anatomical scan. Then, registered spatially concatenated images were concatenated in time to form a movie to assess dynamic patterns of changes.
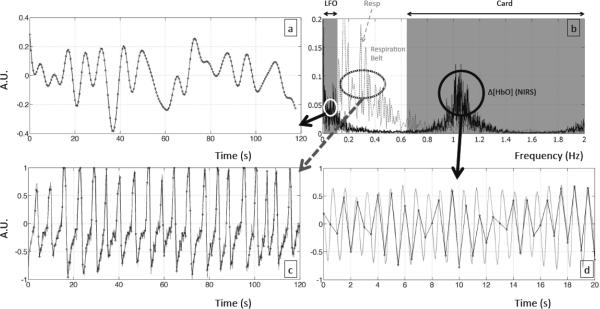
A second modification involved the respiration signal. Little or no signal was observed directly in the fNIRS data in the respiratory frequency band. This could be due to oxygen exchange in the lungs, which is a complicated process and does not couple to the blood signal linearly and directly, like the cardiac pulsation. Therefore, a respiration belt was used to collect the data for the respiration regressor in order to better isolate the direct contribution of respiration to the fMRI BOLD signal. As shown in Figure 3(b), the spectral graphs of Δ[HbO] (solid line) and data recorded from the respiration belt (dotted line) for one subject were plotted together. While the respiration signal was clear in the data collected using the respiration belt, it was hard to identify in the fNIRS data. It was hypothesized that using the temporal signal from the respiration belt (instead of fNIRS data from the same spectral band) as regressors would allow identification of the brain areas affected directly by the respiration process.
Figure 3.
Temporal traces of different frequency components from the Δ[HbO] fNIRS and physiological data recorded in one subject. LFO is shown in (a), the cardiac signal in (d), physiological recording of respiration in (c). Black dots are the resampling points at every 0.5 s. Spectra of fNIRS Δ[HbO] and respiration data of the same subject is in (b) with shaded areas corresponding to LFO, and cardiac responses.
Figure 3(a,c,d) illustrates the downsampling procedures for 3 physiological processes separated by the spectral ranges. Figure 3(a) shows the LFO (0.01 ~ 0.1 Hz) using the data from Δ[HbO] fNIRS. Figure 3(c) shows the respiration (0.1 ~ 0.6 Hz) using the data from the respiration belt. Figure 3(d) shows the cardiac band (> 0.8 Hz) using the data from Δ[HbO] fNIRS.
3. Results
Of the 6 subjects who started the experiment, one subject did not finish the scan due to discomfort caused by excessive tightness of the fNIRS probe. The analyses were conducted on the remaining 5 subjects, and the cardiac and LFO results were averaged based on these 5 subjects. For the respiration data, only 4 subjects' data were averaged, due to misplacement of the respiration belt on one of the remaining subjects, which resulted in unusable data.
3.1 Detailed passage of the pressure wave (cardiac pulsations)
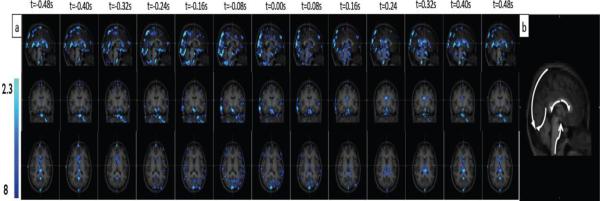
The dynamic patterns generated by the fNIRS regressors from the LFO and cardiac band are depicted for one subject in Figure 4(a) (overlaid on the subject's anatomical scan) and the movie is shown in Video 1. The figure and movie illustrate the time evolution of the correlation of the BOLD signal with the cardiac regressor (in blue) over all time lags and all segments for the whole brain. As comparison, the time evolution of the correlation of the BOLD signal with LFO regressor over the same time lags is also shown as red-yellow in the movie. The green dot in the middle panel of the movie indicates the time shift of the fNIRS regressor relative to the BOLD data. The total time shift is from −8s to +8s in steps of 0.08s. The movie repeats, each time showing different coronal and axial views (marked by the cross on the sagittal image). Even though the data for each segment was derived from an individual fMRI resting state scan, by concatenating the z-statistics maps, we were still able to visualize the blood flow (i.e. blood flowing through the vasculature, rather than CBF) between different sections as if they were scanned at the same time. This is because each dynamic map generated for each section was generated using regressors that were shifted in time relative to the fNIRS recording on the forehead. In other words, the dynamic map assesses the relative arrival time of the cardiac wave or LFO signal at the forehead (where the fNIRS probe was placed) and other locations in the brain. Therefore, as long as the travel time (i.e. the propagation speed of the cardiac wave or blood flow) does not change considerably between segment scans, the maps can be concatenated to assess the blood flow in the whole brain.
Figure 4.
Concatenated z-statistic maps using cardiac regressor and its temporal shifts (indicated at top of each column) from fNIRS. The results are shown in sagittal, coronal and axial view (top, middle and bottom) overlaid on the high-resolution scan in (a). Five apparent passages of cardiac signal from (a) are marked by white arrows in (b).
Both Figure 4(a) and the movie appear to show the propagation of the wave associated with the heart beat in the brain (in blue). The dynamic pattern repeats itself at a rate of about 1 Hz, which matches the subject's heart rate. In the Figure 4(a), only one cycle is shown, corresponding to the fNIRS regressor from −0.48s to +0.48s in steps of 0.08s. The similar dynamic patterns (one cycle) generated by the fNIRS regressors from the cardiac band are depicted for the rest four subjects in Figure 1 in the supplemental material. In the movie, multiple cycles are shown, each corresponding to the fNIRS regressor from −8s to +8s at steps of 0.08s. The result with the highest correlation coefficient is generated using fNIRS regressors with moderate time shifts (± 1s, middle portion of the movie). At longer time shifts, greater temporal mismatches between BOLD and fNIRS result in lower correlations (less significant voxels in the beginning and at the end of the movie). This demonstrates the sensitivity of the current method; even though the cardiac signal is highly periodic, fNIRS measures of cardiac signal become gradually out of phase with respect to the BOLD signal as time shifts in fNIRS regressors increase. Arrows in Figure 4(b) indicate apparent paths of blood flow in a sagittal view of the mid brain, including ascending flow in the basilar artery, descending flow in the straight sinus and superior sagittal sinus, and lateral flow in the posterior vein of the corpus callosum and the anterior cerebral vein. Although these blood paths are consistent with cerebral vasculature, there are two factors that may be misleading in the interpretation of the dynamic patterns. First, it is hard to present blood flow using 2-D images (axial, sagittal and coronal) because the blood vessels have complicated 3-D structures. Some voxels appear to be on the same flow path in 2-D, but they are actually not. Second, because the cardiac waveform is a highly periodic signal, the apparent flow speeds and directions may be the result of a stroboscopic effect. However, this will not affect the total blood vasculature map (i.e. the sum of spatial distribution of the propagating blood) obtained by performing a maximum intensity projection along the time axis (maximum z-statistic map) in Figure 4(a). The averaged result of these maps over all subjects is visualized in Figure 5(a), in which many vascular structures can be identified. Due to the high quality of the cerebral blood map generated by the combined z-statistics map from the cardiac band data, this map was additionally used as a reference (shown in white in Figure 5(b–c)) for studying the locations of activated voxels correlating with other frequencies (LFO and respiration). Figure 6 shows the individual optimum delay maps for the cardiac signal in each subject; the delays can be robustly determined in every subject; moreover, simulations (Figure 2 in the supplemental material) show that because the time delay is estimated by correlation of the entire waveform over the BOLD dataset, delay times as small as 0.08 seconds can be discriminated, despite the BOLD sampling frequency of 0.5 seconds.
Figure 5.
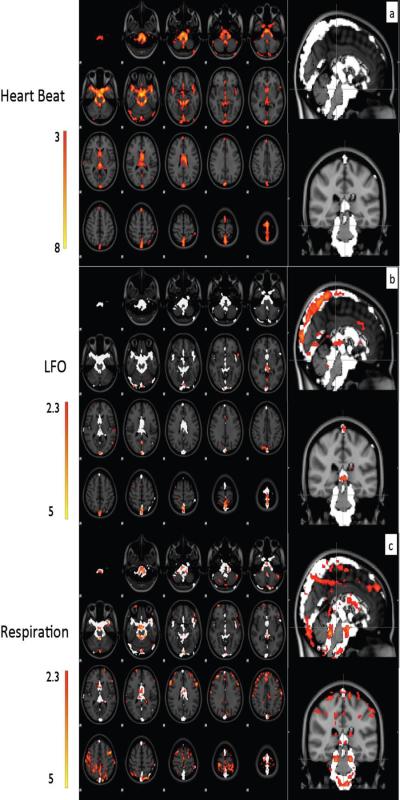
Averaged maximum z-statistic maps over 5 subjects using the cardiac component of fNIRS as regressor are depicted as the axial images in (a). The same maps are shown in the sagittal and coronal view in white color. Averaged maximum z-statistic maps using LFO component of fNIRS as regressors are illustrated in (b), and using the respiration recording from the respiration belt as regressors are shown in (c). The reference white patterns in (b) and (c) are the resulting vasculature maps obtained in (a). All the z-statistic maps are overlaid on the high-resolution scan and transformed into standard (MNI152) space.
Figure 6.
The maps of optimum regressor time delays using the cardiac component of fNIRS as regressor are showed for all 5 subjects.
3.2 LFO
Figure 5(b) shows the averaged z-statistic map (5 subjects) using the LFO regressor from fNIRS (s-d pair B1 in Figure 1), overlaid on the standard brain (MNI_152) as well as the blood map from the cardiac band analysis. Video 1 shows the dynamic pattern of this LFO (in red) together with that of the cardiac data (in blue) for comparison. Two observations are worth noting. First, the dynamic pattern of LFO moves much slower than that of the cardiac pulsations; we hypothesize that the pattern of LFO represents bulk blood flow whereas that of the cardiac wave represents the propagation of a pressure wave; Second, the activated voxels of the LFO are primarily in the large veins, including the superior sagittal sinus and straight sinus. Unlike the cardiac analysis, there are almost no activated voxels of LFO detected in the arteries.
3.3 Respiration pattern
In Figure 5(c), the averaged z-statistics map (4 subjects) from respiration was registered to and overlaid on top of the standard brain, together with the averaged z-statistic map from the cardiac waveform as a reference. The image generated from the respiratory belt data has a greater number of correlated voxels with the fMRI data than the one generated by using the respiration data from the fNIRS data (Tong et al., 2011b). This is due to the fact that the respiration regressors from the respiration belt have a much higher signal to noise ratio, as seen in Figure 2(b); and that the short TR (0.5s) samples the respiration signal in the brain without aliasing (as shown in Figure 2(c)).
4 Discussion
4.1 Map of pulsation
The averaged maximum z-statistic map shown in Figure 5(a) resembles the whole cerebral vasculature, including the large arteries and veins. Among them, the areas with highest correlations (with fNIRS cardiac signal: z>5) are located in regions with large arteries or a high density of arteries, including the basilar artery and the middle cerebral artery. No gray matter was detected at a significance level of 3 (z value). This indicates that the cardiac waveform has a relatively small influence on the regional blood flow at capillary levels in gray matter. In other words, the BOLD from the gray matter is less affected by the heart beat. A possible explanation for why cardiac effects on the BOLD signal are greater in arteries than in veins and gray matter is offered below in the section “Different signals in arteries and veins”. Our result is consistent with previous finding of Dagli (Dagli et al., 1999) using retrospective gating method, in which they found cardiac-induced variations are typically near major blood vessels. In both studies, cardiac pulsation wave was directly mapped. A recent study (Shmueli et al., 2007) suggested that heart rate fluctuations might indirectly contribute to BOLD signal, which are not concentrated entirely around major vessels, but also on gray matter. They also found the heart rate fluctuation was negatively correlated with BOLD signal at one time lag range and positively correlated with BOLD signal at another longer time lag range.
As demonstrated, decreasing TR in the BOLD acquisition increases the accuracy of the cardiac vessel maps compared to previous work using BOLD fMRI with TR of 1.5 seconds (Tong et al, 2011) in term of total voxels affected by the heart beat signal being detected. Although the method is shown to have higher sensitivity with decreasing TRs, short TR scans are not required to map the voxels affected by the cardiac signal. Using the result in Figure 5(a) as a reference, we found the result from the analysis with longer TR (1.5s) yielded similar cerebral blood maps (the maps from the analyses with TR=1.5s and TR=0.5s are shown as Figure 3 in the supplement material).
Physiological contributors to BOLD signal are generally treated as noise and removed. Here we use the physiological signal from a robust and easily measured physiological process (cardiac pulsations) to assess the cerebral vasculature statically and dynamically yielding maps of the cerebral blood vasculature (Figure 5(a)), as well as the dynamic pattern of the cardiac wave through the cerebral vasculature (Figure 4(a) and movie in Video 1), using the cardiac signal recorded by fNIRS. This method does not require additional MRI scans and can be added easily to any fMRI scans with regular TR, either in resting state or functional studies. It only requires simultaneous recording of the cardiac pulsation with high temporal resolution (>5 Hz) either using a dedicated fNIRS instrument, or an oximeter which is widely available on many scanners. These results can be used either to assess cerebral vasculature integrity and changes in different populations, or to separate brain areas where the BOLD signal is known to have a large cardiac or respiratory component.
4.2 Respiration
As shown in Figure 5(c), the voxels affected most strongly by respiration are located in the arteries, overlapping with the results of the cardiac waveform analysis. This could be due to the fact that freshly oxygenated blood from the lungs (modulated by the breathing) quickly reaches these arteries. Activated voxels are also present in the ventricles, tissue edges and in and out of motor cortex, which is likely due to global respiration related B0 changes (Van de Moortele et al., 2002; van Gelderen et al., 2007). There is also evidence for head motion artifacts from the result of one subject, in which the MRI visible marker in the fNIRS probe appears in the resulting z-statistics map, indicating high correlation between the probe and breathing rhythm (data not shown). Other researchers also found respiration-induced effects can translate into head motion (Glover et al., 2000).
As we know, respiration affects blood oxygen level. It also modulates blood flow and volume indirectly. However, it takes time for the alteration of the blood oxygen level, which happens in the lungs, to travel to the brain. Moreover, this modulated blood travels to the brain via several different paths, each with a different path length, thus arrives at different times. These unknowns in the transit times of the modulated blood to different sites of the brain make it very hard to directly identify the respiration signal from BOLD and model it using simultaneous recording of either the respiration belt or End Tidal CO2 monitoring. The RIPTiDe method offers a way to track the modulated blood by time shifting respiration data (from the respiration belt) and correlating it with the BOLD from the whole brain. No prerequisite of transit time is needed, since a wide range of time shifts (16 sec in this case) with small steps (0.08s) are utilized. The analyses should be able to identify all the voxels where BOLD signal has a significant respiratory signal component within the temporal window of 16 s, which is sufficient due to the fact that each breath is about 3~8s.
This study concentrated only on the direct correlation between BOLD and breathing pattern. However, the respiration process may influence the BOLD signal in multiple ways. For instance, Birn and colleagues (Birn et al., 2006) found that a time shifted temporal course of respiration volume per time is significantly correlated with BOLD signals in gray matter and near large blood vessels. Thus there are likely to be other voxels that are indirectly affected by breathing, which could not be identified by our method.
4.3 Different signals in arteries and veins
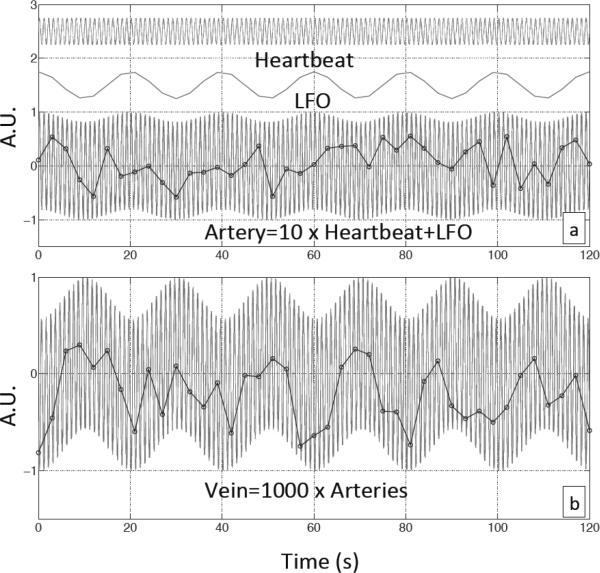
Figures 5(a) and (b) show the difference in the averaged z-statistic maps of 5 subjects using time-shifted fNIRS regressors corresponding to the cardiac spectral band (in Figure 5(a)) the LFO band (in Figure 5(b)). As mentioned before, the cardiac band results resemble the whole cerebral vasculature, including the large arteries and veins. Among them, the areas with highest correlations (with fNIRS cardiac signal: z>5) are located in regions with large arteries or a high density of arteries, including the basilar artery and the middle cerebral artery. In contrast, the results from the LFO band only resemble the distribution of cerebral veins (Figure 5(b)), such as superior sagittal sinus. Based on previous work, it seems likely that similar to the cardiac signal, large fractions of the LFO signal observed in the brain are also generated extracerebrally and carried in the blood (Tong and Frederick, 2010). These oscillations travel along the cerebral blood vessels (e.g. the cardiac signal is a pressure wave, which travels much faster) and both contribute to the BOLD signal. When the blood is pumped out of the heart and carried by the arteries into the brain, the cardiac pulsation is the dominant signal. Therefore, the BOLD fMRI obtained at voxels from these arteries is predominantly affected by the cardiac pulsation. We qualitatively demonstrate this process in Figure 7(a), in which, two sine waves with different oscillation frequencies (~1Hz, ~0.05Hz) were used to simulate the cardiac wave and LFO respectively. To represent the signal in the arteries, where the cardiac signal is comparably strong, we mixed these two signals (cardiac signal 10 times stronger than the LFO). The individual waveforms and the mixed signal are shown in Figure 7(a) as a representation of signals that might be measured in a typical artery. When the mixed signal is sampled every three seconds to simulate a BOLD fMRI acquisition with TR=3 in an arterial voxel, the resulting data set (shown in the black circles) would be mostly affected by the (aliased) cardiac oscillations. The LFO can hardly be seen due to its relatively low power.
Figure 7.
Simulated temporal traces of blood-related signal in arteries and veins. (a) Simulated cardiac and LFO signals and their combination (artery signal) with dominant cardiac signal. (b) The sum of 1000 artery signals, each with small random phase shifts, represents the vein signal. Black dots in both graphs represent the sampling procedure of BOLD fMRI with TR=3s.
After traveling through the arteries, blood travels through various paths made up of arterioles, capillaries and venules. These paths have different lengths and change the phases of the blood signal differently and in proportion to their frequencies. When the blood reaches veins, the sum of these signals with mismatched phase changes would greatly reduce the amplitude of the cardiac signals due to its much higher frequency, while change in the phases of the LFO signal would be much less. Thus, the BOLD fMRI obtained at voxels from the veins are predominantly affected by LFO. To simulate the signal in the veins, we summed 1000 signals after first introducing a small random phase shift (based on the literature) to each in the mixed signal (like the one in Figure 7(a)), to simulate mixing of blood traversing multiple paths through the parenchyma in the veins. Figure 7(b) shows the result of this sum. As predicted, the LFO became the predominant component with the cardiac component making a relatively smaller contribution. When this dataset is sampled every 3 seconds to simulate a BOLD fMRI acquisition with TR=3 in a venous voxel, the resulting data set (shown in the black circles) reflects mostly the shape of the LFO. The parameters used in the model are based on literature(Conroy et al., 2005; Gladdish et al., 2005; Kikuchi et al., 2002) and can be found in the supplement materials. While this model is quite simple, it is able to generate a qualitatively similar result to what is seen in vivo; the BOLD fMRI from arteries is predominantly affected by the cardiac signal, while BOLD fMRI from veins is predominantly affected by the LFO signal. It also explains why few gray matter voxels had high correlations with the cardiac signal. This is because the BOLD signal of the voxel in gray matter, as the summation of all the blood signals within that voxel, reflects a decreased contribution from the cardiac signal due to destructive interference produced by phase mismatches. Further refinement of the model may lead to a closer alignment with the result observed in vivo. In addition, regional blood flow changes caused by neuronal activity in gray matter might surpass the physiological LFO signal, and become the predominant signal (Tong et al., 2011a). The Matlab code used to generate the simulations and parameters can be found in the supplement material.
Concurrent fNIRS fMRI RIPTiDe analysis offers a new method to examine the propagation of blood through the brain in greater detail than has been previously possible. By allowing the parsing of physiological “noise” into various components which can be separately analyzed, this may provide new techniques for the study of cerebral hemodynamics in healthy brain and in pathological conditions. Moreover, we have previously demonstrated that these physiological signals contribute a significant fraction of the variance in resting state BOLD data, and modeling and removing these signals results in a substantial increase in the relative power of resting state networks, allowing more sensitive detection of these connectivity signals (Frederick and Tong, 2011).
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health, Grant No. R21-DA027877
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure/Conflict of Interest I have nothing to declare.
Reference
- Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: the cardiac response function. NeuroImage. 2009;44:857–869. doi: 10.1016/j.neuroimage.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Effects of model-based physiological noise correction on default mode network anti-correlations and correlations. NeuroImage. 2009a;47:1448–1459. doi: 10.1016/j.neuroimage.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. NeuroImage. 2009b;47:1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy DA, Spielman AJ, Scott RQ. Daily rhythm of cerebral blood flow velocity. Journal of circadian rhythms. 2005;3:3. doi: 10.1186/1740-3391-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagli MS, Ingeholm JE, Haxby JV. Localization of cardiac-induced signal change in fMRI. NeuroImage. 1999;9:407–415. doi: 10.1006/nimg.1998.0424. [DOI] [PubMed] [Google Scholar]
- Frederick BD, Tong Y. Physiological noise reduction in BoLD data using optimally delayed simultaneously acquired NIRS data. 17th Annual Meeting of the Organization for Human Brain Mapping; Quebec, Canada. 2011. [Google Scholar]
- Gibson AP, Hebden JC, Arridge SR. Recent advances in diffuse optical imaging. Physics in medicine and biology. 2005;50:R1–43. doi: 10.1088/0031-9155/50/4/r01. [DOI] [PubMed] [Google Scholar]
- Gladdish S, Manawadu D, Banya W, Cameron J, Bulpitt CJ, Rajkumar C. Repeatability of non-invasive measurement of intracerebral pulse wave velocity using transcranial Doppler. Clinical science. 2005;108:433–439. doi: 10.1042/CS20040251. [DOI] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Hillman EM. Optical brain imaging in vivo: techniques and applications from animal to man. Journal of biomedical optics. 2007;12:051402. doi: 10.1117/1.2789693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi Y. Functional near-infrared spectroscopy: current status and future prospects. Journal of biomedical optics. 2007;12:062106. doi: 10.1117/1.2804911. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Murase K, Miki H, Yasuhara Y, Sugawara Y, Mochizuki T, Ikezoe J, Ohue S. Quantitative evaluation of mean transit times obtained with dynamic susceptibility contrast-enhanced MR imaging and with (133)Xe SPECT in occlusive cerebrovascular disease. AJR. American journal of roentgenology. 2002;179:229–235. doi: 10.2214/ajr.179.1.1790229. [DOI] [PubMed] [Google Scholar]
- Obrig H, Villringer A. Beyond the visible--imaging the human brain with light. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2003;23:1–18. doi: 10.1097/01.WCB.0000043472.45775.29. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Dan H, Sakamoto K, Takeo K, Shimizu K, Kohno S, Oda I, Isobe S, Suzuki T, Kohyama K, Dan I. Three-dimensional probabilistic anatomical cranio-cerebral correlation via the international 10–20 system oriented for transcranial functional brain mapping. Neuroimage. 2004;21:99–111. doi: 10.1016/j.neuroimage.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Shmueli K, van Gelderen P, de Zwart JA, Horovitz SG, Fukunaga M, Jansma JM, Duyn JH. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state fMRI BOLD signal. NeuroImage. 2007;38:306–320. doi: 10.1016/j.neuroimage.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siero JC, Petridou N, Hoogduin H, Luijten PR, Ramsey NF. Cortical depth-dependent temporal dynamics of the BOLD response in the human brain. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2011;31:1999–2008. doi: 10.1038/jcbfm.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Suppl 1):S208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Tian P, Teng IC, May LD, Kurz R, Lu K, Scadeng M, Hillman EM, De Crespigny AJ, D'Arceuil HE, Mandeville JB, Marota JJ, Rosen BR, Liu TT, Boas DA, Buxton RB, Dale AM, Devor A. Cortical depth-specific microvascular dilation underlies laminar differences in blood oxygenation level-dependent functional MRI signal. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15246–15251. doi: 10.1073/pnas.1006735107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Frederick BD. Time lag dependent multimodal processing of concurrent fMRI and near-infrared spectroscopy (NIRS) data suggests a global circulatory origin for low-frequency oscillation signals in human brain. NeuroImage. 2010;53:553–564. doi: 10.1016/j.neuroimage.2010.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Hocke LM, Frederick B. Isolating the sources of widespread physiological fluctuations in functional near-infrared spectroscopy signals. Journal of biomedical optics. 2011a;16:106005. doi: 10.1117/1.3638128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Lindsey KP, Frederick BD. Partitioning of physiological noise signals in the brain with concurrent near-infrared spectroscopy and fMRI. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2011b;31:2352–2362. doi: 10.1038/jcbfm.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafyllou C, Hoge RD, Krueger G, Wiggins CJ, Potthast A, Wiggins GC, Wald LL. Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. NeuroImage. 2005;26:243–250. doi: 10.1016/j.neuroimage.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Van de Moortele PF, Pfeuffer J, Glover GH, Ugurbil K, Hu X. Respiration-induced B0 fluctuations and their spatial distribution in the human brain at 7 Tesla. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2002;47:888–895. doi: 10.1002/mrm.10145. [DOI] [PubMed] [Google Scholar]
- van Gelderen P, de Zwart JA, Starewicz P, Hinks RS, Duyn JH. Real-time shimming to compensate for respiration-induced B0 fluctuations. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2007;57:362–368. doi: 10.1002/mrm.21136. [DOI] [PubMed] [Google Scholar]
- Verstynen TD, Deshpande V. Using pulse oximetry to account for high and low frequency physiological artifacts in the BOLD signal. NeuroImage. 2011;55:1633–1644. doi: 10.1016/j.neuroimage.2010.11.090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.