Abstract
Multimodal imaging improves the accuracy of the localization and the quantification of brain activation when measuring different manifestations of the hemodynamic response associated with cerebral activity. In this study, we incorporated cerebral blood flow (CBF) changes measured with arterial spin labeling (ASL), Diffuse Optical Tomography (DOT) and blood oxygen level-dependent (BOLD) recordings to reconstruct changes in oxy- (ΔHbO2) and deoxyhemoglobin (ΔHbR). Using the Grubb relation between relative changes in CBF and cerebral blood volume (CBV), we incorporated the ASL measurement as a prior to the total hemoglobin concentration change (ΔHbT). We applied this ASL fusion model to both synthetic data and experimental multimodal recordings during a 2-sec finger-tapping task. Our results show that the new approach is very powerful in estimating ΔHbO2 and ΔHbR with high spatial and quantitative accuracy. Moreover, our approach allows the computation of baseline total hemoglobin concentration (HbT0) as well as of the BOLD calibration factor M on a single subject basis. We obtained an average HbT0 of 71 μM, an average M value of 0.18 and an average increase of 13 % in cerebral metabolic rate of oxygen (CMRO2), all of which are in agreement with values previously reported in the literature. Our method yields an independent measurement of M, which provides an alternative measurement to validate the hypercapnic calibration of the BOLD signal.
Keywords: fNIRS, fMRI, ASL, CMRO2, BOLD calibration, multimodal imaging
Introduction
Multimodal brain imaging aims to enhance the localization and the quantification of cerebral activity by combining the strengths of each individual modality (Moseley and Donnan, 2004). Moreover, multimodal imaging allows the study of cerebral activity with different physiological contrast, such as the electrical activity of the neurons and the related hemodynamic response. The relationship between these two phenomena is known as neurovascular coupling and has been studied extensively by combining electrophysiological recordings with hemodynamic measurements. Examples include functional Magnetic Resonance Imaging (fMRI) combined with intracortical recordings of neural signals (Logothetis et al., 2001), Intrinsic Optical Imaging with electrophysiological recording (Jones et al., 2005; Devor et al., 2003), Diffuse Optical Tomography (DOT) with electroencephalography (EEG) (Opitz et al., 1999; Kennan et al., 2002), and DOT with magnetoencephalography (MEG) (Ou et al., 2009).
Simultaneous measurements of functional magnetic resonance imaging (fMRI) and diffuse optical tomography (DOT) have been used in animals (Kida et al., 1996; D’Arceuil et al., 2005) and in humans (Kleinschmidt et al., 1994; Toronov et al., 2001; Strangman et al., 2002; Steinbrick et al., 2006; Tak et al., 2010) to study the normal functioning brain, as well as to investigate pathophysiology (Tak et al., 2011). DOT and fMRI elegantly complement one another. On one hand, the hemodynamic response is reconstructed with higher spatio-temporal resolution by combining the high temporal resolution of DOT with the high spatial resolution of fMRI. On the other, the ability of DOT to independently measure absolute changes in oxyhemoglobin (HbO2) and deoxyhemoglobin (HbR) (and therefore absolute changes in total hemoglobin (HbT)) allows the computation of important physiological parameters (e.g., the BOLD calibration factor and relative changes in CMRO2) more robustly than with fMRI alone (Hoge et al., 2005; Huppert et al., 2009; Tak et al., 2010).
Of the various multimodal approaches, bottom-up fusion models have been used extensively in the literature (Riera et al., 2005; Huppert et al., 2008, Huppert et al., 2009). Bottom-up modeling relates the spatio-temporal evolution of physiological parameters to the measurements through an observation model describing the biophysics of each modality. For DOT-fMRI fusions, these physiological parameters are the concentration of HbO2 and HbR. Huppert et al. (2008) successfully constructed such a model integrating DOT and blood oxygen level-dependent (BOLD) fMRI measurements. Their linear model reconstructed the hemoglobin changes in space and time and computed a numerical correspondence between relative changes in the BOLD signal at 3T and absolute changes in HbR. This method has two major limitations: (1) Although the method provides a robust reconstruction of HbR, the reconstructed HbO2 is subject to greater errors; and (2) due to the lack of baseline HbT concentration (HbT0), the numerical correspondence between HbR and the BOLD changes cannot be compared against the traditional hypercapnia calibration factor M (Davis et al., 1998), and furthermore cannot be used to compute relative changes in the cerebral metabolic rate of oxygen (CMRO2) from independent relative changes in BOLD and cerebral blood flow (CBF) measurements.
In this paper, we address these two drawbacks by adding an arterial spin labeling (ASL) fMRI CBF measurement to the fusion model. The CBF measurement constrains the reconstruction of the HbT image, thus informing the reconstruction of HbO2. Moreover, measurements of absolute changes in HbT from DOT and relative changes in CBF from ASL enable the estimation of baseline total hemoglobin concentration (HbT0) from a simple assumption of the flow-volume relationship (Grubb et al., 1974). The estimated HbT0 combined with simultaneous measurements of relative changes in the BOLD signal and absolute changes in HbR allows calibration of the BOLD signal (Davis et al., 1998) in a framework consistent with the hypercapnic calibration method but without the need for a hypercapnic challenge. Finally, the calibrated BOLD signal is used together with a separate measurement of CBF from ASL to compute relative changes in CMRO2 (Davis et al., 1998; Hoge et al., 1999; Buxton et al., 2010) during a motor task in human subjects.
We first quantify the accuracy of the method with numerical simulations over a realistic human head model obtained from anatomical MRI. Reconstructed hemoglobin concentrations from simultaneous NIRS, BOLD and ASL recordings during 2-sec finger-tapping tasks are then presented. The parameter HbT0, the BOLD calibration factor M and relative changes in CMRO2 are then calculated from the reconstructed hemodynamic responses for each subject individually.
Theory
Our bottom-up NIRS-BOLD-ASL fusion model is a straightforward extension of the NIRS-BOLD fusion model described by Huppert et al. (2008). Essentially, it consists of a multidimensional linear model describing HbO2 and HbR, an observation model relating HbO2 and HbR to the measurements, and a least-squares minimization method. The model is briefly summarized here. Further details can be found in (Huppert et al., 2008).
Multidimensional linear model
The changes in HbR and HbO2 are represented as linear combinations of temporal (T) and spatial basis functions (G) in order to reduce the dimensionality of the problem. As in Huppert et al. (2008), we used four distinct pairings of spatial and temporal basis functions to describe the hemodynamic response in the cortex and other hemodynamic fluctuations across the head. The task-evoked hemodynamic response was modeled by a set of overlapping Gaussian spheres with a 6-mm standard deviation. The spheres were separated by 6 mm in a hexagonal pattern (Huppert et al., 2008) and were constrained to the gray and white matter tissue of the contralateral side of the finger tapping stimulus. The temporal basis used to model this evoked hemodynamic response was a linear combination of a gamma-variant function and its derivative with a resolution of 2 Hz 0 to 20 sec after the stimulus onsets. Systemic hemodynamic fluctuations were modeled independently in the brain; the CSF and the skin were modeled using a set of sine and cosine functions (from 1/20 Hz to 1.0 Hz in 1/20-Hz increments) to describe the temporally oscillating systemic physiology. Fluctuations from the brain are reflected in both the DOT and fMRI measurements while skin and CSF fluctuations are reflected only in the DOT measurements. The CSF fluctuations originate mainly from the hemodynamic response occurring in the pial veins present above the surface of the cortex (Gagnon et al., 2012), while skin fluctuations result from respiration and heartbeats. The multidimensional linear model represents the hemoglobin changes as a product of the spatial-temporal basis function (GT) and a vector of unknown coefficients (β).
| (1) |
The reader is referred to (Huppert et al., 2008) for more information about the geometry, the analytical expression and the values of the parameters used in the spatial and temporal basis functions.
DOT observation model
Light propagation in tissue is described by the photon diffusion equation. Under the Rytov approximation (i.e. when the perturbation is smooth spatially), a linear equation is sufficient to predict the changes in DOT measurements produced by a perturbation of the optical properties of the medium (Arridge et al., 1999). The equation takes the following form:
| (2) |
where Y is the vector of DOT measurements for each source-detector pair, the matrix A is the spatial sensitivity of the detected light determined by Monte-Carlo simulations (Boas et al., 2002; Huppert et al., 2006b), ε is the spectral extinction coefficient at the denoted wavelength and for the denoted hemoglobin species, and v is a noise term specific for each source-detector pair and wavelength. The matrix A handles the partial volume error of NIRS by explicitly mapping measurements from the MR space to the source-detector space of NIRS.
BOLD observation model
The BOLD signal is comprised of an intravascular as well as an extravascular component (Lu and van Zijl, 2005; Obata et al., 2004). In order to retain a linear relationship between absolute changes in HbR and relative changes in BOLD, we consider only the extravascular component of the BOLD signal in our fusion model. This assumption is justified by the fact that the extravascular signal composes the majority of the BOLD signal at 3T (Lu and van Zijl, 2005). The change in relaxation rate is correlated to vo, the frequency offset of water at the outer surface of a magnetized vessel; V, venous blood volume-to-tissue fraction; and Y, the average saturation of the venous pool in the activated state as follows (Obata et al., 2004):
| (3) |
The deoxyghemoglobin changes are related to the venous blood volume-to-tissue fraction (V) and the venous oxygen saturation (Y), and the initial venous oxygen saturation is related to Eo, the resting oxygen extraction fraction, with the equations below (Obata et al., 2004).
| (4) |
Substituting the above equations (Eq. (4)) into Eq. (3), we obtain the linear relationship between relative changes in the BOLD signal and absolute changes in HbR as follows (Davis et al., 1998; Hoge et al., 1999; Obata et al., 2004):
| (5) |
where TE is the echo time, and HbR0 is the baseline HbR concentration. All these parameters are lumped into a single parameter (α = −4.3 · v0 · V0 · E0 · TE/HbR0), which is estimated during the least-square minimization routine (see section Fusion model and Bayesian inversion approach). Therefore, the observation model for the BOLD is given by the equation:
| (6) |
where YBOLD is the vector of BOLD measurements in each voxel and vBOLD is a noise term.
The calibration factor M widely measured during hypercapnic BOLD calibration is defined as the maximum BOLD signal obtained if all of the deoxyhemoglobin is washed out of the venous pool (Davis et al., 1998; Hoge et al., 1999). Substituting ΔHbR = −HbR0 in Eq. (5) gives M = 4.3 · v0 · V0 · E0 · TE0. Therefore, the relationship between the parameter α computed with our fusion model and the calibration factor M used in hypercapnic BOLD calibration studies is given by
| (7) |
where HbR0 is obtained from HbT0 (which is computed in the model as described below) and assuming a baseline oxygen saturation of 70 % (E0 = 0.3, Oja et al., 1999; Bolar et al, 2011) in the venous pool.
ASL observation model
Arterial Spin Labeling (ASL) is a non-invasive MRI technique that allows measurement of regional tissue perfusion (Detre et al., 1992). In order to better reconstruct changes in HbO2 and to estimate HbT0, our new model extends the previous model of Huppert et al. (2008) to include ASL measurements in addition to DOT and BOLD measurements.
We use the Grubb relation (Grubb et al., 1974) to compute relative changes in cerebral blood volume (CBV) from relative changes in CBF measured by ASL. This relationship is given by
| (8) |
where g is a parameter called the Grubb exponent. This relation generally holds for steady-state measurements. To take into account the short duration of our stimulus (2 sec), we computed an effective Grubb exponent using a dynamic multi-compartment Windkessel model on the same data set (Huppert et al., 2009). The value of this effective Grubb exponent was found to be g = 0.228. This value, which was used in our analysis, is close to the BOLD-specific flow-volume relationship recently reported by Chen and Pike (2009). Under constant hematocrit, changes in HbT are directly related to changes in CBV. This has been verified experimentally using combined NIRS and MION-fMRI measurements. Siegel et al. have shown that changes in cerebral blood volume measured with fMRI are directly related to relative changes in HbT measured with DOT (Siegel et al., 2003). Therefore, we can substitute CBV with HbT in Eq. (8) to obtain
| (9) |
This is further justified by the experimental fact that relative changes in CBF measured with ASL are highly correlated both temporally (Huppert et al., 2006a) and spatially (Huppert et al., 2006b) with absolute changes in HbT measured by DOT. With Eq. (9) the observation model for ASL is given by
| (10) |
YASL in Eq. (9) is the relative volume change obtained using ASL data, g is the Grubb exponent relating CBF to HbT, HbT0 is the baseline total hemoglobin concentration, and vASL in Eq. (10) is a noise term.
Fusion model and Bayesian inversion approach
The multidimensional linear models and the three observation models (DOT, BOLD and ASL) can be combined in a single linear equation allowing the recovery of ΔHbO2 and ΔHbR simultaneously from all three measurements:
| (11) |
The values for α and HbT0 are initialized to the estimated values using the raw DOT, BOLD and ASL data. Following this, the state vector X is estimated using a Bayesian formulation of the linear inverse operator:
| (12) |
where R−1 and Γ2Q−1 are the inverses of the measurement noise covariance and the state noise covariance respectively. The matrix R is estimated from the data using the same procedure described in (Huppert et al., 2008). To estimate R, we calculated the variance of the residual for each measurement by iteratively solving the state estimate and estimating R to be the variance of the residual of the measurements. The initial seed of R was calculated from linear regression of the data with the temporal basis. An identity matrix was used for the state covariance matrix Q. The value for the regularization tuning parameter Γ was taken from our simulation results (see section Simulations) and was set to the optimal Γ value (Γ = 10−6).
Once Eq. (12) is solved, the predicted DOT, BOLD and ASL signals are re-calculated using the estimated HbO2 and HbR values via the forward model given by Eq. (11). The α and HbT0 values are then estimated by minimizing the residual between the predicted and measured signals. New estimates of ΔHbO2 and ΔHbR are then computed with Eq. (12) using the optimal α and HbT0 values. This procedure is repeated until convergence is obtained for α and HbT0, which is set to be a deviation of less than ± 10 %.
CMRO2 calculations
We used the following equation to calculate changes in CMRO2 during functional activity (Davis et al., 1998; adapted version from Lin et al., 2009):
| (13) |
where the M value is obtained using Eq. (7), ΔHbT is the sum of the ΔHbO2 and ΔHbR changes estimated by the model (Eq. 12), HbT0 is also estimated from the model, ΔCBF/CBF0 is obtained from the ASL data and β = 1.5 is a constant relating the change in transverse relaxation rate (ΔR2*) to the concentration of HbR.
Methods
Subjects
The experimental protocol and the data used in this study have been previously reported (Huppert et al., 2008). A subset of four out of five healthy, right-handed subjects was used in this study (between 20–60 years old). The study included simultaneous measurement of NIRS, BOLD and ASL signals during an event-related 2-sec finger-tapping task. Each experiment consisted of 6 runs. For the final analysis, we kept only the runs for which a clear hemodynamic response could be observed. The number of good runs ranged from 3 to 5 per subject. Each run was 6 minutes long and included 27–32 stimuli, with an inter-stimulus interval ranging from 4 to 20 sec with an average of 12 sec. The study was approved by the Massachusetts General Hospital Institutional Review Board and subjects gave written consent.
DOT
A multichannel continuous wave optical imager (CW4, TechEn Inc., Milford, Massachusetts), consisting of 18 lasers – half of them at 690 nm (18 mW) and the other half at 830 nm (7 mW) – and 16 detectors, was used for measurements as previously described in (Franceschini et al., 2003). Only four sources and eight detectors were used in this study. The DOT probe was made from flexible plastic strips with plastic caps inserted into them to hold fiber optic bundles. It has two rows of four detectors and one row of four sources arranged in a rectangular grid with 2.9 cm between neighboring source-detector pairs (Fig. 1). During the experiment, the probe was placed on the subject’s head where the contralateral motor cortex was located and secured using Velcro straps and foam padding. 10-m fibers were used to reach the MR scanner while their other end was connected to the DOT instrument in the control room. The DOT data was synchronized to the MRI images as previously described (Huppert et al., 2006a) and downsampled to the same 2-Hz frequency of fMRI using a Nyquist filter. Optical density changes were obtained by taking the negative log of the normalized incident light density. The optical measurement sensitivity profiles were obtained using GPU-accelerated Monte Carlo simulations (108 photons) based on a segmented anatomical MP-RAGE volume with five tissue layers (Fang and Boas, 2009). The locations of optodes in the 3D MR volume were determined with the help of fiducial markers.
Figure 1.
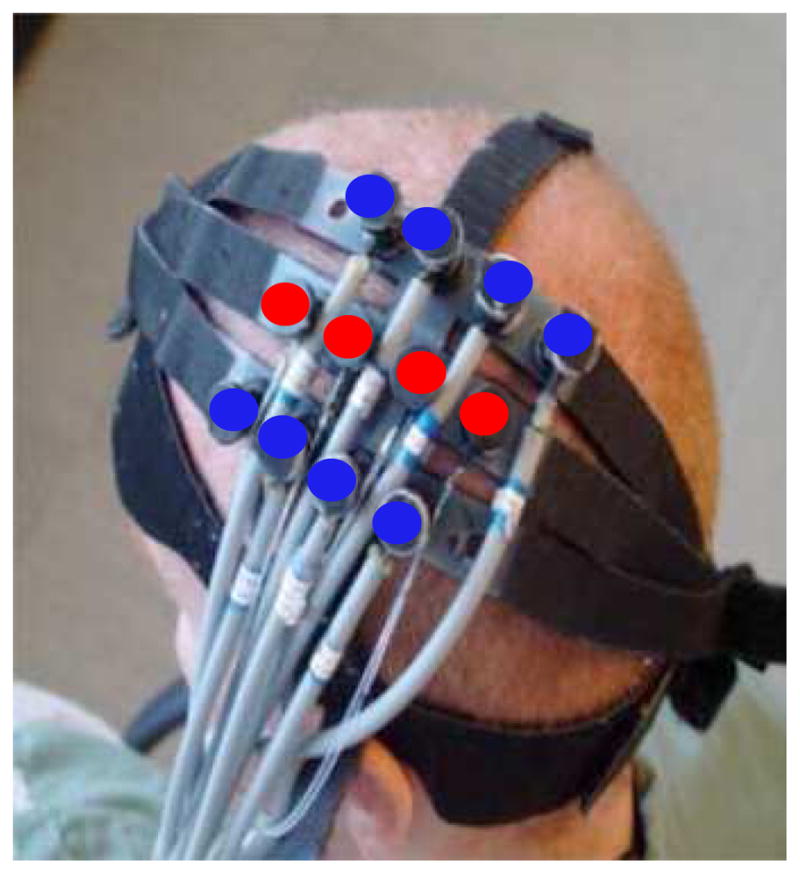
Optical probe positioned over the primary motor area (blue circles: detectors; red circles: sources).
fMRI and ASL
ASL-fMRI measurements were performed on a 3 T Siemens Allegra MR Scanner (Siemens Medical Systems, Erlangen Germany) using PICORE labeling geometry (Wong et al., 1997) with Q2TIPS saturation (Luh et al., 1999) to impose a controlled label duration. The sequence had a post-label delay of 1400 ms, a label duration of 700 ms, a repetition time of 2 s and an echo time of 20 ms (α = 90°). EPI was used to image five 6-mm slices (1-mm spacing) with 3.75-mm in-plane spatial resolution. Structural scans were performed using a T1-weighted MPRAGE sequence (1×1×1.33 mm resolution, TR/TE/α = 2530 ms/3.25 ms/7°). The stimulus was jittered evenly on a 500-ms time step, allowing the response to be calculated at 2 Hz. The functional images were motion corrected (Cox and Jesmanowicz, 1999) and spatially smoothed with a 6-mm Gaussian kernel. The ASL scans were separated into the control (BOLD signal) and negatively labeled tag images, which were then interpolated to upsample to 2 Hz using a cubic spline model (de Boor, 1978). Each negatively labeled tag scan was subtracted from the subsequent control scan to get the flow image series.
The DOT data were downsampled using a Nyquist filter to 2-Hz, while the BOLD and ASL data were upsampled to the same frequency. A third order polynomial was applied to the data set to remove task-unrelated physiological drift.
Simulations
To compare the accuracy of our ASL fusion model with that of the BOLD fusion model previously used by Huppert et al. (2008), a sphere-shaped absorption perturbation was positioned ~25 mm deep in the head and synthetic DOT, BOLD and ASL measurements were generated using the forward model Eq. (11). We added a measurement noise of 5 % for the DOT and the BOLD time courses and a measurement noise of 10 % for the ASL time courses. The noise was higher for ASL to reflect its lower experimental SNR. The maximum values for the simulated ΔHbO2 and ΔHbR were 8 μM and −2.5 μM respectively. The hemoglobin changes were then recovered from the synthetic data using the iterative procedure described in the Fusion Model and Bayesian Inversion Approach section. Different values for the regularization parameter Γ were tested to determine the optimal value for the analysis of the in vivo recordings.
Results
Simulations
The ΔHbO2 and ΔHbR reconstructions obtained using DOT-only model, the BOLD fusion model (DOT and BOLD, Huppert et al, 2008) and the ASL fusion model (DOT, BOLD and ASL) are shown in Fig. 2. The BOLD fusion and the ASL fusion models recovered the HbR response successfully while only the ASL fusion model could reconstruct the HbO2 response with a high accuracy.
Figure 2.
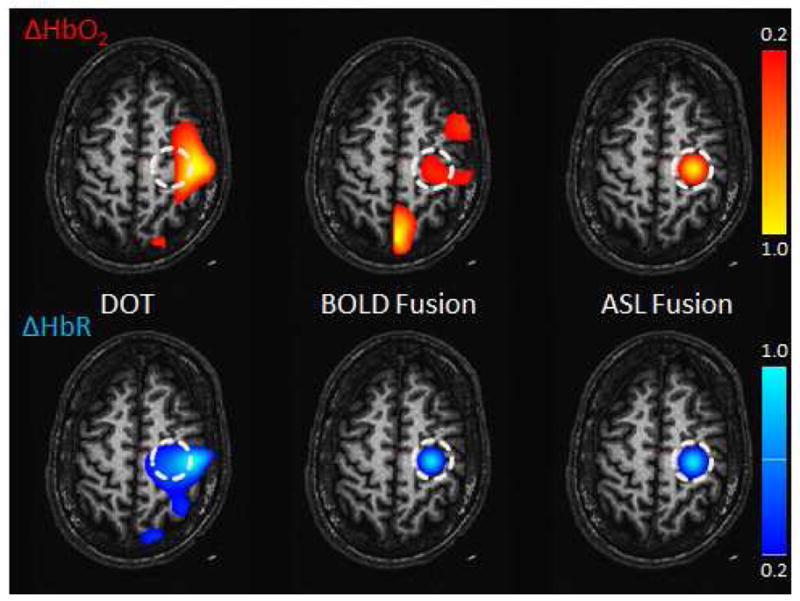
Simulation Results. The reconstructed hemoglobin concentrations are shown for the DOT-only (left), BOLD fusion (middle) and ASL fusion (right) models. (Top) HbO2 (Bottom) HbR. The sphere-shaped inclusion was located at a depth of 25 mm. The contrast-to-noise ratio (CNR) was set to 20:1 for DOT and BOLD and to 10:1 for ASL. The regularization parameter Γ was set to 10−6 (1 μM). Values are normalized to the maximum and minimum for HbO2 and HbR respectively and then thresholded at 20 % of maximum.
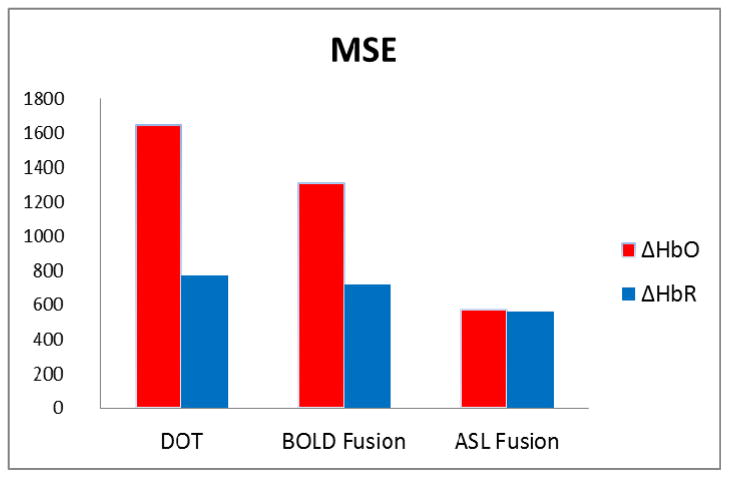
The mean squared errors (MSE) between the simulated and the reconstructed ΔHbO2 and ΔHbR were computed for DOT-only, BOLD fusion and ASL fusion models to compare the accuracy of each model in reconstructing the hemodynamic response. Results are shown in Fig. 3. The ASL fusion model had the lowest MSE values compared to the other two models and was therefore used for the analysis of the finger-tapping data.
Figure 3.
Mean squared error between the simulated and the recovered ΔHbO2 and ΔHbR for the DOT only, BOLD fusion and ASL fusion models.
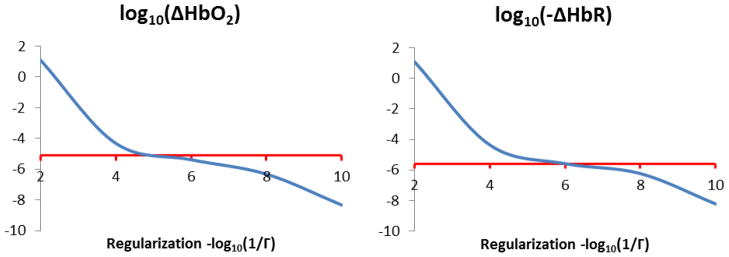
The ΔHbO2 and ΔHbR values obtained for different values of the regularization parameter Γ are shown in Fig. 4. The optimal value of the parameter Γ was roughly 1×10−5 and 1×10−6 for ΔHbO2 and ΔHbR respectively but the reconstruction was still reasonably accurate over the range 1×10−4 to 1×10−8. Therefore, we used a value of 10−6 for both HbO2 and HbR for the analysis of the finger-tapping data.
Figure 4.
Recovered hemoglobin concentrations for different values of the regularization parameter Γ. The simulated concentrations changes are shown in red while the recovered concentration changes are shown in blue for HbO2 (right) and HbR (left).
Hemoglobin reconstruction from in vivo data
The ΔHbO2 and ΔHbR responses obtained with the ASL fusion model from the in vivo recordings are shown in Fig. 5 for all subjects. Results were obtained using a regularization parameter Γ = 10−6 (or 1 μM), which is the optimum regularization value obtained from our simulations. The activation areas were located in the motor and primary somatosensory cortices, which represent finger movement and touch, respectively, during finger tapping.
Figure 5.
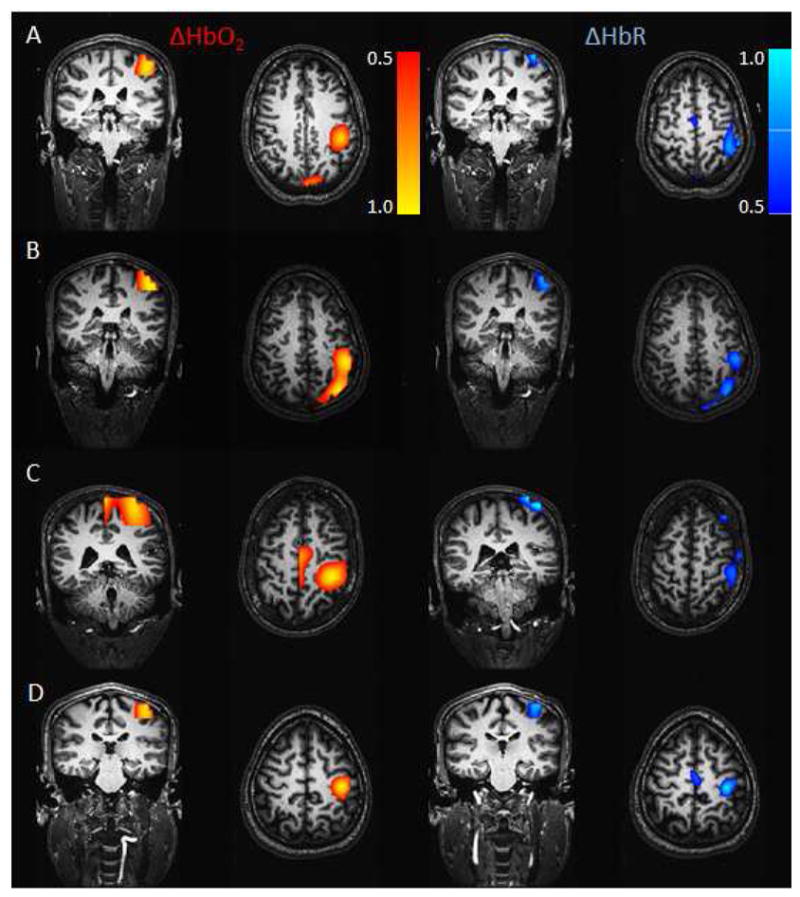
In vivo results. ΔHbR and ΔHbO2 reconstructed from the ASL fusion model for each subject. DOT, BOLD and ASL were acquired simultaneously during a 2-sec finger-tapping task. The regularization parameter was set to Γ = 10−6 (1 μM). The results were normalized and thresholded at half maximum of the response in each case.
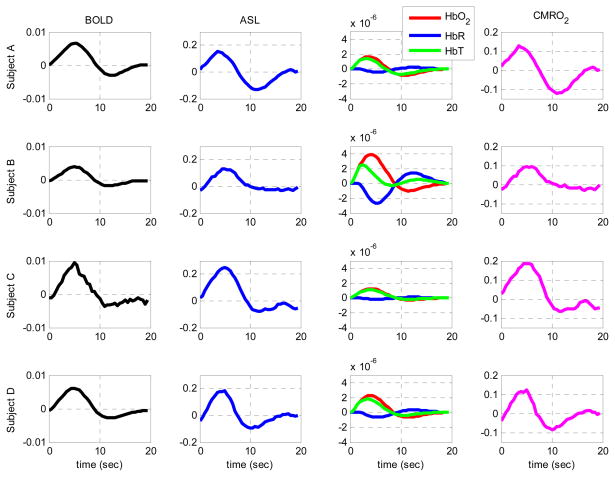
Regions of interest (ROI) were identified for each subject from the BOLD activation t-map due to the higher SNR of BOLD compared to ASL. The HbO2, HbR, BOLD and CBF signals were then averaged over this ROI to produce the time courses shown in Fig. 6.
Figure 6.
Time courses of the changes in BOLD signal, ASL, HbO2, HbR, HbT and CMRO2 during the 2-sec finger-tapping stimulation. BOLD, ASL, HbO2, HbR and HbT traces were obtained by averaging the respective signal over the ROI identified from the BOLD activation t-maps. Hemoglobin changes are shown in molars.
M and HbT0 values and changes in CMRO2
The estimated M and HbT0 values are shown in Table 1 for each subject. The M values were computed using Eq. (5) assuming a baseline oxygen saturation of 70 % in the venous pool (i.e. E0 = 0.3). The average M value obtained was 0.18 and the average HbT0 was 71 μM. The changes in CMRO2 obtained using Eq. (13) are shown in Table 1. The average CMRO2 change and the average flow-consumption ratio observed were 13 % and 1.4 respectively.
Table 1.
Estimated BOLD calibration factor (M), baseline total hemoglobin concentration (HbT0), ΔCMRO2, ΔCBF and flow-consumption ratio (n) from the ASL fusion model. Results are presented individually for each subject as well as for the group average.
| Subject | M value | HbT0 (μM) | ΔCMRO2 (%) | ΔCBF (%) | n value |
|---|---|---|---|---|---|
| A | 0.29 | 70 | 13 | 16 | 1.2 |
| B | 0.09 | 137 | 9 | 13 | 1.4 |
| C | 0.24 | 25 | 19 | 25 | 1.3 |
| D | 0.11 | 51 | 12 | 19 | 1.6 |
| Average (±std) | 0.18 (±0.10) | 71 (±48) | 13 (±4) | 18 (±5) | 1.4 (±0.14) |
Discussion
ASL fusion model improves ΔHbO2 reconstruction
In this study, we extended the fusion model proposed by Huppert et al. (2008) to include ASL data with the help of Grubb’s relation. As shown in Figures 2, 3 and 5, this approach brings great improvement to the previous model in terms of estimating ΔHbO2. The simulation results in Fig. 2 show that the ASL fusion model can estimate both ΔHbO2 and ΔHbR with high spatial accuracy. The lower MSEs obtained with the ASL fusion model (Fig. 3) indicate that the quantification of the reconstruction is more accurate for the ASL fusion model compared to the previous BOLD fusion model. This improvement was larger for HbO2 compared to HbR simply because the simulated changes were three times higher for HbO2 compared to HbR. Overall, the better performance of the ASL fusion model is explained by the fact that the model obtains prior information about the spatial location of HbO2 information from the ASL data, which is correlated with total blood volume changes and hence total hemoglobin changes. Finally, the simulations at various regularization levels presented in Fig. 4 show that the ASL fusion model is robust in estimating the true ΔHbO2 and ΔHbR values for a wide range of the regularization parameter (from 10−4 to 10−8).
Overall, the spatial locations of the increases in HbO2 and decreases in HbR during the motor task (Fig. 5) are very close and mostly overlapping. Nevertheless, slight differences can be observed regarding the spatial extent of the activation for the two chromophores. Physiologically, the spatial extend of BOLD and ASL maps have been shown to be non-coincident. This is because arterial dilation gives rise to venous HbR washout. While there is also HbO2 washout in the veins, our prior work has suggested that HbO2 has a greater contribution from the arterial side than does HbR (Huppert et al. 2006a). This could give rise to the spatial discrepancies observed between HbO2 and HbR reconstruction in Fig. 5.
Baseline physiology and BOLD calibration
The ASL fusion model presented in this paper has the advantage of simultaneously estimating the relationship between changes in HbR and BOLD (α parameter in Eq. (7)) and the baseline total hemoglobin concentration. This allows us to compute an experimental data-driven estimate of the BOLD calibration factor, M, from Eq. (7). This estimate of M can be directly compared with M values obtained from hypercapnic studies. Our M values shown in Table 1 are within a reasonable range compared to the experimental values obtained from hypercapnic and carbogen studies (0.07 to 0.25) (Chiarelli et al., 2007; Gauthier et al., 2011). A potential source of error is the baseline oxygen extraction fraction (E0), which was assumed constant for each subject (E0 = 0.3). This assumption could slightly bias our estimation of M but not enough to explain the inter-subject variability since E0 directly multiplies HbT0 and α to give M. Therefore, an E0 range of 0.25–0.35 can only change the M value by ± 12.5 % (in our case 0.18 ± 0.02) (see Eq. (7)). In future work, simultaneous measurements of DOT, BOLD and ASL during hypercapnic challenge and functional activity will allow comparison of M values obtained by the ASL fusion model with those obtained from hypercapnia calibration. This will provide an independent measure of M to validate the widely used hypercapnic BOLD calibration.
Similarly, our HbT0 values presented in Table 1 are also within the physiological range reported in the literature (60 to 100 μM) (Torricelli et al., 2001; Gagnon et al., 2008). Future studies could include an independent time-resolved spectroscopy measure of HbT0 to compare with those recovered from the ASL fusion model.
CMRO2 estimation and flow-consumption ratio
Our new approach also provided an estimation of CMRO2 changes using the recovered hemoglobin values, estimated M and HbT0 values and the ASL data (Eq. (13)). This approach is unique in that the only parameter assumed in calculating CMRO2 is the baseline oxygen extraction fraction. However, using only the extravascular component of the BOLD signal may result in slight overestimation of CMRO2. The average CMRO2 change we obtained was approximately 13%, which is consistent with previous observations with similar stimulus paradigms (Hoge et al., 2005; Tak et al., 2010; Maciejewski et al., 2004). The CMRO2 time courses we obtained follow the same pattern in CBF time courses. This is because of the fact that we obtain CMRO2 by direct multiplication of the CBF time course which is the highest source of noise among others.
The flow-consumption ratio we get (1.4) is in good agreement with the value of 1.5 obtained with a multicompartment windkessel model by a previous work on the same data set (Huppert et al., 2009). On the other hand, this value is lower than the values obtained using PET or calibrated-BOLD methods (Buxton, 2010). A possible explanation for the difference between the studies would be the shorter duration of our stimulus which does not allow steady state to be reached. This is supported by the fact that longer stimulation is needed to reach steady-state and therefore maximal flow response (Dunn et al., 2005, Drew et al., 2011). The lower flow responses obtained for shorter stimulation combined with similar CMRO2 increases could be a potential explanation for the lower flow-consumption ratio we obtained.
Flow-Volume dynamics
An important assumption we made in this paper was to assume Grubb’s relation between relative changes in CBF and CBV. The value of the Grubb exponent historically has been given as 0.38 for steady-state manipulations of blood flow and volume (Grubb et al., 1974). Recent work has suggested that a smaller value more accurately represents the flow-volume relationship during brain activation. Huppert et al. (2009) estimated the effective Grubb exponent for dynamic activation to 0.228 using a multicompartment windkessel model on the same data set. This value is very close to the BOLD-specific value of 0.23 reported recently by Chen and Pike (2009). As a sanity check, we also reconstructed the hemoglobin concentrations from the in vivo recordings assuming the traditional 0.38 value (data not shown) and there was no difference between the model residuals obtained assuming either of the two values. However, we obtained slightly lower HbT0 and M values with the higher Grubb value, but these values were still within the physiological range of 0.07 to 0.25 for M and 60 to 100 μM for HbT0. In future work, a dynamic relationship between CBF and CBV such as the Balloon model (Buxton et al., 1998) or multicompartment windkessel model (Huppert et al., 2009) could be included in the ASL fusion model to better account for the temporal dynamics of CBF and CBV.
Spatial basis functions
The overlapping Gaussian basis functions used in this paper were very general and allowed us to reconstruct the hemodynamic response without any spatial restriction. However, better reconstruction methods, such as the wavelet based method developed by Abdelnour and Huppert (2009), could be used to reconstruct the hemoglobin changes. The method uses a surface-based wavelet representation to model the anatomical structure of the brain and this reduces the number of parameters to be estimated in the reconstruction. A similar approach could be incorporated into our ASL fusion model to reduce the dimensionality of the problem and potentially provide a more robust reconstruction.
Conclusion
In this paper, we extended the previous DOT-BOLD fusion model of Huppert et al. (2008) by incorporating ASL measurements in the reconstruction of ΔHbO2 and ΔHbR. This resulted in a better reconstruction of ΔHbO2 since the ASL measurement acted as a prior for the ΔHbT reconstruction. Moreover, our ASL fusion model allowed the computation of baseline total hemoglobin concentration as well as of the BOLD calibration factor M individually for each subject. We obtained an average HbT0 value of 71 μM, an average M value of 0.18 and an average CMRO2 change of 13%, all of which agree well with values previously reported in the literature. Our model provides a reasonable alternative to validate with an independent method the celebrated hypercapnic BOLD calibration.
Acknowledgments
The authors are grateful to Rick Hoge and Frédéric Lesage for initial discussions related to this work. We also thank Qianqian Fang and Jay Dubb for their help with the Monte Carlo simulations and Gary Boas for his comments on the draft of this paper. This work was supported by NIH grants P41-RR14075, R01-EB006385 and R90-DA023427. L. Gagnon was supported by the Advanced Multimodal Neuroimaging Training Program at MIT and the Fonds Québecois de la Recherche sur la Nature et les Technologies (FQRNT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelnour F, Schmidt B, Huppert TJ. Topographic localization of brain activation in diffuse optical imaging using spherical wavelets. Phys Med Biol. 2009;54(20):6383–413. doi: 10.1088/0031-9155/54/20/023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boas D, Culver J, Stott J, Dunn A. Three dimensional monte carlo code for photon migration through complex heterogenous media including the adult human head. Opt Express. 2002;10 (3):159–170. doi: 10.1364/oe.10.000159. [DOI] [PubMed] [Google Scholar]
- Bolar DS, Rosen BR, Sorensen AG, Adalsteinsson E. QUantitative Imaging of eXtraction of oxygen and TIssue consumption (QUIXOTIC) using venular-targeted velocity-selective spin labeling. Magn Reson Med. 2011;66(6):1550–62. doi: 10.1002/mrm.22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: the balloon model. Magn Reson Med. 1998;39(6):855–64. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- Buxton RB. Interpreting oxygenation-based neuroimaging signals: the importance and the challenge of understanding brain oxygen metabolism. Front Neuroenergetics. 2010;17(2):8. doi: 10.3389/fnene.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Pike GB. BOLD-specific cerebral blood volume and blood flow changes during neuronal activation in humans. NMR Biomed. 2009;22(10):1054–62. doi: 10.1002/nbm.1411. [DOI] [PubMed] [Google Scholar]
- Chiarelli PA, Bulte DP, Piechnik S, Jezzard P. Sources of systematic bias in hypercapnia-calibrated functional MRI estimation of oxygen metabolism. Neuroimage. 2007;34(1):35–43. doi: 10.1016/j.neuroimage.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Cope M, Delpy DT, Reynolds EO, Wray S, Wyatt J, van der Zee P. Methods of quantitating cerebral near infrared spectroscopy data. Adv Exp Med Biol. 1988;222:183–189. doi: 10.1007/978-1-4615-9510-6_21. [DOI] [PubMed] [Google Scholar]
- Cox RW, Jesmanowicz A. Real-time 3D image registration for functional MRI. Magn Reson Med. 1999;42(6):1014–8. doi: 10.1002/(sici)1522-2594(199912)42:6<1014::aid-mrm4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- D’Arceuil HE, Hotakainen MP, Liu C, Themelis G, de Crespigny AJ, Franceschini MA. Near-infrared frequency-domain optical spectroscopy and magnetic resonance imaging: a combined approach to studying cerebral maturation in neonatal rabbits. J Biomed Opt. 2005;10(1):11011. doi: 10.1117/1.1852554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Kwong KK, Weisskoff RM, Rosen BR. Calibrated functional MRI: mapping the dynamics of oxidative metabolism. Proc Natl Acad Sci. 1998;5(4):1834–9. doi: 10.1073/pnas.95.4.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boor C. A Practical Guide to Splines. Springer-Verlag; Berlin: 1978. [Google Scholar]
- Dehghani H, Delpy DT, Arridge SR. Photon migration in non-scattering tissue and the effects on image reconstruction. Phys Med Biol. 1999;44(12):2897–906. doi: 10.1088/0031-9155/44/12/303. [DOI] [PubMed] [Google Scholar]
- Delpy DT, Cope M, van der Zee P, Arridge S, Wray S, Wyatt J. Estimation of optical pathlength through tissue from direct time of flight measurement. Phys Med Biol. 1988;33(12):1433–1442. doi: 10.1088/0031-9155/33/12/008. [DOI] [PubMed] [Google Scholar]
- Detre JA, Leigh JS, Williams DS, Koretsky P. Perfusion Imaging. MRM. 1992;23:37–45. doi: 10.1002/mrm.1910230106. [DOI] [PubMed] [Google Scholar]
- Devor A, Dunn AK, Andermann ML, Ulbert I, Boas DA, Dale AM. Coupling of total hemoglobin concentration, oxygenation, and neural activity in rat somatosensory cortex. Neuron. 2003;39(2):353–9. doi: 10.1016/s0896-6273(03)00403-3. [DOI] [PubMed] [Google Scholar]
- Drew PJ, Shih AY, Kleinfeld D. Fluctuating and sensory-induced vasodynamics in rodent cortex extend arteriole capacity. Proc Natl Acad Sci U S A. 2011;108(20):8473–8. doi: 10.1073/pnas.1100428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Devor A, Dale AM, Boas DA. Spatial extent of oxygen metabolism and hemodynamic changes during functional activation of the rat somatosensory cortex. Neuroimage. 2005;27(2):279–90. doi: 10.1016/j.neuroimage.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Fang Q, Boas DA. Monte Carlo simulation of photon migration in 3D turbid media accelerated by graphics processing units. Opt Express. 2009;17(22):20178–90. doi: 10.1364/OE.17.020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini MA, Fantini S, Thompson JH, Culver JP, Boas DA. Hemodynamic evoked response of the sensorimotor cortex measured non-invasively with near-infrared optical imaging. Psychophysiology. 2003;42(16):3063–3072. [Google Scholar]
- Gagnon L, Gauthier C, Hoge RD, Lesage F, Selb J, Boas DA. Double-layer estimation of intra- and extracerebral hemoglobin concentration with a time-resolved system. J Biomed Opt. 2008;13(5):054019. doi: 10.1117/1.2982524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon L, Yücel MA, Dehaes M, Cooper RJ, Perdue KL, Selb J, Huppert TJ, Hoge RD, Boas DA. Quantification of the cortical contribution to the NIRS signal over the motor cortex using concurrent NIRS-fMRI measurements. Neuroimage. 2011;59(4):3933–40. doi: 10.1016/j.neuroimage.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier CJ, Madjar C, Tancredi FB, Stefanovic B, Hoge RD. Elimination of visually evoked BOLD responses during carbogen inhalation: Implications for calibrated MRI. NeuroImage. 2011;54(2):1001–11. doi: 10.1016/j.neuroimage.2010.09.059. [DOI] [PubMed] [Google Scholar]
- Grubb RL, Raichle ME, Eichling JO, Ter-Pogossian MM. The Effects of Changes in PaCO2 on Cerebral Blood Volume, Blood Flow, and Vascular Mean Transit Time Stroke. 1974;5(5):630–9. doi: 10.1161/01.str.5.5.630. [DOI] [PubMed] [Google Scholar]
- Hoge RD, Atkinson J, Gill B, Crelier GR, Marrett S, Pike GB. Investigation of BOLD Signal Dependence on Cerebral Blood Flow and Oxygen Consumption: The Deoxyhemoglobin Dilution Model. Magnetic Resonance in Medicine. 1999;42(5):849–863. doi: 10.1002/(sici)1522-2594(199911)42:5<849::aid-mrm4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Hoge RD, Franceschini MA, Covolan RJ, Huppert T, Mandeville JB, Boas DA. Simultaneous recording of task-induced changes in blood oxygenation, volume, and flow using diffuse optical imaging and arterial spin-labeling MRI. Neuroimage. 2005;25(3):701–7. doi: 10.1016/j.neuroimage.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Huppert TJ, Allen MS, Diamond SG, Boas DA. Estimating cerebral oxygen metabolism from fMRI with a dynamic multicompartment Windkessel model. Hum Brain Mapp. 2009;30(5):1548–67. doi: 10.1002/hbm.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert TJ, Diamond SG, Boas DA. Direct estimation of evoked hemoglobin changes by multimodality fusion imaging. J Biomed Opt. 2008;13(5):054031. doi: 10.1117/1.2976432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert TJ, Hoge RD, Diamond SG, Franceschini MA, Boas DA. A temporal comparison of BOLD, ASL, and NIRS hemodynamic responses to motor stimuli in adult humans. Neuroimage. 2006a;29(2):368–382. doi: 10.1016/j.neuroimage.2005.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert TJ, Hoge RD, Dale AM, Franceschini MA, Boas DA. Quantitative spatial comparison of diffuse optical imaging with blood oxygen level-dependent and arterial spin labeling-based functional magnetic resonance imaging. J Biomed Opt. 2006b;11(6):064018. doi: 10.1117/1.2400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Hewson-Stoate N, Martindale J, Redgrave P, Mayhew J. Nonlinear coupling of neural activity and CBF in rodent barrel cortex. Neuroimage. 2004;22(2):956–65. doi: 10.1016/j.neuroimage.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Kennan RP, Horovitz SG, Maki A, Yamashita Y, Koizumi H, Gore JC. Simultaneous recording of event-related auditory oddball response using transcranial near infrared optical topography and surface EEG. Neuroimage. 2002;16(3 Pt 1):587–92. doi: 10.1006/nimg.2002.1060. [DOI] [PubMed] [Google Scholar]
- Kida I, Yamamoto T, Tamura M. Interpretation of BOLD MRI signals in rat brain using simultaneously measured near-infrared spectrophotometric information. NMR Biomed. 1996;9(8):333–8. doi: 10.1002/(SICI)1099-1492(199612)9:8<333::AID-NBM439>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt A, Obrig H, Requardt M, Merboldt KD, Dirnagl U, Villringer A, Frahm J. Simultaneous recording of cerebral blood oxygenation changes during human brain activation by magnetic resonance imaging and near-infrared spectroscopy. J Cereb Blood Flow Metab. 1996;16(5):817–26. doi: 10.1097/00004647-199609000-00006. [DOI] [PubMed] [Google Scholar]
- Lin AL, Fox PT, Yang Y, Lu H, Tan LH, Gao JH. Time-dependent correlation of cerebral blood flow with oxygen metabolism in activated human visual cortex as measured by fMRI. Neuroimage. 2009;44(1):16–22. doi: 10.1016/j.neuroimage.2008.08.029. [DOI] [PubMed] [Google Scholar]
- Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412(6843):150–7. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Lu H, van Zijl PC. Experimental measurement of extravascular parenchymal BOLD effects and tissue oxygen extraction fractions using multi-echo VASO fMRI at 1.5 and 3.0 T. Magn Reson Med. 2005;53(4):808–16. doi: 10.1002/mrm.20379. [DOI] [PubMed] [Google Scholar]
- Luh W, Wong E, Bandettini P, Hyde J. QUIPSS II with thin-slice ti1 periodic saturation: a method for improving accuracy of quantitative perfusion imaging using pulsed arterial spin labeling. Magn Reson Med. 1999;6(41):1246–1254. doi: 10.1002/(sici)1522-2594(199906)41:6<1246::aid-mrm22>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Maciejewski PK, Kida I, Hyder F. Estimating dynamic CMRO2 from dynamic CBF and BOLD fMRI measurements. Proc Intl Soc Mag Reson Med. 2004;11:272. [Google Scholar]
- Moseley M, Donnan G. Multimodality Imaging: Introduction. Stroke. 2004;35:2632–2634. [Google Scholar]
- Obata T, Liu TT, Miller KL, Luh WM, Wong EC, Frank LR, Buxton RB. Discrepancies between BOLD and flow dynamics in primary and supplementary motor areas: application of the balloon model to the interpretation of BOLD transients. Neuroimage. 2004;21(1):144–53. doi: 10.1016/j.neuroimage.2003.08.040. [DOI] [PubMed] [Google Scholar]
- Oja JM, Gillen JS, Kauppinen RA, Kraut M, van Zijl PC. Determination of oxygen extraction ratios by magnetic resonance imaging. J Cereb Blood Flow Metab. 1999;19(12):1289–95. doi: 10.1097/00004647-199912000-00001. [DOI] [PubMed] [Google Scholar]
- Opitz B, Mecklinger A, Von Cramon DY, Kruggel F. Combining electrophysiological and hemodynamic measures of the auditory oddball. Psychophysiology. 1999;36(1):142–7. doi: 10.1017/s0048577299980848. [DOI] [PubMed] [Google Scholar]
- Ou W, Nissilä I, Radhakrishnan H, Boas DA, Hämäläinen MS, Franceschini MA. Study of neurovascular coupling in humans via simultaneous magnetoencephalography and diffuse optical imaging acquisition. Neuroimage. 2009;46(3):624–32. doi: 10.1016/j.neuroimage.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera JJ, Watanabe J, Kazuki I, Naoki M, Aubert E, Ozaki T, Kawashima R. A state-space model of the hemodynamic approach: nonlinear filtering of BOLD signals. Neuroimage. 2004;21(2):547–67. doi: 10.1016/j.neuroimage.2003.09.052. [DOI] [PubMed] [Google Scholar]
- Siegel AM, Culver JP, Mandeville JB, Boas DA. Temporal comparison of functional brain imaging with diffuse optical tomography and fMRI during rat forepaw stimulation. Phys Med Biol. 2003;48(10):1391–403. doi: 10.1088/0031-9155/48/10/311. [DOI] [PubMed] [Google Scholar]
- Steinbrink J, Villringer A, Kempf F, Haux D, Boden S, Obrig H. Illuminating the BOLD signal: combined fMRI-fNIRS studies. Magn Reson Imaging. 2006;24(4):495–505. doi: 10.1016/j.mri.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Strangman G, Culver JP, Thompson JH, Boas DA. A quantitative comparison of simultaneous BOLD fMRI and NIRS recordings during functional brain activation. NeuroImage. 2002;17:719–731. [PubMed] [Google Scholar]
- Tak S, Jang J, Lee K, Ye JC. Quantification of CMRO2 without hypercapnia using simultaneous near-infrared spectroscopy and fMRI measurements. Phys Med Biol. 2010;55(11):3249–3269. doi: 10.1088/0031-9155/55/11/017. [DOI] [PubMed] [Google Scholar]
- Tak S, Yoon SJ, Jang J, Yoo K, Jeong Y, Ye JC. Quantitative analysis of hemodynamic and metabolic changes in subcortical vascular dementia using simultaneous near-infrared spectroscopy and fMRI measurements. NeuroImage. 2011;55(1):176–184. doi: 10.1016/j.neuroimage.2010.11.046. [DOI] [PubMed] [Google Scholar]
- Toronov V, Webb A, Choi JH, Wolf M, Michalos A, Gratton E, Hueber D. Investigation of human brain hemodynamics by simultaneous near-infrared spectroscopy and functional magnetic resonance imaging. Med Phys. 2001;28(4):521–7. doi: 10.1118/1.1354627. [DOI] [PubMed] [Google Scholar]
- Torricelli A, Pifferi A, Taroni P, Giambattistelli E, Cubeddu R. In vivo optical characterization of human tissues from 610 to 1010 nm by time-resolved reflectance spectroscopy. Phys Med Biol. 2001;46(8):2227–2237. doi: 10.1088/0031-9155/46/8/313. [DOI] [PubMed] [Google Scholar]
- Wang L, Jacques SL, Zheng L. MCML - Monte Carlo modeling of light transport in multi-layered tissues. Comput Methods Programs Biomed. 1995;47(2):131–146. doi: 10.1016/0169-2607(95)01640-f. [DOI] [PubMed] [Google Scholar]
- Wong E, Buxton R, Frank L. Implementation of quantitative perfusion imaging techniques for functional brain mapping using pulsed arterial spin labeling. NMR Biomed. 1997;4–5(10):237–249. doi: 10.1002/(sici)1099-1492(199706/08)10:4/5<237::aid-nbm475>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]