Abstract
Gold nanoparticles (AuNPs) protected by self-assembled monolayers (SAMs) are capable of presenting precisely engineered surfaces at the nanoscale, allowing the mimicry of biomacromolecules on an artificial platform. Here we review the generation, characterization, and applications of monolayer-protected AuNPs that have been designed for immunorecognition by the integration of an oligopeptide epitope into the protecting monolayer. The resulting peptide-AuNP conjugate is an effective platform for biomimesis, as demonstrated by multiple studies. Recent work is presented and future directions for this field of research are discussed.
1. Introduction
Because of the intrinsically optimized nature of biochemical systems, the natural order is an excellent starting point for the development of diagnostic and therapeutic tools at the molecular level. Classes of molecules which are uniquely capable of performing robust, highly specific chemistry have come to comprise the basis of biological systems. The virtually infinite range of biomolecular chemistry is derived from the diversity and multifunctionality of these molecules. Through evolutionary optimization, chemistry in vivo has become highly efficient and versatile. However, attempts to transpose biochemistry to experiments in vitro generally result in dysfunction. The advantages of native biochemistry are largely inaccessible because of a limited ability to properly harness it.
Mimicking these optimized systems through scientific research and development has yielded a number of technological advances for sensing,1 tissue engineering,2 catalysis,3 and methods for fabricating and processing two- and three-dimensional materials.4–7 The development of artificial molecules which imitate biomolecules offers the opportunity to couple the advantages of naturally- and artificially-developed chemical methods. This molecular biomimicry is an efficient route to the creation of effective tools for the manipulation of biochemistry in or ex vivo.
To generate biomimetic tools, natural components are often modified or conjugated to artificial components to induce a desired response. Common strategies utilise an artificial scaffold1,4,6 on which biomolecules or fragments can be conjugated to grant specificity and/or functionality. In the same way that native biochemistry generally utilizes molecular classes with structural diversity and functional precision, biomimetic strategies must also be both highly versatile and highly precise. Each biomimetic tool must be precisely tailored to achieve a specific aim. The scaffold should provide a surface which can easily engineered for use in any desired application, allowing for conjugated elements to provide robust specificity.
Gold nanoparticles (AuNPs) fill the role of a scaffold in the creation of biomimetic materials. They possess the well-known bioinert properties of gold,8 occupy a size regime ranging from that of small to large biomacromolecules,9,10 and can be conjugated to nearly any organic compound or mixture of compounds that contain a free thiol,11,12 amine,13 or phosphine.14 Our focus has been on the monolayer-protected AuNP, defined as a nanoscale gold core surrounded by a dense self-assembled monolayer (SAM) of gold-thiolate complexes.15,16 These AuNPs are effectively core-shell nanomaterials, in which the organic shell is responsible for biomimetic interactions. The gold-thiolate monolayer confers additional stability on the AuNP, protecting it sterically and electronically from decomposition, aggregation, and other irreversible reactions.11
AuNPs have been synthesized with a variety of protecting thiolate ligands.11,12,17 The termini of the conjugated ligands extend from the metal core, giving the AuNP the desired solubility and determining other physiochemical interactions with the media. The use of a polar, hydrophilic organic ligand shell yields water-soluble AuNPs in a basic form.17,18 The shell can be made more complex by adding other thiolate ligands during19,20 or after21 the reductive formation of the AuNPs, or by modifying the ligands through chemical reactions.12 This allows for the selective addition of desired properties and chemical functionalities to the AuNP. By precisely engineering the organic shell22,23 the shell can be tuned for selective binding or to avoid binding altogether.24–26
The size of the AuNP core affects interactions between the organic shell and its environment. The size of the AuNP can be made similar in scale to various biological macromolecules such as enzymes, receptors, immunoglobulins, antigens, DNA, and carbohydrates. Choosing an optimal size can create greater specificity toward biomolecules with given secondary structural features.27 The size of the core also determines the projection of the monolayer into its surrounding environment. Smaller monolayer-protected AuNPs (approx. <2 nm core diameter) with higher surface curvature are less protected sterically than larger analogues. The greater exposure of the individual protecting ligands and the core-shell interface leads to increased reactivity compared to larger AuNPs.21,28,29 As the size of the AuNP increases, the surface curvature decreases. More dense ligand shells cause ligand termini to be more responsible for chemical interactions.23
Because of these properties monolayer-protected AuNPs are an ideal biomimetic tool. The use of monolayer-protected AuNPs for biomimicry originated with the concept of integrating biologically active molecules, such as small molecule drugs,30 enzyme cofactors,18 DNA,31,32 saccharides,33 and peptides12,34,35 into the monolayer. The latter has been dominant in biomimetic applications because of the importance of protein interactions in biological systems and the ease of integrating cysteine-bearing synthetic peptides into the monolayer. The following is a review of our research into the development of biomimetic monolayer-protected AuNPs for immunorecognition. Results of some recent work are also presented, and areas for future exploration will be discussed.
2. Synthesis, modification and characterization
As in the field of drug discovery, large numbers of biomimetic variants can be examined to find suitable candidates for specific applications. Considering the number of variants which must be synthesized, modified, and characterized, the use of a scaffold which can be quickly brought to a final product is essential. Monolayer-protected AuNPs are such a scaffold, being generated from a remarkably facile synthesis using commercially available materials, followed by simple purification methods and characterization.
Syntheses of water-soluble monolayer-protected AuNPs are generally adapted from early work by Murray and coworkers.18,36 An alcohol solvent is used to dissolve chloroaurate (AuCl4−) and a thiol ligand, or a mixture of ligands. The mixture is then reduced by sodium borohydride and allowed to stir or sit for several hours. The size of the AuNPs can be controlled by the manipulation of several variables in the synthetic scheme. Specifically, the diameter of AuNPs correlates with the AuCl4−-to-ligand ratio, reaction temperature, and the reductant-to-AuCl4− ratio.37
The AuNP product can be purified by precipitation, centrifugation, solvent washes, electrophoresis, and/or dialysis. The core and shell of the AuNPs can then be modified. The core size can be decreased by adding large quantities of free thiol to the AuNPs,38 or increased by the use of other additives.39 The shell can be modified by simple organic reactions.12 An easier and more common modification route is the addition of free thiol-bearing molecules to promote ligand exchange.21 Integrating biomolecules into the monolayer is facilitated by functionalization with cysteine or another thiol. In some cases an inert spacer, such as a short peptide fragment, oligoethylene glycol, or alkyl chain, is added to allow epitope presentation without interference from the rest of the organic shell.
A number of analytical tools are used to characterize AuNPs. Transmission electron microscopy (TEM) is used to measure the core size and monodispersity.11 Nuclear magnetic resonance (NMR) spectroscopy allows characterization of organic shell composition.40 UV-Vis spectroscopy facilitates core size approximation through the observation of a localized surface plasmon band (λ ≈ 520 nm) indicative of larger AuNPs.41,42 Thermogravimetric analysis (TGA) determines the mass fraction of metal and organic components in the AuNP sample.37 Elemental analysis provides precise stoichiometric information.
Mass spectrometry (MS) is a more recently popularized tool for characterizing AuNPs, being capable of characterizing the composition of the organic shell as well as core size in some cases.43 Mass spectrometry has been used to characterize the abundance of thiolate ligands in the organic shell, including cysteine-terminated peptides.43–48 More recently, gas-phase structural separation by ion mobility-mass spectrometry (IM-MS) has been used to distinguish ligands in the organic shell from free impurities.49 The gas-phase separation allows the determination of ligand shell compositions for mixtures too complex to characterize by NMR.50 Furthermore, IM-MS has been demonstrated to reveal the presence of nanoscale phase segregation within the organic shell.51 This capability is particularly useful for future applications of AuNPs with anisotropic organic shells.
Once the AuNP biomimic is assembled, characterization of AuNP-biomolecule binding interactions is performed. Traditional methods for observing antibody interactions, such as enzyme-linked immunosorbent assay, (ELISA)52 or surface plasmon resonance (SPR)53 can be applied. The unique properties of monolayer-protected AuNPs, such as strong absorbance and scattering of visible and higher-energy electromagnetic radiation, can be helpful in this characterization. Another unique property of these nanomaterials is their massiveness: because the major component of monolayer-protected AuNP is gold, it is much more massive than any organic macromolecule of equivalent size. The massive nature of AuNPs enables the use of miniaturized gravimetric detection methods to sensitively detect relatively small amounts of binding.
The quartz crystal microbalance (QCM) consists of a piezoelectric quartz resonator with metal electrodes that is attached to an oscillator circuit. QCM has become a useful analytical technique because of the linear relationship between mass deposited on the crystal and its resonant frequency.54 Oscillator circuits that permit liquid measurements have made the technique useful in the area of bioanalytical chemistry.55,56 Because of its desirable mechanical, electrical, chemical, and thermal properties, the ideal oscillator for this technology is quartz.55 Specifically, AT-cut quartz crystals have a temperature coefficient that is nearly zero between 0 and 50 °C, making them ideal for QCM measurements at ambient temperatures without sophisticated temperature control.57 This technique is rapid, sensitive, label-free, and relatively low-cost compared to SPR and other alternative techniques. The QCM has been adapted to detect and monitor protein binding,55 diseases58–64 and biomimetic AuNPs.34,35,65,66
Utilizing these strategies for synthesizing, characterizing, and testing, monolayer-protected AuNPs have been generated and optimized as scaffolds for biomimetic organic shells. The work described in the following sections is divided into experiments conducted in vitro and in vivo, respectively.
3. Biomimicry of native antigens on AuNPs in vitro
Our primary focus has been in the generation of biomimetic AuNPs to serve as simulants for viruses of clinical interest. The use of biological materials in research and development carries an intrinsic risk to both the manufacturer and the consumer. This risk can only be mitigated through stringent safety and security protocols that are maintained at high cost. Biomimetic simulants obviate the use of dangerous biological material, allowing study without the need for stringent safety and security protocols. The simulants could be used in assay development, diagnostics, or the prevention and treatment of disease.
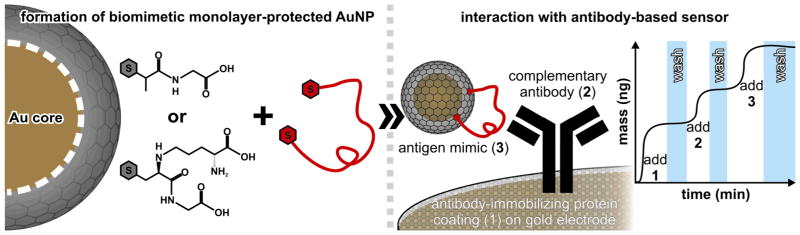
To illustrate the biomimetic capabilities of monolayer-protected AuNPs, we first utilized a glutathione (GSH)-protected AuNP.65 While glutathione is not a traditional antigen, it was intended to serve as a proof of concept that small AuNPs (<3 nm diameter) can be effective scaffolds for projecting antigens that can be recognized and bound by an antibody. A polyclonal anti-GSH antibody was bound to a QCM chip, and the antibody was exposed to GSH AuNPs (Figure 1). Antibody-AuNP binding was observed. The specificity of the binding between the antibody and biomimetic antigen was established by a tiopronin-protected AuNP control. Considering the similarity between the two organic molecules (Figure 1), this specificity is remarkable.65
Figure 1.
Experimental design for in vitro studies described here. A cysteine-appended epitope is integrated into the protecting organic shell of a hydrophilic monolayer-protected AuNP by ligand exchange. The gold electrode on a QCM chip is functionalized with a protein matrix (1) designed to immobilize the selected antibody while resisting non-specific binding to the biomimic. The antibody (2) is added, and subsequently the biomimetic AuNP (3) is brought into contact with the chip. After each addition, the chip is washed to ensure the elimination of non-specifically bound material. Observed mass increase in the final step that is not washed away is indicative of immunorecognition via biomimic-antibody binding.
Improvement over 2D surface (hemagglutinin)
Having established the capability of biomimetic monolayer-protected AuNPs to achieve immunorecognition, the next generation of biomimetic AuNPs was designed. A cysteine-appended 10-amino acid peptide epitope from the hemagglutinin (HA) protein of influenza was integrated into the organic shell.34 The selected peptide sequence has been well-characterized, is present in a neutralizing site for influenza, and has a commercially-available complementary monoclonal antibody (mAb). Having integrated the HA epitope into the shell of a tiopronin-protected AuNP, specific binding to the mAb was observed. The efficacy of the epitope-conjugated AuNP as a biomimetic scaffold was compared to that of an identical epitope bound to a planar gold surface.34 The three-dimensional AuNP yielded a higher ratio of antibody:peptide binding than the two-dimensional planar gold surface. This suggested that a curved three-dimensional surface was more efficient in mimicking the native antigen.
An important consideration for the mimicry of biomolecules using three-dimensional monolayers is the conformation of the conjugated biomolecule. In order to truly mimic a biomolecule, higher-order structure must be preserved. It is assumed that the optimal conformation of an AuNP-bound peptide epitope will be highly similar to that of the epitope in the native antigen. In this context, the ability to utilize bidentate or multidentate attachment schemes is an additional benefit of the monolayer-protected AuNP. Previous studies have suggested that bidentate ligands, each end being in a dynamic equilibrium with the solution phase, will eventually migrate into positions corresponding to the minimal point of the conformational potential energy surface for the epitope.51,67,68 This phenomenon, which could take place through lateral translation of thiol termini or through a series of dissociative and associative steps, should eventually allow for large, multidentate structures to adopt a structure which should be similar to a native structure.
In addition to the ability to constrain bidentate ligands into biologically relevant secondary structures, the availability of numerous binding sites on AuNP surfaces allows for the integration of multiple epitopes or other biologically active ligands into a single scaffold. Continuing the earlier studies of the HA system, tiopronin-protected AuNPs were combined with a FLAG epitope, HA epitope, both epitopes, or neither epitope.64 The peptide epitopes were integrated into the organic shell via a cysteine-terminated hexaethylene glycol linker intended to provide enhanced accessibility to the epitope. QCM using either anti-FLAG or anti-HA IgG-functionalized sensor surfaces were used to evaluate the immunological activity of the mimics synthesized. Each functionalized AuNP was detected by the appropriate antibody, but unmodified tiopronin-protected AuNPs were not recognized.
Bacillus anthracis (protective antigen)
In order to probe the relevance of higher-order structure on biomimicry in a more clinically relevant system, Gerdon and coworkers studied the protective antigen (PA) of B. anthracis, one of three proteins of the anthrax toxin.35 PA was selected because it precedes the other two proteins (edema factor and lethal factor) in their transport for infection, making it an ideal sub-unit target for neutralizing antibodies in commercial vaccines. Three portions of PA were chosen for examination: the C-terminus and two internal loops of the PA protein which were identified as cell receptors. As in the previous study, tiopronin-protected AuNPs were used as a starting material, into which one of the three PA-derived epitopes was integrated. The two internal loops studied were appended with cysteine residues at one or both of their respective N- and C-termini. Peptides with cysteine appended to both termini were expected to attach in a bidentate fashion to a single AuNP, thus preserving the secondary and tertiary structure of the native epitope with the greatest accuracy (Figure 1). Only one cysteine residue was attached to the peptide derived from the C-terminus region of PA, in order to mimic the native terminus.
The biomimetic AuNPs were tested against a commercially-available monoclonal antibody using a QCM platform.35 The loop-mimicking bidentate peptide had a higher affinity (Ka) for the antibody when compared to the linear epitope, especially at physiological pH and ionic strength, confirming that the loop-presenting nanoparticle was more readily recognized by the antibody. Preferential recognition of the loop epitope suggests that the loop better mimicked the native epitope, making critical binding residues more available to the antibody.
Human respiratory syncytial virus (fusion protein)
In the construction an antigenic mimic, it is useful to understand the specific intermolecular forces that are responsible for antibody-antigen binding. Specifically, the goal is to identify the amino acid side chains that are involved in binding and their spatial organization. Given this information, a biomimic can be designed to copy the important parameters without the need to use complicated techniques, expensive and fragile biological systems, or exotic reagents.
An optimal approach to obtaining this information is the use of epitope mapping, in which the affinity of antigens or fragments thereof are tested against a complementary antibody. This yields information about the precise elements of the antigen that are important for interactions with the antibody. Linear epitope mapping refers to techniques which solely interrogate primary structure, while conformational epitope mapping considers the effects of secondary and tertiary structure as well. This approach has been employed previously for the creation of biomimetic AuNPs. Rutledge and co-workers investigated binding “hot spots” in a proposed long C-terminal epitope sequence of Ebola glycoprotein, finding local maxima in an ELISA assay against the mAb 15H10 that appeared to be centred about two separate sequences of three amino acids.66
The success of this approach led us to apply a similar methodology to the human respiratory syncytial virus (HRSV), another clinically relevant disease. We selected the fusion (F) glycoprotein, a common target for monoclonal antibodies, for mimicry. For antibody detection of the mimic we selected Palivizumab (PZ, also known by the trade name Synagis), a commercially available antibody with high affinity for the the HRSV F protein. A linear epitope mapping experiment was then performed using ELISA to find a suitable peptide sequence for binding to an AuNP. The sequence with the highest affinity (NSELLSLINDMPITNDQKKLMSNN) was further probed in order to test the importance of directionality and each of the fragments of the epitope (Table S1, Figure S1). It was found that the amino acids closer to the N-terminus were more important for binding, and that attaching the linker to the N-terminus resulted in much better binding than the equivalent epitope attached at the C-terminus (Figure S2).
Based on this data, multiple antigen mimics composed of tiopronin-protected AuNPs with various peptide epitopes, linkers, and terminal functionalities were then synthesized and evaluated with the QCM immunosensor (Table S2). A QCM immunosensor strategy for SARS-associated coronavirus62 was adapted to specifically and quantitatively detect HRSV or an HRSV mimic. The platform, depicted in Figure S3, was composed of an Fc binding cell (Protein G from Staphylococcus aureus), a blocking agent (bovine serum albumin, BSA), and a monoclonal detection antibody. As a positive control, native HRSV was flowed across the immunosensor. Strong binding was observed (Figure S4, S5).
AuNP antigen mimics and non-mimic negative controls were then tested with the immunosensor. The negative controls and most of the nanoparticle mimics evaluated did not interact with PZ. The two exceptions were designated mimic A and mimic B. It is notable that both mimics used the same epitope that yielded the strongest signal in a peptide ELISA assay (Figure S1). The primary sequence of this epitope contains the same sequence as peptides studied by McLellan and coworkers.69,70
To further test the specificity of the binding, a second experiment was performed in which PZ was left out entirely for the first part of the experiment (Figure 2). The mimic would be brought into contact with a non-sensing QCM chip; any binding observed at this step would be non-specific. The mimic would be removed, PZ added and the mimic brought into contact for a second time. In this experiment, mimic A was found to be highly specific, but mimic B was found to bind non-specifically to the non-sensing proteins on the gold surface in the first part of the experiment. The strong and specific binding of mimic A to the QCM HRSV immunosensor led to further study.
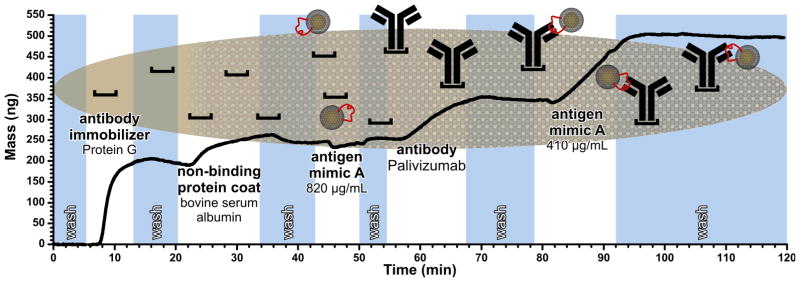
Figure 2.
Detection of mimic A by a QCM immunosensor with and without PZ. Blue sections represent the flow of PBS without analyte. White sections indicate the binding of Protein G (50 μg/mL, 192 ng bound), BSA (1 mg/mL, 53 ng bound), nanoparticle mimic A (820 μg/mL, no detectable amount of material bound), PZ (10 μg/mL, 92 ng bound), and nanoparticle mimic A again (410 μg/mL, 150 ng bound), respectively.
The binding affinity of mimic A against PZ was evaluated in a concentration study with the QCM (Table S3, Figures S7–S10), revealing a dissociation constant (Kd) of 292 ± 177 nM. This is within the range expected for mAb-antigen interaction. The value of the Kd suggests that the affinity of PZ for the mimic is between ~100 and ~300 fold weaker than PZ for the native F protein (Kd = 1.4 nM).71 The binding is competitive with that of epitope scaffolds constructed by McLellan and co-workers (Kd = 87 to 3950 nM) which incorporate the same primary epitope sequence.70 The benefits of biomimetics are apparent in this context: our approach utilizes straightforward laboratory techniques and automated peptide synthesis equipment rather than recombinant genetic techniques and equipment, which are considerably more costly and complex. This provides a significant advantage with respect to the cost of time and materials, as well as the level of experience required of scientists working with this system.
This high-affinity HRSV F protein mimic has been synthesized using straightforward and cost-effective techniques that require relatively little expertise. Mimic A performs nearly as well as the state of the art subunit mimic (Kd = 87 nM),70 but not as well as the native protein antigen (Kd = 1.4 nM), for PZ binding.71 Given this performance, the mimic might serve as a novel subunit vaccine, which is needed in the clinic during the RSV season. This novel material could also be studied as a non-infectious calibration standard for a number of HRSV immunosensors. The performance of this mimic can be improved by further tuning of the AuNP scaffold, specifically by altering the size of the gold core, the primary ligands in the monolayer, and the peptide loading in the monolayer.
4. In vivo: toxicity and biodistribution in murine models
Monolayer-protected AuNPs are clearly capable of mimicking antigens in vitro, thus the next step is to test antigenic effectiveness and toxicity in vivo. As a control experiment, mice were dosed with tiopronin-protected AuNPs equivalent to those used for in vitro studies (Figure 3).72 It was found that dosages above ~23 mg/kg were sufficiently toxic to be fatal. Drawing from research by Brust and co-workers73 and Feldheim and coworkers,74 we integrated a short thioalkylated polymer, mercaptoundecyltetraethylene glycol (MUTEG) into the monolayer in order to enhance biocompatibility. The mixed-ligand Tio:MUTEG AuNPs yielded drastically reduced toxicity,72 indicating that the tiopronin ligand is responsible for the toxicity. It was suggested that the carboxylate termini produce a strong negative charge at the surface of the ligand shell at physiological pH, inducing unintended interactions with biomolecules.26 Reducing the charge by replacing carboxylate- terminated ligands with alcohol-terminated ligands was thought to reduce the charge sufficiently to eliminate the toxic effects.
Figure 3.
Experimental design for in vivo experiments. Mice are injected with monolayer-protected AuNPs. Blood and urine are collected regularly, and organs are collected at the end of the study. The concentration of gold in each is measured by inductively-coupled plasma-optical emission spectroscopy (ICP-OES), and any immune response is observed by an increase in white blood cell counts.
Subsequent studies demonstrated the effectiveness of shorter mercaptotetraethylene glycol (MTEG) ligands and a carboxylic acid-terminated analogue (MTEG acid) for the elimination of AuNP toxicity.75 This finding in the latter case is remarkable: despite the fact that the ligand shell preserved roughly the same number of carboxylate termini, the effect in vivo was altered significantly. This indicates more complex surface effects: either the carboxylate alone was not responsible for toxicity, or the added MTEG acid altered aggregation or serum protein adsorption behaviour. These biologically-induced processes have not yet been studied for these AuNPs, but are known to affect interaction with macrophages and other downstream effects.76 Tetraethylene glycol ligands are roughly double the length of tiopronin ligands: the ligand tails which extend beyond the tiopronin shell may effectively shield the tiopronin carboxylates, reducing the interactions which lead to toxicity.
Another effect of the integration of thiolated tetraethylene glycol ligands is an alteration in biodistribution. Localization in the liver, kidney, and spleen were the most common, but the integration of tetraethylene glycol ligands decreased the concentration in the organs by nearly an order of magnitude.72,75 Circulation in the blood is also increased for these AuNPs, with the concentration of gold in the blood remaining remarkably constant over 4 weeks.75
Glutathione-protected AuNPs
For studies in vivo, the organic shell should simply provide a bioinert surface from which a peptide can be projected in a biomimetic fashion. In order to construct a maximally bioinert monolayer, it is sensible to choose protecting ligands which are natively present in vivo. This is especially relevant considering the susceptibility of the monolayer to ligand exchange with thiols natively present in biological systems. If the AuNP persists for any considerable length of time, it is reasonable to assume that some significant portion of the original monolayer will have been replaced with biomolecules.
Glutathione is the most abundant thiol in vivo, and has been observed at millimolar concentration ranges.77,78 This makes glutathione the most likely molecule to exchange into the monolayer. By synthesizing AuNPs with glutathione as a protecting ligand, this effect can be negated. We hypothesized that, despite the large number of carboxylate termini, glutathione-protected AuNPs would have high biocompatibility and resistance to long-term chemical alteration, reducing the number of unintended interactions and thus the toxicity.
Glutathione-protected AuNPs showed no signs of toxicity at up to 80 mg/kg in a murine model, confirming our hypothesis. Localization in organs was similar to that of the mixed-monolayer AuNPs discussed previously, though the AuNPs were entirely absent from the blood after ~24 hr (Figures S12–S14). The rapid clearance of GS AuNPs confirms the findings of Zheng and co-workers.79 However, there was a remarkable AuNP concentration increase in the blood between 3 and 4 weeks post-injection (Figure S13). This finding was reproduced in a second, longer study. This second study found that, after 4 weeks, the AuNP concentration in blood returned to basal levels, later returning to elevated levels at ~7 weeks post-injection (Figure S14).
In a third study, the synthetic method used for the production of glutathione-protected AuNPs was slightly altered, yielding AuNPs of a virtually identical size but with an apparent decrease in ligand packing density in the organic shell. NMR characterization revealed differences in signal which are consistent with alterations in monolayer structure (Figure S11).80 This second AuNP product induced a remarkably different response in vivo; the delayed circulation observed with the first glutathione-protected AuNP product was not observed in this case.
These findings raise a number of interesting questions. Can AuNPs that have been localized to organs be returned to circulation via an intrinsic or extrinsic trigger? Can variations in surface structure among “isomeric” homoligand AuNPs cause differences in biological function similar to that observed for heteroligand AuNPs?25 This is a fertile area for research, with many relevant questions waiting to be probed. The answers to these questions will shed light on the chemical and biochemical processes that take place during interactions with nanomaterial surfaces.
5. Conclusion
Biomimetic, nanostructured organic shells: a holistic approach
Recent advances in the generation of anisotropic monolayer-protected AuNPs24,51,81 allow biomimetic applications in which large portions of the organic monolayer are involved in biological interactions.23 This stands in contrast to the studies described here, in which an inert organic shell with a single integrated bioactive component is favoured. In the case of structured organic shells, the complex surface chemistry which has produced remarkably variable effects in vivo, as described in the previous section, can be harnessed to tune its biological effects. These effects can range from reduction of non-specific protein interactions24 to modification of cell entry pathways,25 creation of controllable pores in cell membranes,82 and development of specific binding sites.23 It is also possible to begin conformational epitope mapping, in which three-dimensional sites can be mimicked using multiple ligands spatially oriented in the monolayer in a reproducible way.
It is possible to conceptualize a number of new approaches to achieving and applying biomimicry based on combining these advances with developed technologies. To name a few examples: computer-aided molecular design could be adapted to assist in conformational epitope mapping in order to design surface structures which project desired functionalities in a spatially-controlled fashion in order to optimally imitate native antigens. High-throughput screening (HTS) methods utilizing variable size and surface structure could be used to rapidly assess the effectiveness of the various nanoscale structures which are best suited for a given target. Structure-based drug design principles could be applied to maximize affinity and specificity, leading to the development of entirely artificial antigen mimics for use in biological systems. Some of these approaches already exist in a slightly different form,22,23 and have not been specifically applied for antigen mimicry by structured organic shells.
Another advantage of structured organic shells is the ability to build controlled AuNP superstructures.83 This has the potential to lead to a new type of biomimicry, in which individual anisotropic AuNPs become equivalent to protein subunits. The ability to create stable organic-organic interfaces could yield an interesting array of biomimetic chemistry on an AuNP scaffold.
Conclusion
Through the numerous studies described here it has become apparent that monolayer-protected AuNPs can generate a variety of biological responses which are strongly dependent on the surface chemistry of the organic shell. Biomimicry, specifically in the context of antigenicity, is clearly achievable. Though the optimization of these biomimetic scaffolds for applications in vivo is ongoing, the number of tools and variables available for optimization are bringing end goals much closer to reality. The ability to harness nanoscale structure in the organic shell provides a greatly expanded versatility which can both accelerate success and open up new avenues of research into biologically useful nanomaterials. The practically endless possibilities for complex chemistry lead to applications in areas much more diverse than what has been detailed here. Because of their versatility and ease of use, the future of monolayer-protected AuNP-based applications consists of numerous ideas waiting to be explored.
Supplementary Material
Table 2.
Monolayer-protected AuNPs that have been tested in vivo, including those protected by various mercaptan tetraethylene glycols (MTEG), such as ligands with an alkyl linker region (mercaptoundecyl-TEG, or MUTEG) and those with carboxylic acid termini (MTEG acid). Organ retention is listed in order of highest to lowest Au concentration. Only tiopronin-protected AuNPs have been found to be toxic, and most of the variants have safely cleared the system without generating an immune response in se. This is a desirable trait, indicating the potential to be made antigenic with specificity by the integration of an epitope into the organic shell.
| Monolayer composition | Toxicity | Blood and urine clearance time | Immune response | Organ retention |
|---|---|---|---|---|
| Tiopronin (Tio) | >23 mg/kg | 8 hr | none | liver, spleen, kidney |
| Tio:MUTEG | none | >4 wk | at high MUTEG abundance | spleen, kidney, heart, liver |
| Tio:MTEG acid | none | 24 hr | none | liver, spleen, kidney, heart |
| Tio:MTEG | none | 24 hr | none | liver, spleen, kidney, heart |
| Glutathione | none | 24 hr (and later, see text) | none | liver, spleen, heart, kidney |
Acknowledgments
Thanks to Prof. David Wright, Prof. James Crowe, Prof. R. Gerald Keil, and Prof. Leslie Hiatt for helpful discussions in the execution of the research presented here. We acknowledge Prof. Aren Gerdon, Prof. Brian Huffman, and Dr. Carrie Simpson, whose past work has been recounted here. Financial support for this work was provided by the National Institutes of Health (GM 076479 to D.E.C. and RC2DA028981 to J.A.M.), the Defense Threat Reduction Agency (HDTRA1-09-0013), the Vanderbilt Institute for Nanoscale Science & Engineering, the Vanderbilt College of Arts and Sciences, and the Vanderbilt Institute of Chemical Biology, and the Vanderbilt Institute for Integrative Biosystems Research and Education.
Footnotes
Electronic Supplementary Information (ESI) available: Synthetic, characterization, and experimental details. See DOI: 10.1039/b000000x/
Notes and references
- 1.Haupt K, Mosbach K. Chem Rev. 2000;100:2495–2504. doi: 10.1021/cr990099w. [DOI] [PubMed] [Google Scholar]
- 2.Shin H, Jo S, Mikos AG. Biomaterials. 2003;24:4353–4364. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 3.Breslow R. Acc Chem Res. 1995;28:146–153. [Google Scholar]
- 4.Cha JN, Stucky GD, Morse DE, Deming TJ. Nature. 2000;403:289–292. doi: 10.1038/35002038. [DOI] [PubMed] [Google Scholar]
- 5.Bunker BC, Rieke PC, Tarasevich BJ, Campbell AA, Fryxell GE, Graff GL, Song L, Liu J, Virden JW, McVay GL. Science. 1994;264:48–55. doi: 10.1126/science.264.5155.48. [DOI] [PubMed] [Google Scholar]
- 6.Heuer A, Fink D, Laraia V, Arias J, Calvert P, Kendall K, Messing G, Blackwell J, Rieke P, Thompson D, Wheeler A, Veis A. Science. 1992;255:1098–1105. doi: 10.1126/science.1546311. [DOI] [PubMed] [Google Scholar]
- 7.Naik RR, Stringer SJ, Agarwal G, Jones SE, Stone MO. Nat Mater. 2002;1:169–172. doi: 10.1038/nmat758. [DOI] [PubMed] [Google Scholar]
- 8.Cobley CM, Chen J, Cho EC, Wang LV, Xia Y. Chem Soc Rev. 2011;40:44–56. doi: 10.1039/b821763g. [DOI] [PubMed] [Google Scholar]
- 9.Negishi Y, Nobusada K, Tsukuda T. J Am Chem Soc. 2005;127:5261–5270. doi: 10.1021/ja042218h. [DOI] [PubMed] [Google Scholar]
- 10.Zheng N, Fan J, Stucky GD. J Am Chem Soc. 2006;128:6550–6551. doi: 10.1021/ja0604717. [DOI] [PubMed] [Google Scholar]
- 11.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. J Chem Soc, Chem Commun. 1994:801–802. [Google Scholar]
- 12.Templeton AC, Hostetler MJ, Warmoth EK, Chen S, Hartshorn CM, Krishnamurthy VM, Forbes MDE, Murray RW. J Am Chem Soc. 1998;120:4845–4849. [Google Scholar]
- 13.Leff DV, Brandt L, Heath JR. Langmuir. 1996;12:4723–4730. [Google Scholar]
- 14.Weare WW, Reed SM, Warner MG, Hutchison JE. J Am Chem Soc. 2000;122:12890–12891. [Google Scholar]
- 15.Walter M, Akola J, Lopez-Acevedo O, Jadzinsky PD, Calero G, Ackerson CJ, Whetten RL, Gronbeck H, Hakkinen H. Proc Natl Acad Sci U S A. 2008;105:9157–9162. doi: 10.1073/pnas.0801001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jadzinsky PD, Calero G, Ackerson CJ, Bushnell DA, Kornberg RD. Science. 2007;318:430–433. doi: 10.1126/science.1148624. [DOI] [PubMed] [Google Scholar]
- 17.Ackerson CJ, Jadzinsky PD, Kornberg RD. J Am Chem Soc. 2005;127:6550–6551. doi: 10.1021/ja046114i. [DOI] [PubMed] [Google Scholar]
- 18.Templeton AC, Chen S, Gross SM, Murray RW. Langmuir. 1999;15:66–76. [Google Scholar]
- 19.Chen S, Murray RW. J Phys Chem B. 1999;103:9996–10000. [Google Scholar]
- 20.Choo H, Cutler E, Shon YS. Langmuir. 2003;19:8555–8559. [Google Scholar]
- 21.Hostetler MJ, Templeton AC, Murray RW. Langmuir. 1999;15:3782–3789. [Google Scholar]
- 22.Bresee J, Maier KE, Melander C, Feldheim DL. Chem Commun. 2010;46:7516–7518. doi: 10.1039/c0cc02663h. [DOI] [PubMed] [Google Scholar]
- 23.Hung A, Mwenifumbo S, Mager M, Kuna JJ, Stellacci F, Yarovsky I, Stevens MM. J Am Chem Soc. 2011;133:1438–1450. doi: 10.1021/ja108285u. [DOI] [PubMed] [Google Scholar]
- 24.Jackson AM, Myerson JW, Stellacci F. Nat Mater. 2004;3:330–336. doi: 10.1038/nmat1116. [DOI] [PubMed] [Google Scholar]
- 25.Verma A, Uzun O, Hu Y, Hu Y, Han HS, Watson N, Chen S, Irvine DJ, Stellacci F. Nat Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 27.Tsoli M, Kuhn H, Brandau W, Esche H, Schmid G. Small. 2005;1:841–844. doi: 10.1002/smll.200500104. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Acevedo O, Kacprzak KA, Akola J, Hakkinen H. Nature Chem. 2010;2:329–334. doi: 10.1038/nchem.589. [DOI] [PubMed] [Google Scholar]
- 29.Templeton AC, Hostetler MJ, Kraft CT, Murray RW. J Am Chem Soc. 1998;120:1906–1911. [Google Scholar]
- 30.Gibson JD, Khanal BP, Zubarev ER. J Am Chem Soc. 2007;129:11653–11661. doi: 10.1021/ja075181k. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Malicka J, Gryczynski I, Lakowicz JR. Anal Biochem. 2004;330:81–86. doi: 10.1016/j.ab.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jewell CM, Jung JM, Atukorale PU, Carney RP, Stellacci F, Irvine DJ. Angew Chem Int Ed. 2011;50:12312–12315. doi: 10.1002/anie.201104514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manea F, Bindoli C, Fallarini S, Lombardi G, Polito L, Lay L, Bonomi R, Mancin F, Scrimin P. Adv Mater (Weinheim, Ger) 2008;20:4348–4352. [Google Scholar]
- 34.Gerdon AE, Wright DW, Cliffel DE. Biomacromolecules. 2005;6:3419–3424. doi: 10.1021/bm050475o. [DOI] [PubMed] [Google Scholar]
- 35.Gerdon AE, Wright DW, Cliffel DE. Angew Chem Int Ed. 2006;45:594–598. doi: 10.1002/anie.200503328. [DOI] [PubMed] [Google Scholar]
- 36.Templeton AC, Cliffel DE, Murray RW. J Am Chem Soc. 1999;121:7081–7089. [Google Scholar]
- 37.Hostetler MJ, Wingate JE, Zhong CJ, Harris JE, Vachet RW, Clark MR, Londono JD, Green SJ, Stokes JJ, Wignall GD, Glish GL, Porter MD, Evans ND, Murray RW. Langmuir. 1998;14:17–30. [Google Scholar]
- 38.Schaaff TG, Whetten RL. J Phys Chem B. 1999;103:9394–9396. [Google Scholar]
- 39.Kanehara M, Sakurai J-i, Sugimura H, Teranishi T. J Am Chem Soc. 2009;131:1630–1631. doi: 10.1021/ja8058167. [DOI] [PubMed] [Google Scholar]
- 40.Terrill RH, Postlethwaite TA, Chen CH, Poon CD, Terzis A, Chen AD, Hutchison JE, Clark MR, Wignall G, Londono JD, Superfine R, Falvo M, Johnson CS, Samulski ET, Murray RW. J Am Chem Soc. 1995;117:12537–12548. [Google Scholar]
- 41.Qian H, Zhu Y, Jin R. Proc Natl Acad Sci. 2012;109:696–700. doi: 10.1073/pnas.1115307109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dass A. J Am Chem Soc. 2011;133:19259–19261. doi: 10.1021/ja207992r. [DOI] [PubMed] [Google Scholar]
- 43.Harkness KM, Cliffel DE, McLean JA. Analyst. 2010;135:868–874. doi: 10.1039/b922291j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tracy JB, Kalyuzhny G, Crowe MC, Balasubramanian R, Choi JP, Murray RW. J Am Chem Soc. 2007;129:6706–6707. doi: 10.1021/ja071042r. [DOI] [PubMed] [Google Scholar]
- 45.Tracy JB, Crowe MC, Parker JF, Hampe O, Fields-Zinna CA, Dass A, Murray RW. J Am Chem Soc. 2007;129:16209–16215. doi: 10.1021/ja076621a. [DOI] [PubMed] [Google Scholar]
- 46.Dass A, Holt K, Parker JF, Feldberg SW, Murray RW. J Phys Chem C. 2008;112:20276–20283. [Google Scholar]
- 47.Gies AP, Hercules DM, Gerdon AE, Cliffel DE. J Am Chem Soc. 2007;129:1095–1104. doi: 10.1021/ja0639057. [DOI] [PubMed] [Google Scholar]
- 48.Dass A, Stevenson A, Dubay GR, Tracy JB, Murray RW. J Am Chem Soc. 2008;130:5940–5946. doi: 10.1021/ja710323t. [DOI] [PubMed] [Google Scholar]
- 49.Harkness KM, Fenn LS, Cliffel DE, McLean JA. Anal Chem. 2010;82:3061–3066. doi: 10.1021/ac100251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harkness KM, Hixson BC, Fenn LS, Turner BN, Rape AC, Simpson CA, Huffman BJ, Okoli TC, McLean JA, Cliffel DE. Anal Chem. 2010;82:9268–9274. doi: 10.1021/ac102175z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harkness KM, Balinski A, McLean JA, Cliffel DE. Angew Chem Int Ed. 2011;50:10554–10559. doi: 10.1002/anie.201102882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engvall E, Perlmann P. Immunochem. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 53.Liedberg B, Nylander C, Lunström I. Sensors and Actuators. 1983;4:299–304. [Google Scholar]
- 54.Sauerbrey G. Z Phys A. 1959;155:206–222. [Google Scholar]
- 55.Janshoff A, Galla HJ, Steinem C. Angew Chem Int Ed. 2000;39:4004–4032. doi: 10.1002/1521-3773(20001117)39:22<4004::aid-anie4004>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 56.Nomura T, Okuhara M. Anal Chim Acta. 1982;142:281–284. [Google Scholar]
- 57.Meixner H, Jones R, editors. Micro- and Nano-Sensor Technology/Trends in Sensor Markets. VCH; Weinheim, Germany: 1995. [Google Scholar]
- 58.Davis KA, Leary TR. Anal Chem. 1989;61:1227–1230. doi: 10.1021/ac00186a010. [DOI] [PubMed] [Google Scholar]
- 59.Koenig B, Graetzel M. Anal Chem. 1994;66:341–344. doi: 10.1021/ac00075a005. [DOI] [PubMed] [Google Scholar]
- 60.Muramatsu H, Dicks JM, Tamiya E, Karube I. Anal Chem. 1987;59:2760–2763. doi: 10.1021/ac00150a007. [DOI] [PubMed] [Google Scholar]
- 61.Uttenthaler E, Kößlinger C, Drost S. Anal Chim Acta. 1998;362:91–100. [Google Scholar]
- 62.Zuo B, Li S, Guo Z, Zhang J, Chen C. Anal Chem. 2004;76:3536–3540. doi: 10.1021/ac035367b. [DOI] [PubMed] [Google Scholar]
- 63.Yu JS, Liao HX, Gerdon AE, Huffman B, Scearce RM, McAdams M, Alam SM, Popernack PM, Sullivan NJ, Wright D, Cliffel DE, Nabel GJ, Haynes BF. J Virol Methods. 2006;137:219–228. doi: 10.1016/j.jviromet.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 64.Miller S, Hiatt L, Keil R, Wright D, Cliffel D. Anal Bioanal Chem. 2011;399:1021–1029. doi: 10.1007/s00216-010-4419-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerdon AE, Wright DW, Cliffel DE. Anal Chem. 2005;77:304–310. doi: 10.1021/ac048953t. [DOI] [PubMed] [Google Scholar]
- 66.Rutledge RD, Huffman BJ, Cliffel DE, Wright DW. J Mater Res. 2008;23:3161–3168. doi: 10.1557/JMR.2008.0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ionita P, Volkov A, Jeschke G, Chechik V. Anal Chem. 2007;80:95–106. doi: 10.1021/ac071266s. [DOI] [PubMed] [Google Scholar]
- 68.Caragheorgheopol A, Chechik V. Phys Chem Chem Phys. 2008;10:5029–5041. doi: 10.1039/b805551c. [DOI] [PubMed] [Google Scholar]
- 69.McLellan JS, Chen M, Kim A, Yang Y, Graham BS, Kwong PD. Nat Struct Mol Biol. 2010;17:248–250. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McLellan JS, Correia BE, Chen M, Yang Y, Graham BS, Schief WR, Kwong PD. J Mol Biol. 2011;409:853–866. doi: 10.1016/j.jmb.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, Dormitzer M, O’Grady J, Koenig S, Tamura JK, Woods R, Bansal G, Couchenour D, Tsao E, Hall WC, Young JF. J Infect Dis. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 72.Simpson CA, Huffman BJ, Gerdon AE, Cliffel DE. Chem Res Toxicol. 2010;23:1608–1616. doi: 10.1021/tx100209t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanaras AG, Kamounah FS, Schaumburg K, Kiely CJ, Brust M. Chem Commun. 2002:2294–2295. doi: 10.1039/b207838b. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y, Shipton MK, Ryan J, Kaufman ED, Franzen S, Feldheim DL. Anal Chem. 2007;79:2221–2229. doi: 10.1021/ac061578f. [DOI] [PubMed] [Google Scholar]
- 75.Simpson CA, Agrawal AC, Balinski A, Harkness KM, Cliffel DE. ACS Nano. 2011;5:3577–3584. doi: 10.1021/nn103148x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW. J Am Chem Soc. 2011;134:2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 77.Moron MS, Depierre JW, Mannervik B. Biochim Biophys Acta, Gen Subj. 1979;582:67–78. doi: 10.1016/0304-4165(79)90289-7. [DOI] [PubMed] [Google Scholar]
- 78.Akerboom TPM, Sies H. In: Methods Enzymol. William BJ, editor. Vol. 77. Academic Press; 1981. pp. 373–382. [DOI] [PubMed] [Google Scholar]
- 79.Zhou C, Long M, Qin Y, Sun X, Zheng J. Angew Chem Int Ed. 2011;50:3168–3172. doi: 10.1002/anie.201007321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song Y, Harper AS, Murray RW. Langmuir. 2005;21:5492–5500. doi: 10.1021/la0503606. [DOI] [PubMed] [Google Scholar]
- 81.Pradhan S, Xu L, Chen S. Adv Funct Mater. 2007;17:2385–2392. [Google Scholar]
- 82.Alexeev A, Uspal WE, Balazs AC. ACS Nano. 2008;2:1117–1122. doi: 10.1021/nn8000998. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Z, Glotzer SC. Nano Lett. 2004;4:1407–1413. doi: 10.1021/nl0493500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.