Abstract
Modern neuroscientific research stands on the shoulders of countless giants. PubMed alone contains more than 21 million peer-reviewed articles with 40–50,000 more published every month. Understanding the human brain, cognition, and disease will require integrating facts from dozens of scientific fields spread amongst millions of studies locked away in static documents, making any such integration daunting, at best. The future of scientific progress will be aided by bridging the gap between the millions of published research articles and modern databases such as the Allen Brain Atlas (ABA). To that end, we have analyzed the text of over 3.5 million scientific abstracts to find associations between neuroscientific concepts. From the literature alone, we show that we can blindly and algorithmically extract a “cognome”: relationships between brain structure, function, and disease. We demonstrate the potential of data-mining and cross-platform data-integration with the ABA by introducing two methods for semiautomated hypothesis generation. By analyzing statistical “holes” and discrepancies in the literature we can find understudied or overlooked research paths. That is, we have added a layer of semi-automation to a part of the scientific process itself. This is an important step toward fundamentally incorporating data-mining algorithms into the scientific method in a manner that is generalizable to any scientific or medical field.
1. Introduction
The scientific method begins with a hypothesis about our reality that can be tested via experimental observation. Hypothesis formation is iterative, building off prior scientific knowledge. Before one can form a hypothesis, one must have a thorough understanding of previous research to ensure that the path of inquiry is founded upon a stable base of established facts. But how can a researcher perform a thorough, unbiased literature review when over one million scientific articles are published annually (Björk et al., 2009)? The rate of scientific discovery has outpaced our ability to integrate knowledge in an unbiased, principled fashion. One solution may be via automated information aggregation (Akil et al., 2011). In this manuscript we show that, by calculating associations between concepts in the peer-reviewed literature, we can algorithmically synthesize scientific information and use that knowledge to help formulate plausible low-level hypotheses.
Neuroscience is a particularly complex discipline that relies upon expertise from many disparate fields (Akil et al., 2011). The aim of neuroscience is to understand relationships between brain, behavior, and disease; yet, no one person or group can possibly unify all neuroscientific understanding into a coherent framework. In this paper, we show that the literature contains a hidden network of connected facts that, by definition, recapitulate known neuroscientific relationships. Neuroanatomical, behavioral, and disease associations can be quantified and visualized to speed research and education or to discover understudied research paths (Yarkoni et al., 2010; Wren et al., 2004; Bilder et al., 2009). Rather than allowing our limited ability to review the entire scientific literature bias our hypotheses, we can algorithmically integrate millions of scientific research papers in a principled fashion.
To accomplish this, we used a co-occurrence algorithm to calculate the pair-wise association index (AI) between neuroscientific terms (and their synonyms) contained within more than 3.5 million papers indexed in PubMed (see Methods). The primary assumption is that the frequency with which terms appeared together across the titles or abstracts of manuscripts is proportional to their probability of association. That is, we assumed an underlying structure within the peer-reviewed neuroscientific literature that we could leverage to our advantage. We conceive of our system as a proof-of-concept tool for knowledge discovery limited only by the size and quality of the inputs. We believe that, in its current state, when combined with the website search and visualization system we created to accompany it (http://www.brainscanr.com), it acts as a more sophisticated complement to normal PubMed searches. Furthermore, it provides, for the first time, a method for quantifying the relationship between disparate neuroscientific concepts, paving the way for researchers to incorporate statistical decision making into their future research.
2. Methods
2.1. Data collection
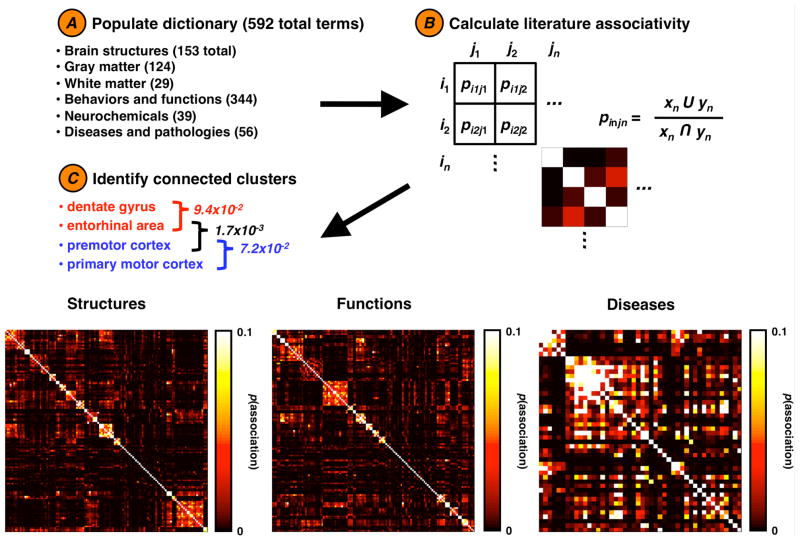
We populated a dictionary with phrases for 124 brain regions, 291 cognitive functions, and 47 diseases. Brain region names and associated synonyms were selected from BrainInfo (2007) (Bowden et al., 2007), Neuroscience Division, National Primate Research Center, University of Washington (Bowden and Dubach, 2003). Cognitive functions were obtained from (http://www.cognitiveatlas.org/) (Poldrack et al., 2011). Disease names are from (http://www.ninds.nih.gov/). The initial population of the dictionary was meant to represent the broadest, most plausibly common search terms that are also relatively unique (and thus likely not to lead to spurious connections). The full list of terms and their synonyms are included in the Supporting List 1.
2.2. Association probabilities
We quantified the association between two terms using a weighted co-occurrence algorithm (Jaccard index) that highlights the unique relationship between term pairs. For any given pair of terms i and j, we define the association index,
where the intersection between ci,j and di,j was calculated using the following query of the PubMed database using the ESearch utility and the count return type (using the example c is “prefrontal cortex” and d is “striatum”):
The union was calculated using the sum of two separate queries:
Note that for these searches, all synonyms for a given term are included within the parentheses and each term is individually surrounded by quotation marks to limit the search to each exact phrase. Furthermore, the “field=word” modifier limits the search to the article’s title and abstract. This reduces instances of false associations due to name or journal title homographs (e.g., author name “Fear” or journal name “Language” as opposed to the behavioral terms “fear” or “language”).
2.3. Data visualization and website creation
The brainSCANr website was created using the Google App Engine (Google, Inc.) framework. Graph connectivity plotting (see Figure 3C for an example) was performed using the JavaScript InfoVis Toolkit (Nicolas Garcia Belmonte, http://thejit.org/). The full association database used in this study is available at that site for download. For Figure 2 the graph was plotted using the GraphViz (AT&T Research Labs) radial plot function.
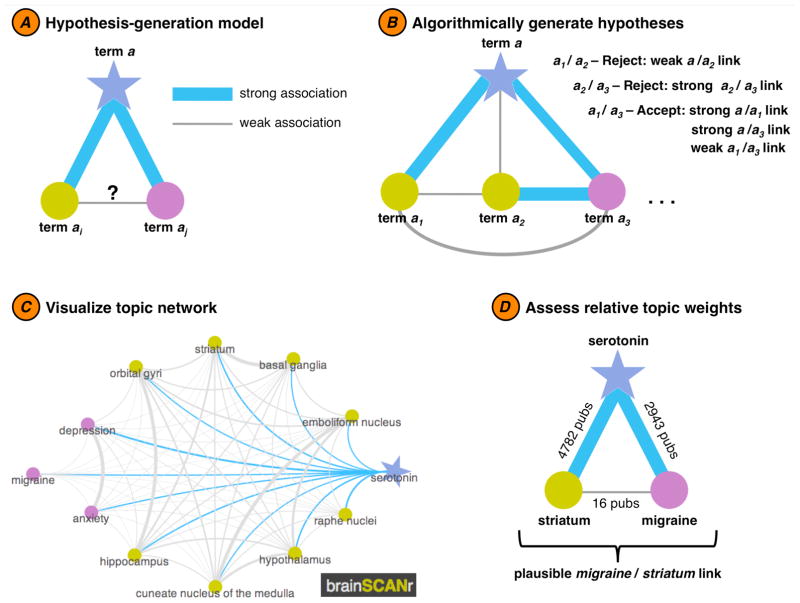
Figure 3.
Semi-automated hypothesis generation. A simple algorithm is used to evaluate possible novel or under-studied research topics based upon statistical discrepancies in the scientific literature. In the example above, we algorithmically determine a possible relationship between migraine and serotonin-related brain regions such as the striatum (for a full list of possible hypotheses, see Supporting List 2). (A) The hypothesis generation model is based on a simple “friend-of-a-friend should be a friend” concept: if term a is strongly associated with two terms (ai and aj), yet the association between ai and aj is weak, then perhaps we are (scientifically) missing a relationship between ai and aj. (B) In order for the algorithm to flag a relationship as a plausible hypothesis, three conditions must be met: terms ai and aj each need to have a strong relationship with their parent term, a, and the relationship between ai and aj should be weak. (C) The topic network for term a can be visualized (here using http://www.brainSCANr.com) to highlight relative associations between terms (main term a: blue star; brain regions: gold circles; diseases: purple circles). (D) An example of an algorithmically-defined hypothesis. Here, the term serotonin is strongly associated with two terms: striatum (2943 joint publications) and migraine (4782 joint publications). In contrast, however, there are only 16 publications (at the time of this writing) that jointly mention striatum and migraine. Given that serotonin is so strongly related to these two topics, perhaps there is a missing association between migraines and the striatum.
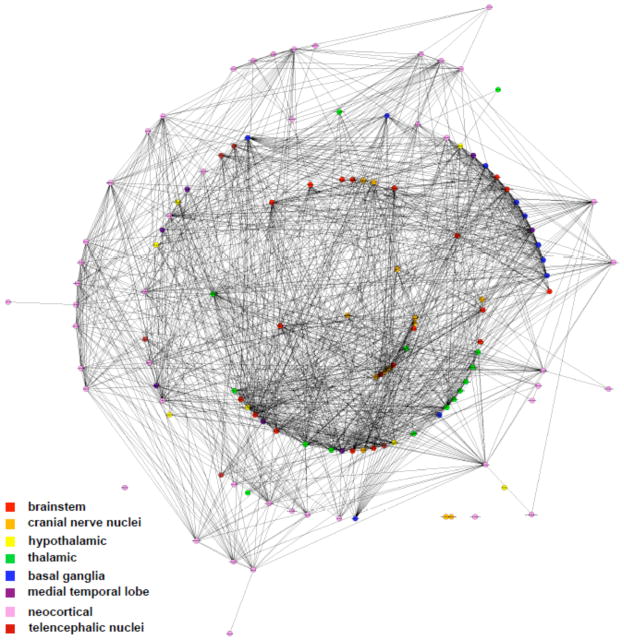
Figure 2.
Inferred systems-level connectome. Based upon a pre-defined dictionary of 124 brain regions and their 703 synonyms, we calculated the probability of association between all pairs of brain regions based upon their co-occurrence in the scientific literature indexed via PubMed. This method recovers known neuroanatomical relationships (see Supporting Table 1). In the center rings, brainstem structures cluster together, with telencephalic/neocortical structures arranged in the outside rings. Note the clustering of thalamic and basal ganglia structures in the middle rings. Graphic visualization was performed using GraphViz (AT&T Research Labs) with a connectivity threshold of 0.095.
Clustering was performed using an iterative (k-means) clustering algorithm (MATLAB® R2009b, Natick, MA; kmeans.m) and hierarchical clustering (linkage.m). For the brain structure and functions analyses, we used 20 clusters, and for the disease analysis we used 5 clusters. It is important to note that there are many techniques for clustering data (see Parsons et al., 2004), but the actual resulting clusters and dendrogram presented herein do not affect the results, but rather are included for display purposes.
2.4. Allen Brain Atlas
Gene expression data from the Allen Brain Atlas (ABA) were taken from subject H0351.2001 and visualized (Figure 4A) on the ABA website. In order to allow for cross-database comparisons, raw expression intensity values for each gene g in brain region b was normalized across all n brain regions B in the ABA by converting them to a z-score such that,
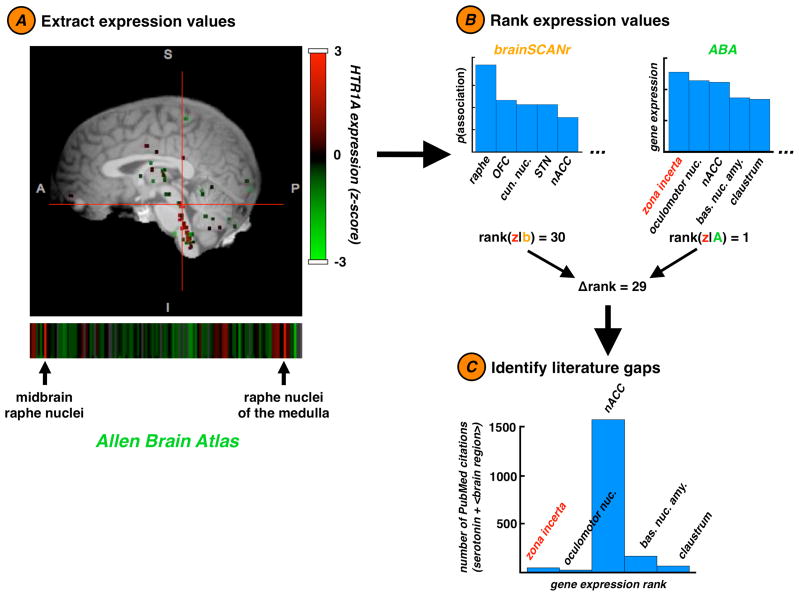
Figure 4.
Allen Brain Atlas integration. A second approach toward semi-automated hypothesis generation is accomplished via integrating our data with the Allen Brain Atlas (ABA). (A) From the ABA we extract real gene expression values for a sub-selection of human brain regions (60 were used in our ABA analyses). Here we show an expression map for HTR1A, the gene that encodes the 5-HT1A receptor. (B) We begin by ranking the brain regions that most strongly express genes related to a specific neurochemical (here, serotonin). According to the ABA (A, green), serotonin-related genes are most strongly expressed in the zona incerta (z, red). However according to our data (b, orange), serotonin is most strongly associated with the brain region raphe nuclei; the zona incerta ranks 30th out of 60 brain regions. (C) When we examine the number of publications in PubMed that discuss serotonin with the 5 brain regions that most strongly express serotonin-related genes, we find that the nucleus accumbens has orders of magnitude more publications than the other regions (1584 publications), whereas only 42 papers discuss serotonin and the zona incerta, despite the fact that the zona incerta expresses serotonin-related genes most strongly. This discrepancy suggests that the role of the zona incerta in serotonergic processes and serotonin-related functions is poorly understood. Our method demonstrates that such holes in our understanding may be identified automatically and algorithmically.
To calculate a single expression value for broad neurochemical such as serotonin, which may have many genes coding receptors and transporters, we averaged normalized gene expression values across all relevant genes by searching the ABA genetic probe ontology for the neurochemical name. So, for example, the gene expression deviation value we report for serotonin represents the average across 208 serotonin-related genes. Because both the ABA and this manuscript use the same neuroanatomical naming ontology (Bowden and Dubach, 2003), we could compare brain region and gene expression data between databases.
For the analyses in Figure 5, gene expression deviation is defined as the absolute value of average gene expression z-scores, as both gene under- and over-expression are heavily researched. For the ABA/AI correlation analysis, we only correlated data where AI > 0 for a given neurochemical/brain region pair, yielding 1131 correlated pairs out of 1500 possible (25 neurochemicals times 60 brain regions). For surrogate correlation analyses (Figure 5B), 104 surrogate correlations were calculated by extracting the correlation coefficient r between gene expression deviation and a random permutation of the AI data. This gave a distribution of possible r values against which the real correlation could be compared by calculating the z-score and associated p-value. For the surrogate difference analyses (Figure 5D) the same technique was used as in the surrogate correlation analysis, however we compared the real difference with 104 surrogate differences, rather than correlation data. Surrogate difference scores were calculated by first combining all gene expression deviation values (for AI > 0 and for AI = 0) and then randomly drawing 1131 values (to represent data where AI > 0), calculating the mean, and comparing that against the mean of the remaining 369 values.
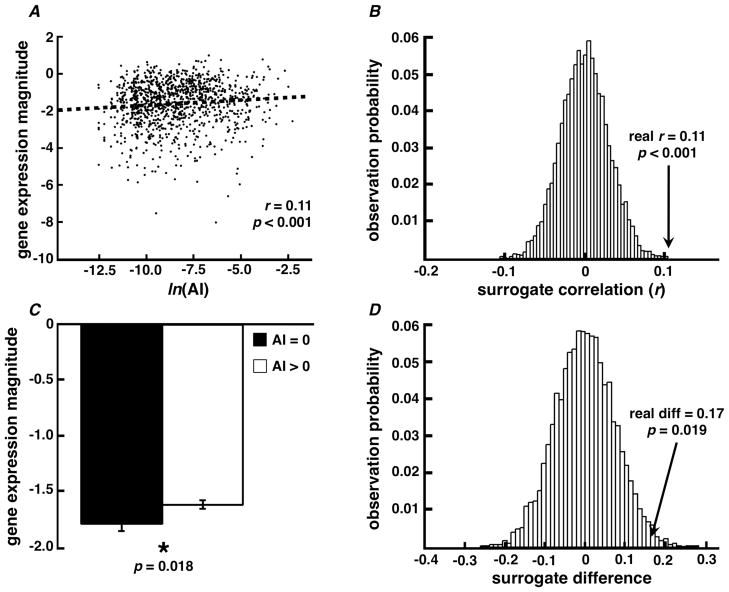
Figure 5.
Allen Brain Atlas / brainSCANr validation. We validated PubMed association probabilities by comparing them to brain/gene expression data from the ABA. (A) We find a significant correlation between association index (AI) and gene expression magnitude from the ABA for 1131 brain/gene expression relationships. (B) Resampling statistics suggest that the observed correlation (r = 0.11) is unlikely due to an artifact caused by AI or gene expression magnitude calculation techniques (see Methods). (C) ABA gene expression magnitude is lower when the PubMed association probability equals zero. That is, when there are no published manuscripts relating a given brain region with a specific neurochemical, the magnitude of actual gene expression is likely to be lower than for cases where brain/neurochemical relationships do exist in PubMed (two-sample t-test, p = 0.018). (D) Resampling statistics suggest that the observed difference between the mean gene expression magnitudes when AI equals zero (n = 369) versus when AI is greater than zero (n = 1131; real diff = 0.17) is also unlikely due to an artifact caused by a difference in the number of trials between groups.
2.5. Semi-automated hypothesis generation
We introduce two methods for semi-automated hypothesis generation. The first relies on a simple “friend-of-a-friend should be a friend” concept wherein we assume that two terms that each strongly relate to a parent term should relate to one another. If they do not, then that relationship is flagged as a possible hypothesis. More technically, we considered each “parent” term and looked to find the terms the parent is strongly related to (more than 1000 joint publications between parent and “child”). If the parent had two or more such relationships we then examined the relations between each of these strong children. Any child/child pair that had a weak relationship (fewer than 30 publications) was flagged as a possible hypothesis. While the values used to define “strong” and “weak” relationships are somewhat arbitrary, we sought to keep the number of hypothesis candidates low. This choice of cutoffs yielded 896 hypotheses out of 175528 total term pairs (0.51% rate).
The second method for hypothesis generation looks for discrepancies between actual gene expression data extracted from the ABA and the calculated neurochemical/brain relationship from PubMed. ABA gene expression values for each of the 25 neurochemicals were first sorted to find in which brain regions they are most strongly expressed. We then identified cases where brain regions were found to strongly express a given gene, yet had relatively few publications mentioning that region and gene.
3. Results
3.1. Cognome Construction
In order to reconstruct this cognome, we calculated the probability of association between each term and every other term, giving an association matrix of size (Figure 1 and Methods). Once the full association matrix was calculated we 2 constructed a full brain connectivity graph (Figure 2 and Supporting Figure 1), limited only by the dictionary used to define the search terms (see Supporting List 1 for the full list). We find relatively strong associations between all brain region terms. For visualization purposes we classified each brain region as belonging to one of 8 predefined macroscale clusters and colored each node according to group membership. This coloring highlights the clustering of brain regions by type; cortical regions form distinct groups farther from the central brainstem structures while thalamic and basal ganglia structures cluster together nearer the brainstem.
Figure 1.
Calculating brain structure, function, and disease relationships. We begin by (A) populating a database with search terms and their synonyms. From this we calculate (B) the probability of association (p) between a term (in) and all other terms (j), giving us (C) a series of association matrices. Each row/column index in this matrix represents the probability of association between two terms as calculated from PubMed. For each matrix, data are sorted according to the clusters identified via k-means clustering using 20 structure clusters, 20 function clusters, and 5 disease clusters. This method highlights several within-cluster associations along the diagonal (see Supporting Tables for clusters).
We then blindly clustered structures based upon their association weights (Figure 1 and Table 1). These clusters—defined by their PubMed associations—recapitulate known neuroanatomical circuits. Several of these circuits are anatomically diffuse; for example, one cluster, a “visual” circuit, associates the lateral geniculate nucleus and pulvinar, the superior colliculus, and the primary visual and visual extrastriate cortices. We observe clusters of brainstem auditory and prosencephalic auditory circuits as well as oculomotor nuclei. These results show that there is an inherent structure to the peer-reviewed literature that can be algorithmically recovered. Note that differences in clustering methods yield different results, and that the clusters shown here are meant as illustrative examples of the structure inherent to the data. The full hierarchical dendrogram can be viewed in Supporting Figure 2.
Table 1.
Brain structure clusters. Clusters identified via k-means clustering of the brain region association matrix. Cluster titles were defined post-hoc based upon author’s interpretation.
| Unknown | Speech/Motor | Basal Ganglia | Thalamic |
| cerebellum | Broca’s area | caudate nucleus | centromedian nucleus |
| cuneate nucleus of the medulla | insula | globus pallidus | intralaminar nuclear group |
| emboliform nucleus | operculum | nucleus accumbens | reticular nucleus of the thalamus |
| hippocampus | premotor cortex | putamen | ventral anterior nucleus |
| hypothalamus | primary motor cortex | substantia nigra | ventral posterior nucleus |
| orbital gyri | supplementary motor cortex | ventral tegmental area | ventral posterolateral nucleus |
| thalamus | Wernicke’s area | zona incerta | |
| Basal Ganglia II | Cingulate | Prefrontal | Heschls gyrus |
| basal ganglia | anterior cingulate gyrus | medial prefrontal cortex | planum temporale |
| striatum | cingulate gyrus | prefrontal cortex | transverse temporal gyrus |
| subthalamic nucleus | posterior cingulate gyrus | ||
| Visual | Hippocampal | Amygdalar | Hypothalamic |
| lateral geniculate nucleus | dentate gyrus | amygdala | dorsomedial nucleus of the hypothalamus |
| primary visual cortex | entorhinal area | basal forebrain nucleus | medial dorsal nucleus |
| pulvinar | perirhinal area | basal nucleus of the amygdala | posterior nucleus of the hypothalamus |
| superior colliculus | subiculum | diagonal band | ventral posteromedial nucleus |
| visual extrastriate | stria terminalis | ||
| Prosencephalic Auditory | Brainstem Auditory | Oculomotor | Cranial Nerve Nuclei |
| inferior colliculus | cochlear nuclei | abducens nucleus | dorsal motor nucleus of the vagus nerve |
| medial geniculate body | superior olive | interstitial nucleus of Cajal | hypoglossal nucleus |
| primary auditory area | trapezoid body | oculomotor nuclear complex | nucleus ambiguus |
| trochlear nucleus | solitary nucleus | ||
| Brainstem | Cortical | ||
| locus ceruleus | angular gyrus | ||
| medulla | fusiform gyrus | ||
| periaqueductal | inferior frontal gyrus | ||
| pons | inferior parietal lobule | ||
| raphe nuclei | inferior temporal gyrus | ||
| lingual gyrus | |||
| medial frontal gyrus | |||
| medial parietal gyrus | |||
| middle frontal gyrus | |||
| middle temporal gyrus | |||
| parahippocampal gyrus | |||
| postcentral gyrus | |||
| precuneus | |||
| superior frontal gyrus | |||
| superior parietal lobule | |||
| superior temporal gyrus |
Just as we can indentify clusters of associated brain regions, we can also cluster functions or diseases (see Figure 1). Conceptually, clustering cognitive and behavioral functions provides a quantification of the relationship between two cognitive tasks or behavioral states (Yarkoni et al., 2011; Poldrack et al., 2009). For example, there is a known relationship between “visual working memory” and “delayed match to sample” tasks (Voytek and Knight, 2010; Voytek et al., 2010) that is recovered by our algorithm (AI = 8.5*10−3); similarly there is a weak association between “visual working memory” and “stress” (AI = 6.1*10−6), suggesting the two concepts are relatively unrelated. As can be seen in Table 2, there are two clusters of tasks identified as “executive functions” and “monitoring and control” clusters. The former contains 9 tasks such as the Stroop and Wisconsin card sorting tasks, as well as working memory. The latter cluster contains tasks such as go/no-go, stop signal, and antisaccade. These tasks are known to be functionally related and interdependent. In Table 3 we outline clusters of diseases identified from their associations. This results in a cluster of psychiatric disorders including bipolar disorder, schizophrenia, and obsessive-compulsive disorder, as well as a cluster of agnosias such as Broca’s and Wernicke’s aphasia, apraxia, and prosopagnosia.
Table 2.
Functional clusters. Clusters identified via k-means clustering of the functions association matrix. Cluster titles were defined post-hoc based upon author’s interpretation.
| Social/Emotional | Attention | Cogntion/Consciousness | Language |
| affect recognition | attentional capacity | anticipation | language comprehension |
| emotional perception | attentional shifting | arousal | language processing |
| emotional recognition | attentional blink | association learning | language production |
| face perception | attentional resources | awareness | lexical processing |
| face recognition | auditory attention | classical conditioning | lexical retrieval |
| facial expression | divided attention | cognition | phonological encoding |
| familiarity | focused attention | consciousness | picture naming |
| happiness | inhibition of return | decision making | semantic processing |
| long term memory | oddball | fear | sentence comprehension |
| memory consolidation | selective attention | intelligence | sentence production |
| memory storage | spatial attention | pain | syntactic processing |
| memory trace | sustained attention | Pavlovian conditioning | word comprehension |
| object recognition | target detection | reward | word production |
| recognition memory | target processing | stress | |
| reconsolidation | visual attention | uncertainty | |
| social cognition | visual search | ||
| spatial memory | |||
| theory of mind | |||
| Monitoring and Control | Executive Functioning | Learning and Memory | Learning and Memory II |
| antisaccade | delayed recall | declarative memory | autobiographical memory |
| behavioral inhibition | digit span | habit learning | episodic memory |
| cognitive control | executive function | habit memory | memory retrieval |
| deductive reasoning | set shifting | procedural learning | semantic knowledge |
| error detection | Stroop | procedural memory | semantic memory |
| executive control | trail making | skill learning | |
| go/no-go | verbal memory | ||
| inductive reasoning | visual memory | ||
| performance monitoring | Wisconsin card sorting | ||
| stop signal | |||
| task switching | |||
| Working Memory | Phonological Processes | Knowledge | Perception |
| central executive | phonological buffer | declarative knowledge | auditory perception |
| phonological loop | phonological discrimination | nondeclarative knowledge | color perception |
| short term memory | phonological working memory | nondeclarative memory | form perception |
| spatial working memory | word repetition | procedural knowledge | visual perception |
| working memory | |||
| Speech | Implicit/Explicit Learning | Analogical Processes | Implicit/Explicit Memory |
| articulation | explicit knowledge | analogical problem solving | explicit memory |
| speech perception | explicit learning | analogical reasoning | implicit memory |
| speech production | implicit knowledge | ||
| Eye Movement | Sequence Learning | Intelligence | |
| eye movement | motor sequence learning | crystallized intelligence | |
| saccade | sequence learning | fluid intelligence |
Table 3.
Disease clusters. Clusters identified via k-means clustering of the disease association matrix. Cluster titles were defined post-hoc based upon author’s interpretation.
| Psychiatric Disorders | Agnosias | Alzheimer’s | Eating Disorders |
| anxiety | agnosia | Alzheimer’s disease | anorexia |
| bipolar disorder | aphasia | dementia | bulimia |
| depression | apraxia | ||
| obsessive compulsive disorder | Broca’s aphasia | ||
| panic disorder | prosopagnosia | ||
| schizophrenia | Wernicke’s aphasia | ||
| social phobia |
While these classifications provide important data on within-category clustering, by combining structural, functional, and disease terms in a unified matrix we can calculate cross-category clusters (see Table 4). We observe cross-category relationships for language terms including language comprehension, Wernicke’s area, and Wernicke’s aphasia. We also find a Parkinson’s disease cluster containing terms such as Parkinson’s disease, caudate nucleus, and substantia nigra. Such cross-category clustering demonstrates the utility of this method for integrating and unifying complex interrelationships across a broad range of neuroscientific fields.
Table 4.
Cross-category clusters. Clusters identified via k-means clustering of the entire association matrix. Cluster titles were defined post-hoc based upon author’s interpretation.
| Visual Cognition | Language | Learning and Reward | Speech Prodcution |
| antisaccade | language comprehension | association learning | articulation |
| attentional shifting | sentence comprehension | classical conditioning | speech perception |
| auditory attention | syntactic processing | reward | speech production |
| cognitive control | Broca’s aphasia | medial prefrontal cortex | word recognition |
| divided attention | Wernicke’s aphasia | nucleus accumbens | aphasia |
| executive control | Broca’s area | prefrontal cortex | apraxia |
| focused attention | Wernicke’s area | ventral tegmental area | dyslexia |
| inhibition of return | |||
| selective attention | |||
| spatial attention | Consciousness | Parkinson’s | Facial Perception |
| sustained attention | consciousness | Parkinson’s disease | face perception |
| task switching | ataxia | caudate nucleus | face recognition |
| visual attention | coma | globus pallidus | agnosia |
| visual search | cerebellum | putamen | prosopagnosia |
| frontal eye field | medulla | substantia nigra | |
| visual extrastriate | pons | ||
| TMS | Alzheimer’s | fMRI | Medical EEG |
| transcranial magnetic stimulation | cognition | functional magnetic resonance imaging | electroencephalography |
| premotor cortex | Alzheimer’s disease | inferior frontal gyrus | epilepsy |
| primary motor cortex | dementia | insula | |
| supplementary motor cortex |
3.2. Hypothesis Generation
While known relationships can be captured automatically, we can identify statistical “holes” in the literature using a method we call “semi-automated hypothesis generation”. In Figure 3 we outline the algorithm used to find these statistical discrepancies, based on a simple “friend-of-a-friend should be a friend” concept. For example, in Figure 3A we show a hypothetical relationship between a parent term a and its two children: ai and aj. In this case, both ai and aj are strongly related to their parent, but weakly related to one another. This is the basis for our hypothesis-finding algorithm (Figure 3C). In Figure 3D we show one real-world example (out of 896 identified) wherein both striatum and migraine are strongly related to serotonin (>2900 publications for each relationship), yet the striatum and migraines have few shared publications (only 16). While the lack of association may be due to a publication bias wherein null results go unreported, there may be a true association between the two concepts that is understudied. Using this method, the process of uncovering new research paths could drastically speed knowledge discovery (the full list of identified hypotheses is available in Supporting List 2).
We can extend this hypothesis discovery technique by incorporating gene expression data from the ABA (Lein et al., 2007). In Figure 4 we show comparisons between regional human gene expression data for serotonin-related genes versus the relationship between serotonin and those brain regions in PubMed. We find a significant discrepancy between actual gene expression data and literature associations. For example, serotonin-related genes are most highly expressed in the zona incerta, yet there are only 42 papers that discuss the zona incerta in relation to serotonin. In contrast, there are 1584 papers that discuss the nucleus accumbens and serotonin. While the first method finds statistical holes in the literature, this method identifies biases in neurochemical research.
3.3. Data Validation
We sought to validate our data by correlating the AI calculated from PubMed associations with real data. Because the Allen Brain Atlas uses the same neuroanatomical naming ontology as used by our database, we could more easily compare these two datasets than we could with other external sources. We find that the calculated AI for neurochemical/brain region relationships is significantly correlated (r1131 = 0.11, p < 0.001) with the gene expression magnitude for the same gene expression/brain region pairings (Figure 5A), and that this correlation is unlikely due to an artifact of our calculation and integration methods (Figure 5B). Furthermore, as would be expected, actual gene expression magnitude is lower for pairings where we find no neurochemical/brain region relationship in the literature (AI = 0) than for when there is a relationship (AI > 0) (t1498 = 2.36, p = 0.018), and that this effect is unlikely due to differences in the amount of data between the two groupings (Figure 5D).
4. Discussion
In this manuscript we demonstrate that, by mining the peer-review literature for associations between neuroscientific terms, we can recapitulate known scientific relationships. Furthermore, we introduce an algorithm for semi-automated hypothesis generation that can be used to speed research discovery. Although the current analysis is restricted to a limited dictionary of terms, the association and visualization methods are applicable to any search term or phrase found in PubMed, meaning that our method can be more broadly generalized to any scientific field using any peer-review database. Of course, there are limitations to our method. While we show that calculated AI does significantly correlate with real gene expression data, the correlation is relatively weak and explains only about 1% of the variance. This may be caused by several underlying factors, including, but not limited to, a loss of specificity as a result of averaging across a wide range of genes, inaccuracies in our text-mining approach or in the literature itself, the reliance upon gene expression data from a single human subject, or an incompatibility between the IA metric and gene expression data. Nevertheless the significance suggests that literature mining can capture real relationships (Figure 5A).
Furthermore, our calculations are by definition based upon the existing literature, thus associations may reflect publication biases (though there is a well-described publication bias such that negative results are underreported (Begg and Berlin, 1989; Dirnagl and Lauritzen, 2010; Ioannidis et al., 1997; Stern and Simes, 1997). Furthermore, our method does not differentiate positive from negative results: if a paper states that the amygdala does not relate to fear, that paper is weighted equally to a paper that finds a positive relationship. Despite these limitations, our associations map onto known relationships with remarkable accuracy.
Given the matrix of association values for each brain region, we show that we can recreate known neuroanatomical circuits using blind, automated clustering algorithms. These algorithms identify physically diffuse but functionally associated networks such as subcortical-cortical visual pathways, brainstem auditory nuclei, and even behavioral circuits involved in speech and other high-level cognitive processes. By clustering cognitive functions, we can quantify the relationship between a variety of cognitive tasks commonly used in neuroscientific research, such as the relationships between tasks used to study executive functioning or cognitive control, similar to automated meta-analytic methods (Yarkoni et al., 2011). Finally, searching across all brain structures, functions, and diseases, we show that we can uncover statistical discrepancies in the literature to aid in scientific discovery. While we cannot confirm that such algorithmically identified hypotheses are correct without conducting actual experiments, the search space in the biomedical sciences is so vast that we believe that any principled methods to reduce that space can only help speed discovery, as even ruling out incorrect relationships is helpful. That is, these hypotheses are not meant to represent a teleological endpoint, but rather a stepping-stone for researchers to help unmask possibly hidden mediating factors.
There is currently a massive scientific effort to identify the human connectome (Sporns et al., 2005; Modha and Singh, 2010; Editors, 2010). Even at the relatively macroscopic scale of systems and networks, the intricacies of neuroanatomical interconnectivity and how brain regions give rise to cognition and relate to disease are difficult to comprehend and visualize. Often these connectivity data are spread across dozens of research manuscripts, brain atlases, websites, and other repositories in static formats not openly accessible to all researchers. While fields such as genetics have put great effort into ontological projects (Zhang et al., 2010), the adoption of ontologies for neuroanatomy and cognition has been slow (but see (Bowden et al., 2007; Bohland et al., 2009; Larson and Martone, 2009; Stephan et al., 2000)). While the semantic associations we present herein appear to work well for the biomedical and psychological sciences, they may have more limited use in the physical sciences, for example, where the ontological weight is carried less by textual relationships.
Nevertheless, we can leverage the power of millions of publications to bootstrap informative relationships (Michel et al., 2010) and uncover scientific “metaknowledge” (Evans and Foster, 2011). Furthermore, the use of network mapping of textual relationships has recently been used in a variety of psychological and neuroscientific domains, including an analysis of comorbidity in psychiatric disorders based upon DSM-IV diagnostic criteria (Borsboom et al., 2011) and relationships between genes (Alako et al., 2005). By mining these relationships, we show that it is possible to add a layer of intelligent automation to the scientific method as has been demonstrated for the data modeling stage (Schmidt and Lipson, 2009). By implementing a connection-finding algorithm, we believe we can speed the process of discovering new relationships. So while the future of scientific research does not rely on these tools, we believe it will be greatly aided by them. This is a small step toward a future of semi-automated, algorithmic scientific research.
Supplementary Material
Acknowledgments
We thank Curtis Chambers for technical assistance, and Leon Deouell, Amitai Shenhav, Avgusta Shestyuk, Kirstie Whitaker, and many brainSCANr beta testers for technical discussions. BV is funded by the National Institute of Neurological Disorders and Stroke (NS21135-22S1) and a National Institutes of Health Institutional Research and Academic Career Development Award (IRACDA)(GM081266-05).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akil H, Martone M, Van Essen D. Challenges and Opportunities in Mining Neuroscience Data. Science. 2001;331:708–12. doi: 10.1126/science.1199305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alako BT, et al. CoPub Mapper: Mining MEDLINE based on search term co-publication. BMC Bioinformatics. 2005;6:51–66. doi: 10.1186/1471-2105-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg CB, Berlin JA. Publication bias and dissemination of clinical research. J Natl Cancer Inst. 1989;81:107–15. doi: 10.1093/jnci/81.2.107. [DOI] [PubMed] [Google Scholar]
- Bilder, et al. Cognitive ontologies for neuropsychiatric phenomics research. Cogn Neuropsychiat. 2009;4:419–50. doi: 10.1080/13546800902787180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk B, Roos A, Lauri M. Global annual volume of peer reviewed scholarly articles and the share available via different Open Access options. The International Conference on Electronic Publishing (ELPUB 2008). [Google Scholar]
- Bohland JW, et al. A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS Comput Biol. 2009;3:1–9. doi: 10.1371/journal.pcbi.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsboom D, Cramer AOJ, Schmittmann VD, Epskamp S, Waldorp LJ. The Small World of Psychopathology. PLoS ONE. 2011;11:27407. doi: 10.1371/journal.pone.0027407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden DM, Dubach MF. NeuroNames 2002. Neuroinformatics. 2003;1:43–59. doi: 10.1385/NI:1:1:043. [DOI] [PubMed] [Google Scholar]
- Bowden DM, Dubach M, Park J. Creating neuroscience ontologies. Methods Mol Biol. 2007;401:67–87. doi: 10.1007/978-1-59745-520-6_5. [DOI] [PubMed] [Google Scholar]
- Dirnagl U, Lauritzen M. Fighting publication bias: introducing the Negative Results section. J Cereb Blood Flow Metab. 2010;30:1263–64. doi: 10.1038/jcbfm.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editors. A critical look at connectomics. Nat Neurosci. 2010;13:1441. doi: 10.1038/nn1210-1441. [DOI] [PubMed] [Google Scholar]
- Evans JA, Foster JG. Metaknowledge. Science. 2011;331:721–25. doi: 10.1126/science.1201765. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Cappelleri JC, Sacks HS, Lau J. The relationship between study design, results, and reporting of randomized clinical trials of HIV infection. J Control Clin Trials. 1997;18:431–44. doi: 10.1016/s0197-2456(97)00097-4. [DOI] [PubMed] [Google Scholar]
- Larson SD, Martone ME. Ontonologies for neuroscience: What are they and what are they good for? Front Neurosci. 2009;3:60–67. doi: 10.3389/neuro.01.007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Michel JB, et al. Quantitative analysis of culture using millions of digitized books. Science. 2010;331:176–82. doi: 10.1126/science.1199644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modha D, Singh R. Network architecture of the long-distance pathways in the macaque brain. Proc Natl Acad Sci USA. 2010;107:13485–90. doi: 10.1073/pnas.1008054107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons L, Haque E, Liu H. Subspace clustering for high dimensional data: a review. ACM SIGKDD Explorations Newsletter. 2004;6:90–105. [Google Scholar]
- Poldrack R, Halchenko Y, Hanson S. Decoding the large-scale structure of brain function by classifying mental states across individuals. Psychol Sci. 2009;20:1364–72. doi: 10.1111/j.1467-9280.2009.02460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, et al. The Cognitive Atlas: Towards a Knowledge Foundation for Cognitive Neuroscience. Front Neuroinformatics. 2011;5:1–11. doi: 10.3389/fninf.2011.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Lipson H. Distilling Free-Form Natural Laws from Experimental Data. Science. 2009;324:81–85. doi: 10.1126/science.1165893. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The Human Connectome: A Structural Description of the Human Brain. PLoS Comp Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Hilgetag CC, Burns GA, O’Neill MA, Young MP, Kötter R. Computational analysis of functional connectivity between areas of primate cerebral cortex. Philos Trans R Soc Lond B, Biol Sci. 2000;355:111–26. doi: 10.1098/rstb.2000.0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JM, Simes RJ. Publication bias: evidence of delayed publication in a cohort study of clinical research projects. B M J. 1997;315:640–45. doi: 10.1136/bmj.315.7109.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytek B, Knight RT. Prefrontal cortex and basal ganglia contributions to visual working memory. Proc Natl Acad Sci USA. 2010;107:18167–72. doi: 10.1073/pnas.1007277107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytek B, et al. Shifts in gamma phase-amplitude coupling frequency from theta to alpha over posterior cortex during visual tasks. Neuron. 2010;68:401–8. doi: 10.3389/fnhum.2010.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren J, Bekeredjian R, Stewart J, Shohet R, Garner H. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004;20:389–98. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Essen DCV, Wager TD. Cognitive neuroscience 2. 0: building a cumulative science of human brain function. Trends Cogn Sci. 2010;14:489–96. doi: 10.1016/j.tics.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 2011;8:665–70. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Systematic analysis, comparison, and integration of disease based human genetic association data and mouse genetic phenotypic information. BMC Med Genomics. 2010;3:1–22. doi: 10.1186/1755-8794-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.