Abstract
BCR-ABL transforms bone marrow progenitor cells and promotes genome instability, leading to development of chronic myelogenous leukemia (CML). The tyrosine kinase inhibitor imatinib effectively treats CML, but acquired resistance can develop due to BCR-ABL mutations. Mechanisms for acquisition of BCR-ABL mutations are not fully understood. Using a novel culture model of CML acquired resistance, we show that inhibition of SIRT1 deacetylase by small molecule inhibitors or gene knockdown blocks acquisition of BCR-ABL mutations and relapse of CML cells on tyrosine kinase inhibitors. SIRT1 knockdown also suppresses de novo genetic mutations of HPRT (hypoxanthine phosphoribosyl transferase) gene in CML and non-CML cells upon treatment with DNA damaging agent camptothecin. Although SIRT1 can enhance cellular DNA damage response, it alters functions of DNA repair machineries in CML cells and stimulates activity of error-prone DNA damage repair, in association with acquisition of genetic mutations. These results reveal a previously unrecognized role of SIRT1 for promoting mutation acquisition in cancer, and have implication for targeting SIRT1 to overcome CML drug resistance.
INTRODUCTION
Chronic myelogenous leukemia (CML) is a lethal hematopoietic malignancy caused by oncogenic fusion gene BCR-ABL that activates multiple signaling pathways for cell proliferation and alters DNA damage repair pathways.1 Development of BCR-ABL tyrosine kinase inhibitor imatinib mesylate (Gleevec) was a major milestone in CML treatment that dramatically increased the 5-year survival of chronic CML patients.2 However, acquired resistance through genetic mutations of BCR-ABL remains a challenge for CML treatment. In the accelerated and blast crisis phases of CML, imatinib treatment has poor response and suffers high frequency of relapse in the patients having response.3 Clinical resistance in these patients is mediated primarily by genetic mutations of the BCR-ABL kinase domain.4,5 Among them, T315I mutation is especially problematic because of its frequent occurrence and failure to respond to treatment with first and second generation tyrosine kinase inhibitors.6–10 Even in the chronic phase CML, once imatinib is discontinued, the disease can relapse rapidly with development of BCR-ABL mutations.11 In spite of significant effort to develop more potent tyrosine kinase inhibitors to overcome resistance, mechanisms of acquiring BCR-ABL mutations are not fully clear.
To help address resistance mechanisms, we have developed a novel culture model for acquired resistance using blast crisis CML cell line KCL-22.12 We have shown that acquisition of BCR-ABL mutations for imatinib resistance does not require pre-existing mutations or involve aberrant chromosomal rearrangement and mutator phenotype of the cells. Instead, mutation acquisition is a dynamic process that is influenced by BCR-ABL gene expression and the native BCR-ABL translocation locus.12 Our study suggests possible involvement of epigenetic elements on the BCR-ABL translocation locus in deriving the mutations.
SIRT1 is a mammalian nicotinamide adenine dinucleotide dependent histone/protein deacetylase, and a homologue of yeast silent information regulator 2 that is required for replicative lifespan extension upon calorie restriction.13 SIRT1 plays direct or indirect roles in epigenomic regulation by deacetylating histones and chromatin modifiers such as Suv39h1.14–16 In response to DNA damage, SIRT1 is recruited to DNA double strand break sites, remodeling local chromatin structure presumably to help repair.17 Multiple DNA damage repair factors themselves are modified by SIRT1 through deacetylation, including Ku70,18 Nijmegen Breakage Syndrome protein (NBS1),19 Werner syndrome protein(WRN),20 and xeroderma pigmentosum c protein 21 for various repair mechanisms. Loss of SIRT1 results in chromosomal abnormality and translocation in mouse embryonic cells.18,22 These studies suggest that one important function of SIRT1 is involved in epigenetic modifications of both local chromatin structure and DNA repair machineries for facilitating DNA damage repair.
While appropriate DNA damage repair restores cellular functions, cells with excessive damage and unable to repair properly may undergo apoptosis. In this regard, it is important to note that SIRT1 promotes mammalian cell survival under oxidative and genotoxic stresses through deacetylation of multiple substrates including p53,23,24 Ku70 25 and FOXO proteins 26–28. It is plausible that the ability of SIRT1 to promote cell survival and DNA damage repair may interplay to ensure the survival of cells undergoing DNA damage repair. However, it is unknown whether SIRT1 may play a role in deriving rare genetic mutations for cancer drug resistance.
We have shown that tumor suppressor HIC1 (hypermethylated in cancer 1) represses SIRT1 expression to modulate DNA damage response.29 HIC1 is progressively inactivated by promoter hypermethylation towards blast crisis CML and relapsed leukemia from chemotherapy.30 We hypothesized that SIRT1 could be activated in CML cells to promote chemoresistance. We have recently shown that SIRT1 is over-expressed in both primary CML samples and blast crisis CML cell lines, and that SIRT1 is activated by BCR-ABL in hematopoietic progenitor cells and this activation is essential for BCR-ABL mediated leukemogenesis.31 Here we demonstrate that SIRT1 promotes DNA damage repair in CML cells, but surprisingly, inhibition of SIRT1 suppresses acquisition of BCR-ABL mutations upon imatinib treatment. SIRT1 knockdown also suppresses de novo genetic mutations of HPRT gene upon acute DNA damage. The ability of SIRT1 to promote mutation acquisition is associated with its ability to alter cellular DNA damage repair pathways and increase error-prone DNA damage repair.
RESULTS
Pharmacological inhibition of SIRT1 blocked acquired resistance of CML cells to imatinib
To examine roles of SIRT1 in CML acquired resistance, we used the KCL-22 cell model that we have developed.12 KCL-22 cells, originated from a blast crisis CML patient, undergoes initial apoptosis upon imatinib treatment, but cells re-grow after two weeks with acquisition of T315I BCR-ABL mutation. For most experiments in this study, the original KCL-22 cells that have not acquired BCR-ABL mutations or been treated with imatinib were used as the starting material, referred to as “KCL-22 cells” hereafter. In one subset of experiments, however, four clonal KCL-22 lines (L1, L7, Ag 3 and Ag 11) that have not acquired BCR-ABL mutations or been treated with imatinib were used as starting materials. These clonal KCL-22 lines can acquire different BCR-ABL mutations as described before,12 and they were designated as “clonal KCL-22 cells” or “KCL-22 cell clone(s)” in the text.
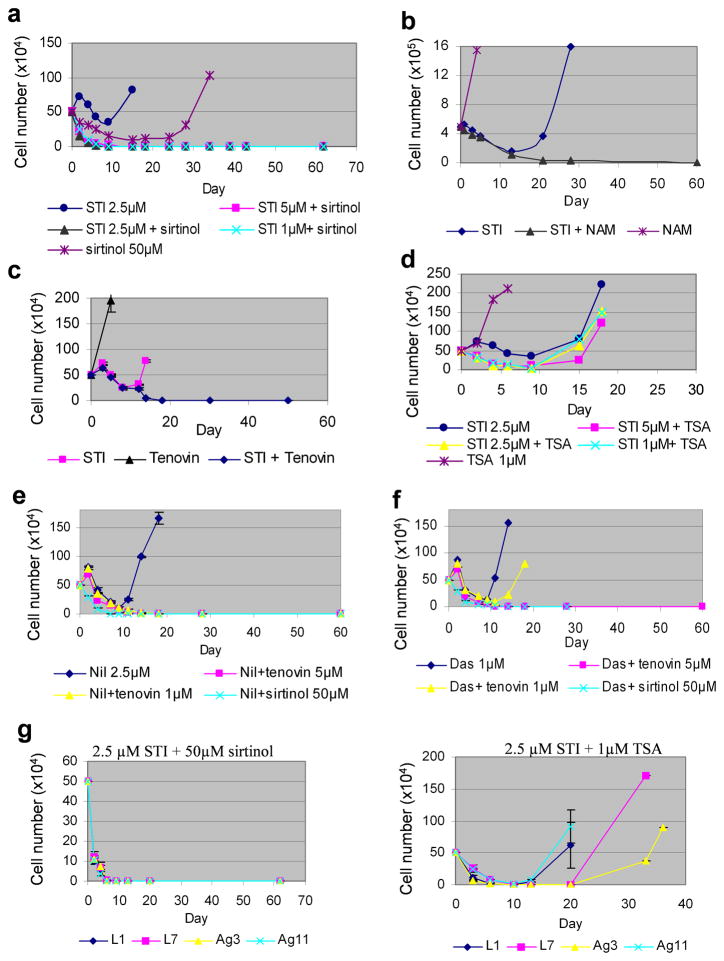
We first treated KCL-22 cells with SIRT1 inhibitors in the presence or absence of imatinib. We found that SIRT1 inhibitors, sirtinol, nicotinamide and tenovin-6, all blocked CML cell relapse when combined with imatinib (Figure 1a–c). Whereas combination of sirtinol with imatinib increased cell death, tenovin-6 blocked the relapse at as low as 1 μM that was below the concentrations to increase imatinib-mediated cell killing (Figure 1c and Supplementary Figure 1). Similarly, nicotinamide blocked cell relapse without enhancing cell death (Figure 1b).
Figure 1. Pharmacological inhibition of SIRT1 blocked acquired resistance of CML cells on tyrosine kinase inhibitors.
(a) KCL-22 cells were treated with 50 μM sirtinol or imatinib (STI) alone at the concentrations indicated or in combination of the two drugs. Cells for STI treatment alone all relapsed, and for simplicity, only the 2.5 μM curve was shown. (b) KCL-22 cells were treated with 15 mM nicotinamide (NAM) and 5 μM imatinib alone or in combination. (c) KCL-22 cells were treated with 1μM tenovin-6 and 2.5 μM imatinib alone or in combination. (d) KCL-22 cells were treated with 1μM trichostatin A (TSA) without or with STI at the concentrations indicated. (e) KCL-22 cell relapsed on 2.5 μM Nilotinib (Nil) with T315I mutation, but combination with tenovin-6 or sirtinol blocked relapse. (f) KCL-22 cell relapsed on 1 μM Dasatinib (Das) with T315I mutation. Combination with sirtinol or 5 μM tenovin-6 blocked relapse, and combination with 1 μM tenovin-6 delayed the relapse. (g) Left, sirtinol blocked clonal cells relapse on STI treatment. Right, relapse of clonal KCL-22 cells (L1, L7, Ag3 and Ag11) on STI plus TSA treatment.
Inhibitors for class I and II histone deacetylases (HDACs) have been used for treatment of hematopoietic malignancies.32 We found that the HDAC inhibitor trichostatin A (TSA) at 2.5 μM and higher concentrations depleted BCR-ABL and killed KCL-22 cells in the absence of imatinib (Supplementary Figure 2). TSA at low concentrations (1 μM or less) were insufficient to kill KCL-22 cells, but sensitized KCL-22 cells to imatinib-mediated cell killing (Figure 1d and Supplementary Figure 2), similar to previous observations of this class of compounds.33–35 However, TSA at low concentrations failed to prevent relapse of KCL-22 cells from imatinib and the relapsed cells developed T315I mutation (Figure 1d and not shown).
Treatment with second generation BCR-ABL inhibitors nilotinib and dasatinib also resulted in relapse and acquisition of T315I mutation in KCL-22 cells, but their combination with tenovin-6 or sirtinol blocked the recurrence (Figure 1e,f and not shown). We have derived four KCL-22 cell clones, three of which can acquire different BCR-ABL mutations upon imatinib treatment, i.e. E255K (clone L1), Y253H (clone L7) and T315I (clone Ag11), whereas clone Ag3 develops resistance without BCR-ABL mutations.12 We found that combination of imatinib with sirtinol, but not 1 μM TSA, blocked relapse of all clonal KCL-22 cells (Figure 1g). Together, these results suggest that SIRT1 inhibitors and HDAC inhibitors may function distinctly, and combination of SIRT1 inhibition with BCR-ABL inhibition may be a powerful approach to overcome acquired resistance through BCR-ABL mutations.
SIRT1 knockdown reduced BCR-ABL mutations and inhibited CML acquired resistance
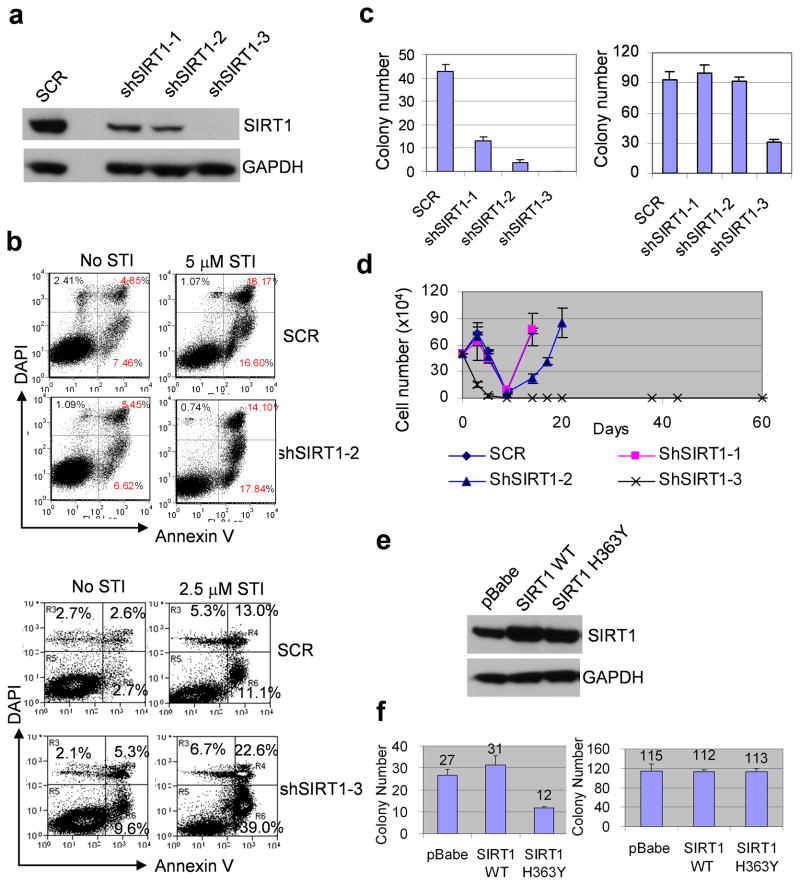
We designed three sets of SIRT1 shRNA to knock down the gene in KCL-22 cells (Figure 2a). The shSIRT1-3 vector exhibited the most robust SIRT1 knockdown that increased cell apoptosis in the absence or presence of imatinib, whereas shSIRT1-1 and shSIRT1-2 vectors did not (Figure 2a, b and not shown). By clonogenic assay, all three sets of SIRT1 shRNA suppressed formation of resistant colonies from KCL-22 cells treated with imatinib and the efficiency of suppression was proportional to the levels of gene knockdown, with shSIRT1-3 being the most effective (Figure 2c). Mutant colonies derived from scrambled shRNA, shSIRT1-1 or shSIRT1-2 knockdown were sequenced and confirmed to harbor T315I mutation (Supplementary Figure 3). In liquid culture, KCL-22 cell relapse on imatinib was delayed by shSIRT1-2 and completely blocked by shSIRT1-3 (Figure 2d). These findings are in line with the effect of SIRT1 inhibitors described above and suggest SIRT1 inhibition suppresses acquisition of BCR-ABL mutations and CML cell relapse from imatinib treatment.
Figure 2. SIRT1 specific inhibition suppressed acquisition of BCR-ABL mutations.
(a) SIRT1 protein levels in KCL-22 cells after knockdown with 3 sets of SIRT1 shRNA. SCR, scrambled shRNA for control. (b) Effect of apoptosis induction in KCL-22 cells after SIRT1 knockdown using shSIRT1-2 or shSIRT1-3 with or without imatinib (STI). (c) Left, three days after shRNA transduction, one million SCR or shSIRT1 knockdown KCL-22 cells per plate were seeded in soft agar in triplicate with 5μM imatinib. At day 21, resistant colonies were scored. Right, plating control with 500 cells per well seeded in soft agar without imatinib. (d) Three days after shRNA transduction, one half million of SCR or shSIRT1 knockdown KCL-22 cells were treated with 5μM STI in triplicate and viable cells were counted at indicated days. (e) Over-expression of wild type or H363Y mutant SIRT1 in KCL-22 cells. The transduced cells were enriched by puromycin selection. (f) Left, Wild type or H363Y SIRT1 transduced cells were analyzed for BCR-ABL mutation frequency on imatinib by clonogenic assay as in c. Right, plating control as in c. pBabe was an empty vector control.
To examine if SIRT1 deacetylase activity is required for BCR-ABL mutagenesis, we over-expressed wild type or H363Y deacetylase-deficient SIRT1 24 in KCL-22 cells (Figure 2e). The expression of either wild type or H363Y SIRT1 did not affect cell growth (Supplementary Figure 4). Over-expression of wild type SIRT1 elevated mutations but H363Y SIRT1 expression significantly reduced BCR-ABL mutations (Figure 2f). Together, our results suggest that high levels of SIRT1 in CML cells promote BCR-ABL mutation acquisition for acquired resistance, a process that is dependent on SIRT1 deacetylase function.
SIRT1 promoted de novo genetic mutations of cancer cells upon DNA damage
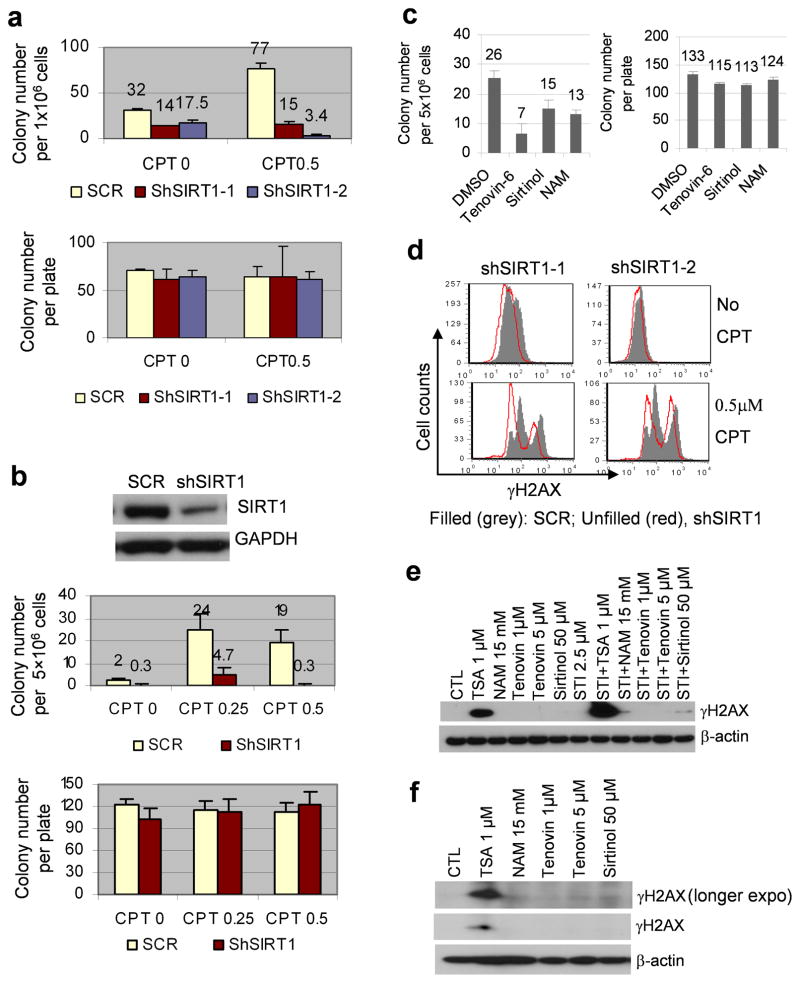
We have shown that acquisition of BCR-ABL mutations under imatinib therapeutic stress is distinct from formation of spontaneous HPRT mutations that are not required for cell survival.12 To determine if SIRT1 is involved in regulating mutations under a different stress condition, we examined effects of SIRT1 knockdown on camptothecin (CPT)-induced de novo HPRT mutations. CPT is a chemotherapeutic agent that induces DNA damage by inhibiting topoisomerase I.36 Mock or SIRT1 knockdown KCL-22 cells were cultured with HAT (hypoxanthine aminopterin thymidine) to remove cells bearing pre-existing HPRT mutations. HAT-selected KCL-22 cells were then treated with CPT to induce DNA damage. We found that SIRT1 knockdown using shSIRT1-1 or shSIRT1-2 vector drastically suppressed CPT-induced de novo HPRT mutations in KCL-22 cells (Figure 3a). SIRT1 knockdown also reduced, but to a lesser extent, spontaneous HPRT mutations when these cells were allowed to grow continuously for one month in culture (Figure 3a). Similarly, SIRT1 knockdown robustly suppressed CPT-induced de novo HPRT mutations in prostate cancer PC3 cells (Figure 3b), in which SIRT1 is over-expressed.37,38 Furthermore, we found that transient exposure of PC3 cells to tenovin-6, sirtinol or nicotinamide for two days was sufficient to reduce CPT-induced de novo HPRT mutations (Figure 3c). These results indicate that SIRT1 promotes HPRT mutations under acute CPT-induced stress in a similar manner to its promotion of BCR-ABL mutations under imatinib therapeutic stress, and that SIRT1-mediated mutagenesis is independent of cancer cell types.
Figure 3. SIRT1 knockdown inhibited camptothecin-induced HPRT mutations in cancer cells.
(a) Effects of SIRT1 knockdown on de novo HPRT mutations in CML cells. Top: HAT-selected SCR or shSIRT1 knockdown KCL-22 cells were treated with 0.5 μM CPT. After recovery, cells were seeded at 1 million/plate for clonogenic assay with 6-thiaguinine selection for HPRT mutations. HAT treated cells were also grown in normal medium without CPT for one month, and then used for clonogenic assay to detect newly occurring spontaneous mutations. Bottom: plating control with 500 cells/plate seeded without 6-thiaguinine. (b) Effects of SIRT1 knockdown on de novo HPRT mutations in PC3 cells. Top panel, SIRT1 knockdown using shSIRT1-1. Middle panel, five million HAT-selected and CPT treated SCR or shSIRT1 knockdown PC3 cells per plate were analyzed for 6-thioguanine resistance. Bottom panel, plating control with 200 cells/well seeded without drug. (c) HAT-selected PC3 cells were treated DMSO, 2.5 μM tenovin-6, 25 μM sirtinol or 15 mM NAM for 6h, followed by exposure to 0.5μM CPT for 1h. Cells were then cultured with DMSO, tenovin-6, sirtinol or NAM, respectively, for two days, followed by recovery without drugs for two weeks. HPRT mutations (left) and plating control (right) were analyzed as in b. (d) Flow cytometry analysis of γH2AX in SCR or shSIRT1 knockdown KCL-22 cells with or without CPT treatment. (e, f) Western blot analysis of γH2AX in KCL-22 (e) and PC3 (f) cells after 24h treatment with drugs indicated.
The above findings are surprising given that SIRT1 can promote DNA damage repair, presumably to suppress genetic mutations.18,22 It has been shown that SIRT1 knockout impairs DNA damage response in mouse fibroblasts with reduction of γH2AX foci formation in response to radiation.22 For comparison, we found that SIRT1 knockdown also reduced γH2AX staining upon CPT treatment in KCL-22 cells (Figure 3d). Accordingly, we found that SIRT1 inhibitors only slightly increased γH2AX expression in KCL-22 cells in the presence of imatinib, in contrast to TSA that robustly induced γH2AX expression in the presence or absence of imatinib (Figure 3e). TSA also more effectively induced γH2AX expression than SIRT1 inhibitors in prostate cancer cells (Figure 3f). The potent induction of γH2AX by TSA may be due to its ability to cause actual DNA damage.39 Our results indicate that suppression of mutation acquisition by SIRT1 inhibition in cancer cells can not be simply explained by γH2AX change; however, SIRT1 inhibitors do differ from class I/II HDAC inhibitors for inducing γH2AX expression and suppression of mutation acquisition.
SIRT1 altered DNA damage repair pathways for acquired resistance of CML cells
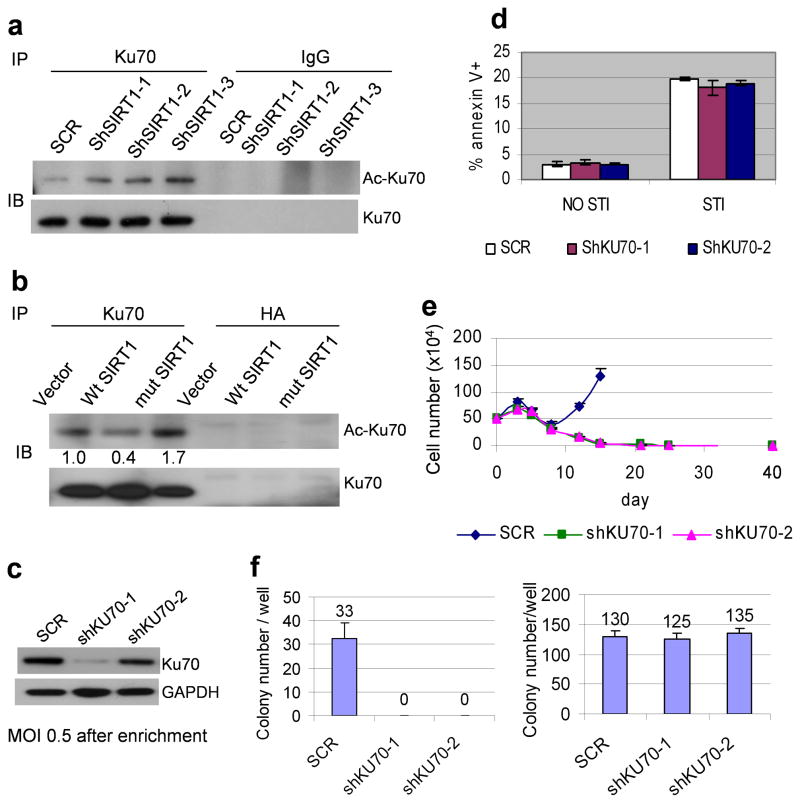
BCR-ABL alters functions of both homologous recombination (HR) and error-prone, non-homologous end joining (NHEJ) DNA repair machineries. BCR-ABL increases expression of RAD51 that abnormally stimulates HR repair and CML drug resistance.40 BCR-ABL promotes DNA damage repair but increases genetic mutations in association with the compromised fidelity of both HR and NHEJ repairs.41 We have shown that acquisition of BCR-ABL mutations in KCL-22 cells depends on BCR-ABL expression,12 and that BCR-ABL activates SIRT1 expression.31 To determine if SIRT1 may act as a key downstream effecter of BCR-ABL to regulate repair machineries for CML acquired resistance, we first examined the roles of Ku70 that is a key component of NHEJ repair.42 Ku70 is deacetylated and activated by SIRT1, which promotes both cell survival under stress 25 and DNA damage repair.18 We found that Ku70 acetylation levels in CML cells was increased in proportion to the levels of SIRT1 knockdown (Figure 4a), which may inactivate Ku70 functions.25,43 Similarly, over-expression of H363Y deacetylase mutant SIRT1 increased Ku70 acetylation, whereas over-expression of wild type SIRT1 reduced Ku70 acetylation (Figure 4b).
Figure 4. SIRT1 regulated NHEJ repair for CML acquired resistance.
(a) Acetylation of Ku70 after SIRT1 knockdown in KCL-22 cells. Ku70 was immunoprecipitated from total cell lysate of mock or SIRT1 knockdown KCL-22 cells. Western blots were probed with anti-acetylated lysine antibody followed by Ku70 antibody. (b) Ku70 acetylation after exogenous expression of wild type or H363Y mutant SIRT1 was analyzed as in a. HA antibody was used for immunoprecipitation control. The numbers were densitometry results of acetylated Ku70 that was normalized to total Ku70 and compared to the vector control. (c) Moderate Ku70 knockdown in KCL-22 cells using a low MOI of 0.5. Cells were enriched by puromycin selection. (d) No change of KCL-22 cell apoptosis was observed after moderate Ku70 knockdown in the absence or presence of 2.5 μM imatinib. Cells were selected by puromycin for 4 days followed by recovery for 5 days in normal medium before analysis. (e) Moderate Ku70 knockdown blocked KCL-22 cells relapse from 5μM imatinib. (f) Left, moderate Ku70 knockdown eliminated BCR-ABL mutant soft agar colony formation on 5 μM imatinib. One million cells per well were seeded in triplicate in 6-well plates with imatinib. Right, plating control with 500 cells seeded per well.
Because the efficient Ku70 knockdown induced apoptosis of KCL-22 cells,31 to segregate roles of Ku70 in BCR-ABL mutations from apoptosis, we minimally knocked down Ku70 using a multiplicity of infection (MOI) of 0.5 and enriched the transduced cells by puromycin selection. Low MOI transduction resulted in moderate Ku70 knockdown that did not increase apoptosis (Figure 4c,d and Supplementary Figure 5), but blocked acquisition of BCR-ABL mutations in both liquid culture and soft agar assays (Figure 4e,f), suggesting a crucial role of Ku70 in acquisition of BCR-ABL mutations.
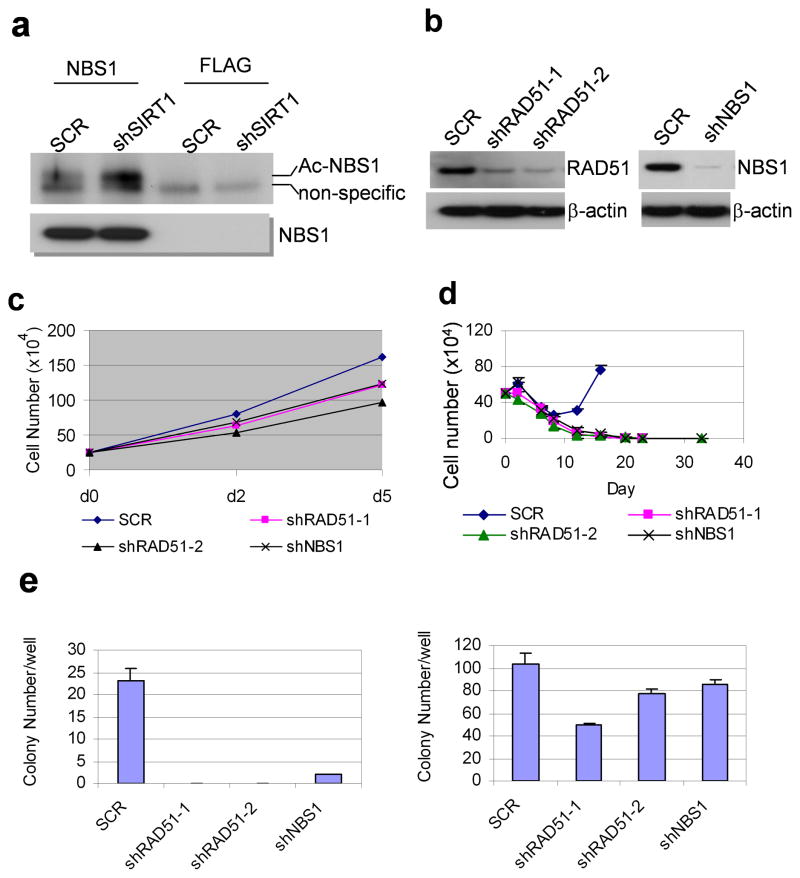
SIRT1 also deacetylates HR repair factor NBS1 (Nijmegen Breakage Syndrome), a component of MRN (MRE11-RAD50-NBS1) complex,19 and regulates recruitment of NBS1 and RAD51 to DNA damage foci for repair.18 We found that SIRT1 knockdown increased NBS1 acetylation in KCL-22 cells (Figure 5a). Knockdown of NBS1 or RAD51 moderately affected cell growth (Figure 5b,c), but suppressed BCR-ABL mutations and CML cell relapse on imatinib (Figure 5d,e). These results suggest that SIRT1 also regulates HR repair machineries for BCR-ABL mutation acquisition.
Figure 5. Influence of HR repair factors for CML acquired resistance.
(a) NBS1 acetylation upon SIRT1 knockdown. NBS1 was immunoprecipitated and analyzed by Western blot with anti-acetylated lysine antibody, followed by NBS1 antibody. FLAG antibody was used for immunoprecipitation control. (b) NBS1 and RAD51 knockdown in KCL-22 cells. (c) Effects of RAD51 and NBS1 knockdown on KCL-22 cell growth. (d) NBS1 and RAD51 knockdown KCL-22 cells were enriched with puromycin selection and subjected to relapse assay on 2.5 μM imatinib. (e) RAD51 and NBS1 knockdown reduced BCR-ABL mutation by soft agar clonogenic assay. Left panel, resistant colonies with imatinib treatment. Right panel, plating control without imatinib.
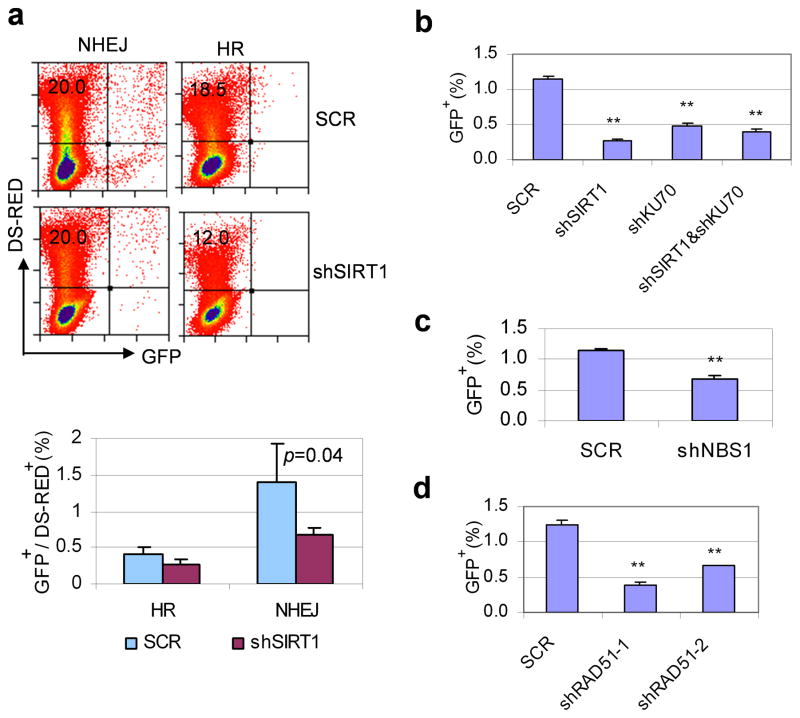
To further determine the role of SIRT1 in DNA damage repair in CML cells, we generated stable KCL-22 cell clones bearing a NHEJ reporter construct EJ5-GFP,44 and clones carrying a HR reporter DR-GFP.45 After introducing DNA damage on these reporters with the endonuclease I-SceI by electroporation, we found that NHEJ was more efficient than HR for DNA damage repair in KCL-22 cells, and that SIRT1 knockdown suppressed both NHEJ and HR activity with more prominent effect on NHEJ (Figure 6a).
Figure 6. SIRT1 altered functions of DNA damage repair in CML cells.
(a) Flow cytometry analysis of HR and NHEJ repair after SIRT1 knockdown using stably integrated reporter constructs in KCL-22 cells. I-SceI was introduced by electroporation and DS-Red was used for transfection control. The repair rate was normalized to DS-Red. (b–d) NHEJ repair assay using inducible I-SceI expression. Gene knockdown was carried out in inducible I-SceI expressing EJ5-GFP KCL-22 cells. 48 hrs after gene knockdown, I-SceI expression was induced by doxycycline for 72 hrs, and GFP positive cells were analyzed by flow cytometry. b, effect of Ku70 and SIRT1 knockdown; c, effect of NBS1 knockdown; d, effect of RAD51 knockdown. ** indicates P<0.05.
Because the procedure of electroporation to introduce I-SceI killed many cells and increased experimental variation, to improve the assay, we generated an inducible I-SceI expression lentiviral vector and established stable EJ5-GFP and DR-GFP reporter cell lines with inducible I-SceI expression. The DNA repair rates for EJ5-GFP and DR-GFP cells after doxycycline induction were similar to those with I-SceI electroporation, but cell death remained at the background level (data not shown). Using this system, we found that knockdown of either SIRT1 or Ku70 reduced NHEJ repair, and simultaneous knockdown of both genes did not further reduce the repair rate (Figure 6b), further supporting the role of SIRT1 for activating Ku70 for NHEJ repair. Interestingly, knockdown of NBS1 or RAD51 also reduced NHEJ repair (Figure 6c,d). Although precise mechanisms for such an effect remain to be elucidated, functional overlap among repair machineries is becoming more appreciated, for instance, MRN complex is shown to increase NHEJ activity.46–48 Together, our findings suggest that over-expression of SIRT1 promotes CML acquired resistance through acquisition of BCR-ABL mutations, which is associated with its ability to modulate functions of DNA damage repair machineries.
DISCUSSION
Although SIRT1 is known to enhance DNA damage repair,18,22 our study reveals another surprising facet of SIRT1 in cancer cells in which it promotes acquisition of genetic mutations. In CML cells, BCR-ABL promotes genome instability through altering functions of multiple repair machineries and increasing erroneous DNA damage repair.49 We have shown that acquisition of BCR-ABL mutations for imatinib resistance is dependent on BCR-ABL expression,12 and that BCR-ABL transcriptionally activates SIRT1 expression in hematopoietic progenitor cells.31 Our current study supports that SIRT1 is a key intermediate of BCR-ABL to alter functions of DNA damage repair machineries and promote acquisition of genetic mutations for CML drug resistance.
Promoting genetic mutations by SIRT1-regulated DNA damage repair could be a unique feature of cancer cells. In normal cells, repair fidelity is typically high unless cells are aging or senescent.50 SIRT1 may function to facilitate DNA damage repair and promote cell survival. In CML cells, however, BCR-ABL transformation leads to the substantially increased production of reactive oxygen species and DNA double strand breaks.41 BCR-ABL alters repair machinery functions to allow rapid repair of DNA damage with reduced fidelity,41,51 which may critically involve SIRT1, and result in significant increase of repair errors. The rare genetic mutations generated after erroneous repair in cancer cells may render survival advantage for resistance when cancer cells are treated with chemotherapeutic agents. Alternatively, chemotherapeutic stress itself may influence DNA damage repair process in CML cells, accentuating erroneous repair and promoting de novo acquisition of mutations for resistance. It remains to be determined how SIRT1 over-expression in cancer cells may alter DNA damage repair fidelity and how that may cause acquisition of genetic mutations for cancer drug resistance. We have proposed that the local epigenome of BCR-ABL translocation locus may play a role in sensing therapeutic stress signals for mutation acquisition.52 Given that SIRT1 is a stress-response gene and epigenome regulator, it would be of interest to determine if and how SIRT1 may modulate this process through epigenome in future.
NHEJ is the major repair pathway in higher eukaryotes and also the main contributor for repair errors.42 Ku70/86 heterodimer is essential for the increased activity of NHEJ and concomitant DNA misrepair in myeloid leukemia cells.53,54 Consistently, we found that NHEJ is more effective than HR repair in KCL-22 cells and Ku70 is essential for BCR-ABL mutation acquisition, which is regulated by SIRT1. Our results also support likely a crosstalk between NHEJ and other repair machineries. NBS1 is part of the MRN complex that facilitates the end processing of double strand breaks for HR and NHEJ.46–48 Our results that knockdown of NBS1 inhibits BCR-ABL mutations and NHEJ repair are in line with those findings. It is interesting that RAD51 knockdown also reduces BCR-ABL mutation and NHEJ activity. Although the precise mechanism for this reduction of NHEJ is not clear, there is evidence of a crosstalk between HR and NHEJ. For example, in response to DNA damage, HR factor c-ABL that phosphorylates RAD51 42 also phosphorylates NHEJ factor DNA-PKcs, and competes with Ku70/86 for binding DNA-PKcs.55,56 HR and NHEJ repair pathways can be even coupled in certain types of damage repair.57
Our KCL-22 cell resistance model provides a new tool for uncovering mechanisms of CML acquired resistance. We demonstrated a proof-of-concept of novel SIRT1 functions in acquisition of genetic mutations. Although discovered in one cell line, we believe the conclusion would not be restricted to this cell line or BCR-ABL. First, our mutation analyses are carried out in two independent systems involving ABL signaling for BCR-ABL and de novo nucleotide synthesis for HPRT. Second, effects of SIRT1 on HPRT mutations are also observed in prostate cancer PC3 cells upon DNA damage induced by CPT. Therefore, we speculate that SIRT1 may have a broader role in acquired resistance of cancer. Acquired resistance through genetic mutations occurs in targeted therapy of several types of cancer 58 and SIRT1 is over-expressed in many types of human cancer.37 It would be interesting to determine if SIRT1 may play a role in acquisition of genetic mutations in other settings in future.
This study improves our understanding of roles of SIRT1 in cancer drug resistance. This study also suggests potential therapeutic application of inhibiting aberrant DNA repair activities through SIRT1 inhibition for cancer therapy. Towards this end, it is important to note that SIRT1 inhibition can also induce cancer cell apoptosis,23,24,37 and that we have found that SIRT1 inhibition suppresses BCR-ABL transformation of bone marrow progenitor cells and leukemogenesis.31 Meanwhile, SIRT1 inhibition is well tolerated by normal human CD34+ progenitor cells and normal mice.31 SIRT1 (class III deacetylase) and HDACs (class I and II deacetylases) may have overlapping functions, and inhibitors for SIRT1 and HDACs can both induce apoptosis. However, we show that they bear important difference. TSA can cause actual DNA damage and induce robust damage response that may actually stimulate mutations, whereas SIRT1 inhibitors do not or at least to a much lesser extent, and therefore, SIRT1 inhibitors have an added advantage to block acquisition of BCR-ABL mutations. It is likely that inhibition of CML cell survival by SIRT1 inhibitors may work in concert with blocking BCR-ABL mutations for preventing relapse of CML cells upon treatment with tyrosine kinase inhibitors. In conclusion, our study suggests that SIRT1 deacetylase promotes acquisition of genetic mutations in CML cells, and targeting SIRT1 may be valuable for improving CML treatment to overcome resistance.
MATERIALS AND METHODS
Cell lines, drugs and DNA constructs
KCL-22 cells were purchased from German Collection of Cell Cultures, Braunschweig, Germany. PC3 cells were purchased from American Typed Cell Culture. Imatinib (STI-571) was kindly provided by Novartis, Basel, Switzerland. Sirtinol, splitomicin, nicotinamide, trichostatin A, 6-thioguanine, and HAT were from Sigma. Lentiviral shRNA vectors pSicoR PGK-puro and CMV-GFP, wild type and H363Y SIRT1 expressing retroviral vectors 24 were from Addgene. Tenovin-6 was purchased from Cayman Chemical.
Drug resistance, BCR-ABL mutation, apoptosis and clonogenic assays
These assays were performed as described previously.12 Briefly, for drug resistance assay, one half million KCL-22 cells were seeded in 1 ml medium per well in 24-well plates, and treated with different concentrations of tyrosine kinase inhibitors and monitored over time. For sequencing ABL kinase domain, we amplified the ABL kinase domain by RT-PCR of total RNA or by PCR of genomic DNA with a high fidelity DNA polymerase (Stratagene). Apoptosis was analyzed with Annexin V Assay kit (BD Pharmingen). For clonogenic assay, a standard two-layer soft agar culture was performed with bottom layer of 0.6% agarose and top layer of 0.35% agarose. Colonies were scored after staining with 0.005% Crystal Violet.
Gene knockdown using lentiviral vectors
The knockdown vector design and production were carried out as previously described12. Sequences for shRNAs are available upon request. High titer lentiviral stocks, typically 1 to 3 ×107 infectious units/ml, were used for transduction. Unless specified, a multiplicity of infection (MOI) around 5 was typically used for infection so that nearly complete transduction was achieved.
Protein analysis
The following antibodies were used for Western blots: rabbit monoclonal anti-human SIRT1 (Epitomics), mouse monoclonal anti-c-ABL (BD Pharmingen), mouse monoclonal anti-Ku70 (Neomarker), and rabbit monoclonal anti-NBS1 antibody (Epitomics). To analyze Ku70 acetylation, we pulled down Ku70 from total cell lysate with anti-Ku70 and protein A-agarose beads (Upstate Biotech) followed by acetylation detection with rabbit anti-acetyl lysine antibody (Cell Signaling). To analyze NBS1 acetylation, NBS1 was pulled down using with rabbit anti-NBS1 polyclonal antibody (Bethyl Laboratories) and protein A/G plus-agarose beads (Santa Cruz Biotech). The blot was probed with anti-acetyl lysine antibody followed by rabbit monoclonal anti-NBS1 antibody (Epitomics).
DNA damage induction by camptothecin and de novo mutation analysis
Cells were pre-selected for four days in HAT medium to remove pre-existing HPRT mutations. The efficiency of HAT selection was confirmed by plating these cells on soft agar with 2.5 μg/ml 6-thioguanine, which produced zero colony. HAT-selected cells were then treated with 0.5 μM CPT for 1 hour, and then cultured for at least 10 days to expand before being used for soft agar clonogenic assay with 6-thioguanine selection. The rest of HAT-selected cells were cultured in medium without selection. The change of γH2AX was analyzed with γH2AX Assay Kit (Upstate Biotech).
DNA damage repair assays
Five million KCL-22 cells were transfected with 15 μg linearized repair reporter construct DR-GFP or EJ5-GFP by electroporation, and cells were selected for puromycin resistance. Individual clones were plucked from soft agar and expanded to screen for clones carrying an intact copy of the reporter constructs by Southern blotting as described previously.44,45 The clones with an intact copy of reporters were transduced by shSIRT1, shKu70, shRAD51 or shNBS1 for 24h followed by electroporation with 50 μg I-SceI encoding plasmid plus 10 μg Ds-Red. After another 48 h culture, the cells were analyzed by flow cytometry for GFP and Ds-Red expression to determine the repair efficiency. GFP+ cells were the successfully repaired cells, and repair rate was normalized to Ds-Red transfection efficiency.
To generate inducible HR and NHEJ reporter KCL22 cell lines, clonal DR-GFP or EJ5-GFP cells were infected with a lentiviral vector (pITA.Bleo.rtTA) carrying EF1α promoter-driven rtTA (reverse tetracycline transactivator) with a bleomycin selection cassette. Cells were selected with 150μg/mL of zeocin (Invitrogen). We generated another lentiviral vector (pTZG.puro.iSCE-RFP) expressing mRFP (monomeric red fluorescent protein) and I-SceI fusion protein driven by tetracycline response element (Tet-on). The zeocin-selected cells were then infected with pTZG.puro.iSCE-RFP vector. Cells were seeded on soft agar and clonal cells were plucked and expanded for screening for RFP expression by addition of 1μg/mL doxycycline (Fisher) and analyzed by fluorescent microscopy. Clones with the highest RFP expression were used for repair assay. Doxycycline treatment induced RFP expression in most cells and did not significantly affect cell viability. No GFP expression was detected in the absence of doxycycline. The repair rate was determined as the percentage of GFP+ cells over viable cells without normalization to RFP expression. The GFP expression with HR repair in such system was too low and not used for further analysis.
Statistical analysis
Two-tailed t-test analysis was used in all cases and P < 0.05 is considered statically significant.
Supplementary Material
Acknowledgments
This study was supported by the following grants: W81XWH-06-1-0268 from the US Department of Defense, a career development award from the STOPCANCER Foundation, a translational research grant from the V-Foundation and R01 CA143421 to W.Y.C. R.B. was supported by R01 CA95684, and JMS was supported by R01 CA120954. The core facilities used in this study were supported by NCI P30 CA033572. The contents are solely the responsibility of the authors and do not represent the official views of the National Cancer Institute or NIH.
Footnotes
Conflict of interest disclosure: Parts of this manuscript were used for a patent application filed by City of Hope.
References
- 1.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 3.Deininger MW, Druker BJ. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol Rev. 2003;55:401–423. doi: 10.1124/pr.55.3.4. [DOI] [PubMed] [Google Scholar]
- 4.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 5.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 6.Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 7.von Bubnoff N, Manley PW, Mestan J, Sanger J, Peschel C, Duyster J. Bcr-Abl resistance screening predicts a limited spectrum of point mutations to be associated with clinical resistance to the Abl kinase inhibitor nilotinib (AMN107) Blood. 2006;108:1328–1333. doi: 10.1182/blood-2005-12-010132. [DOI] [PubMed] [Google Scholar]
- 8.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 9.Kantarjian H, Giles F, Wunderle L, Bhalla K, O’Brien S, Wassmann B, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 10.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 11.Michor F, Hughes TP, Iwasa Y, Branford S, Shah NP, Sawyers CL, et al. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 12.Yuan H, Wang Z, Gao C, Chen W, Huang Q, Yee JK, et al. BCR-ABL gene expression is required for its mutations in a novel KCL-22 cell culture model for acquired resistance of chronic myelogenous leukemia. J Biol Chem. 2010;285:5085–5096. doi: 10.1074/jbc.M109.039206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 14.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 15.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 16.Bosch-Presegue L, Raurell-Vila H, Marazuela-Duque A, Kane-Goldsmith N, Valle A, Oliver J, et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell. 2011;42:210–223. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 17.O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, et al. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. 2008;283:7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 21.Ming M, Shea CR, Guo X, Li X, Soltani K, Han W, et al. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc Natl Acad Sci U S A. 2010;107:22623–22628. doi: 10.1073/pnas.1010377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 24.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 25.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 26.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 27.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 28.Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Issa JP, Zehnbauer BA, Kaufmann SH, Biel MA, Baylin SB. HIC1 hypermethylation is a late event in hematopoietic neoplasms. Cancer Res. 1997;57:1678–1681. [PubMed] [Google Scholar]
- 31.Yuan H, Wang Z, Li L, Zhang H, Modi H, Horne D, et al. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 2011 doi: 10.1182/blood-2011-06-361691. e-pub ahead of print December 29, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quintas-Cardama A, Santos FP, Garcia-Manero G. Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia. 2011;25:226–235. doi: 10.1038/leu.2010.276. [DOI] [PubMed] [Google Scholar]
- 33.Yu C, Rahmani M, Almenara J, Subler M, Krystal G, Conrad D, et al. Histone deacetylase inhibitors promote STI571-mediated apoptosis in STI571-sensitive and -resistant Bcr/Abl+ human myeloid leukemia cells. Cancer Res. 2003;63:2118–2126. [PubMed] [Google Scholar]
- 34.Fiskus W, Pranpat M, Balasis M, Bali P, Estrella V, Kumaraswamy S, et al. Cotreatment with vorinostat (suberoylanilide hydroxamic acid) enhances activity of dasatinib (BMS-354825) against imatinib mesylate-sensitive or imatinib mesylate-resistant chronic myelogenous leukemia cells. Clin Cancer Res. 2006;12:5869–5878. doi: 10.1158/1078-0432.CCR-06-0980. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Strauss AC, Chu S, Li M, Ho Y, Shiang KD, et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell. 2010;17:427–442. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Hasan MK, Alvarado E, Yuan H, Wu H, Chen WY. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30:907–921. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- 38.Jung-Hynes B, Nihal M, Zhong W, Ahmad N. Role of sirtuin histone deacetylase SIRT1 in prostate cancer. A target for prostate cancer management via its inhibition? J Biol Chem. 2009;284:3823–3832. doi: 10.1074/jbc.M807869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaymes TJ, Padua RA, Pla M, Orr S, Omidvar N, Chomienne C, et al. Histone deacetylase inhibitors (HDI) cause DNA damage in leukemia cells: a mechanism for leukemia-specific HDI-dependent apoptosis? Mol Cancer Res. 2006;4:563–573. doi: 10.1158/1541-7786.MCR-06-0111. [DOI] [PubMed] [Google Scholar]
- 40.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 41.Nowicki MO, Falinski R, Koptyra M, Slupianek A, Stoklosa T, Gloc E, et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104:3746–3753. doi: 10.1182/blood-2004-05-1941. [DOI] [PubMed] [Google Scholar]
- 42.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 43.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 44.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 47.Dinkelmann M, Spehalski E, Stoneham T, Buis J, Wu Y, Sekiguchi JM, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perrotti D, Jamieson C, Goldman J, Skorski T. Chronic myeloid leukemia: mechanisms of blastic transformation. J Clin Invest. 2010;120:2254–2264. doi: 10.1172/JCI41246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci U S A. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slupianek A, Nowicki MO, Koptyra M, Skorski T. BCR/ABL modifies the kinetics and fidelity of DNA double-strand breaks repair in hematopoietic cells. DNA Repair (Amst) 2006;5:243–250. doi: 10.1016/j.dnarep.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen WY, Yuan H, Wang Z. De novo acquisition of BCR-ABL mutations for CML acquired resistance. In: Koschmieder S, Krug U, editors. Myeloid Leukemia: Basic Mechanisms of Leukemogenesis. INTECH; 2011. pp. 69–84. [Google Scholar]
- 53.Brady N, Gaymes TJ, Cheung M, Mufti GJ, Rassool FV. Increased error-prone NHEJ activity in myeloid leukemias is associated with DNA damage at sites that recruit key nonhomologous end-joining proteins. Cancer Res. 2003;63:1798–1805. [PubMed] [Google Scholar]
- 54.Gaymes TJ, Mufti GJ, Rassool FV. Myeloid leukemias have increased activity of the nonhomologous end-joining pathway and concomitant DNA misrepair that is dependent on the Ku70/86 heterodimer. Cancer Res. 2002;62:2791–2797. [PubMed] [Google Scholar]
- 55.Kharbanda S, Pandey P, Jin S, Inoue S, Bharti A, Yuan ZM, et al. Functional interaction between DNA-PK and c-Abl in response to DNA damage. Nature. 1997;386:732–735. doi: 10.1038/386732a0. [DOI] [PubMed] [Google Scholar]
- 56.Jin S, Kharbanda S, Mayer B, Kufe D, Weaver DT. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J Biol Chem. 1997;272:24763–24766. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 57.Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175–1178. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.