Abstract
Heterozygous in-frame mutations (p.E2207del and p.R2308_M2309dup) in the α-II subunit of spectrin (SPTAN1) were recently identified in two patients with intellectual disability (ID), infantile spasms (IS), hypomyelination, and brain atrophy. These mutations affected the C-terminal domain of the protein, which contains the nucleation site of the α/β spectrin heterodimer. By screening SPTAN1 in 95 patients with idiopathic ID, we found a de novo in-frame mutation (p.Q2202del) in the same C-terminal domain in a patient with mild generalized epilepsy and pontocerebellar atrophy, but without IS, hypomyelination, or other brain structural defects, allowing us to define the core phenotype associated with these C-terminal SPTAN1 mutations. We also found a de novo missense variant (p.R566P) of unclear clinical significance in a patient with non-syndromic ID. These two mutations induced different patterns of aggregation between spectrin subunits in transfected neuronal cell lines, providing a paradigm for the classification of candidate variants.
Keywords: intellectual disability, epilepsy, SPTAN1, pontocerebellar atrophy
Introduction
The targeting and maintenance of groups of proteins at specific membrane domains of neurons, such as the synapse and the axon, are critical for the brain function and development. Not surprisingly, mutations in proteins that participate in the assembling of these domains, such as the spectrins, have been shown to cause neurological disorders. Spectrins consist of α- and β-subunits that form antiparallel heterodimers, which are assembled to form heterotetramers in a head-to-head configuration.1, 2 Heterozygous mutations in the β-III subunit (SPTBN2) were shown to cause spinocerebellar ataxia type-5.3 Recently, de novo in-frame mutations in the α-II spectrin subunit (SPTAN1) were identified in patients with severe intellectual disability (ID), infantile spasms (IS) with hypsarrhythmia, hypomyelination, and atrophy of various regions of the brain, including the cerebellum and brainstem.4 These mutations are located in a C-terminal domain that includes the nucleation site involved in α/β spectrin heterodimer formation.5 Biochemical studies indicated that these mutations destabilized this interaction, leading to aggregation of α-II(mut)/β-II and α-II(mut)/β-III spectrin heterodimers in mouse cortical neurons and in lymphoblastoid cells from the patients.4
Recent studies suggest that de novo point mutations in the germ line, which in humans are estimated to occur at a rate of ∼1.1 × 10−8 per base, may explain a large fraction of ID cases.6, 7, 8 In the course of a project aimed to identify such mutations in candidate synaptic genes, we sequenced SPTAN1 in patients with idiopathic ID. We identified de novo mutations in SPTAN1 in a patient with non-syndromic ID and in a patient with ID, epilepsy and pontocerebellar atrophy, further expanding the phenotypical spectrum associated with mutations in SPTAN1. Moreover, we found that these mutations induce different patterns of aggregations of spectrin subunits in cultured neuronal cells, providing a paradigm to validate candidate variants and to establish correlations between genotypes and phenotypes.
Methods
Subjects and DNA sequencing
Ninety-five cases of idiopathic ID (46 males and 49 females), mostly of French–Canadian ethnicity, without growth abnormalities or specific dysmorphic features, as well as 190 French–Canadians controls were studied. A subset of the ID cases (25/95) also displayed epilepsy. Genomic DNA was extracted from blood samples (Qiagen/Gentra, Toronto, ON, Canada) and paternity/maternity was confirmed using six informative unlinked microsatellite markers (Supplementary Table S1). Polymerase chain reaction amplification of all SPTAN1-coding exons and their splice junctions (Supplementary Table S2 for primers), DNA sequencing, and mutation analyses are detailed in the Supplementary Information.
Expression vectors
SPTAN1 mutants (c.1697G>C/p.R566P, c.6605_6607del/p.Q2202del, c.6619_6621del/p.E2207del; positions based on Refseq NM_001130438.2) were generated by site-directed mutagenesis using KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan), and verified by sequencing. A full-length human SPTAN1 cDNA (α-II spectrin) with a C-terminal Flag-tag was cloned into a CAG-promoter vector to express C'-Flag-tagged α-II spectrin.
Cell culture, transfection, and immunofluorescence
Mouse neuroblastoma 2A (N2A) cells and primary cultures of mouse cortical neurons were grown, transfected, and tested by immunofluorescence for the expression of wild type and mutant spectrin subunits, as previously described,4, 9 and detailed in the Supplementary Information.
Results
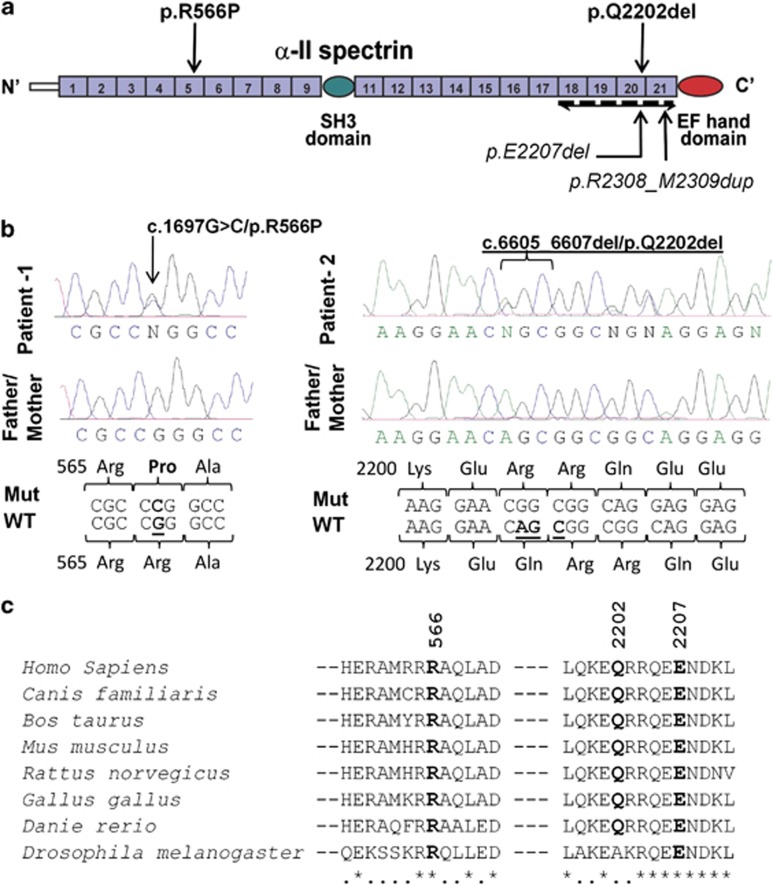
We identified three heterozygous variants in SPTAN1 in different patients, including two missense (c.1679A>G/p.E560G and c.1697G>C/p.R566P) and one in-frame amino-acid deletion (c.6605_6607del/p.Q2202del). p.E560G was transmitted from a healthy mother and is predicted not to affect protein function by various algorithms (Supplementary Table S3). In contrast, p.R566P (patient-1) and p.Q2202del (patient-2) were absent from the blood DNA of the corresponding healthy parents, indicating that they were de novo, and were not detected in 190 French–Canadian controls, nor reported in dbSNP134. p.R566P affects a well-conserved residue that lies at the N-terminus of SPTAN1 and is predicted to damage protein function by SIFT10 and polyphen.11 The deleted p.Q2202 residue is part of a C-terminal domain required for α/β spectrin subunit heterodimer formation.5 Interestingly, p.Q2202del is in close proximity to the recently reported SPTAN1 in-frame mutations, p.E2207del and p.R2308_M2309dup, identified in patients with ID, IS, and structural brain defects (Figure 1).4
Figure 1.
De novo mutations identified in SPTAN1. (a) Schematic representation of the SPTAN1 protein consisting of 22 domains, including 20-spectrin repeats, an SH3 domain, and an EF hand domains. The last four spectrin repeats required for α/β spectrin heterodimer associations are indicated (bidirectional arrow). Mutations identified in this study (p.R566P and p.Q2202del) and those by Saitsu et al4 (p.E2207del and p.R2308_M2309dup) are shown. (b) Chromatograms of the de novo p.R566P and p.Q2202del mutations (Mut) and wildtype (WT) SPTAN1 sequences. (c) Amino-acid conservation of the SPTAN1 residues affected with de novo mutations (p.R566P, p.Q2202del and p.E2207del). Amino-acid alignments were generated by homologene (NCBI) and the amino-acid sequences flanking the mutations are shown.
The phenotypes of the patients with p.R566P and p.E2207del were different (Supplementary Information). Patient-1 (p.R566P), a 9-year-old boy, shows mild non-syndromic ID without epilepsy. The brain CT-scan did not reveal any abnormality. He has a sister who also shows non-syndromic ID but who does not carry the mutation. Patient-2 (p.Q2202del), an 11-year-old boy, shows severe ID. He did not present with IS but developed mild generalized epilepsy. The brain MRI showed severe atrophy of the cerebellum and mild atrophy of the brainstem, without any hypomyelination or other structural defects (Figure 2).
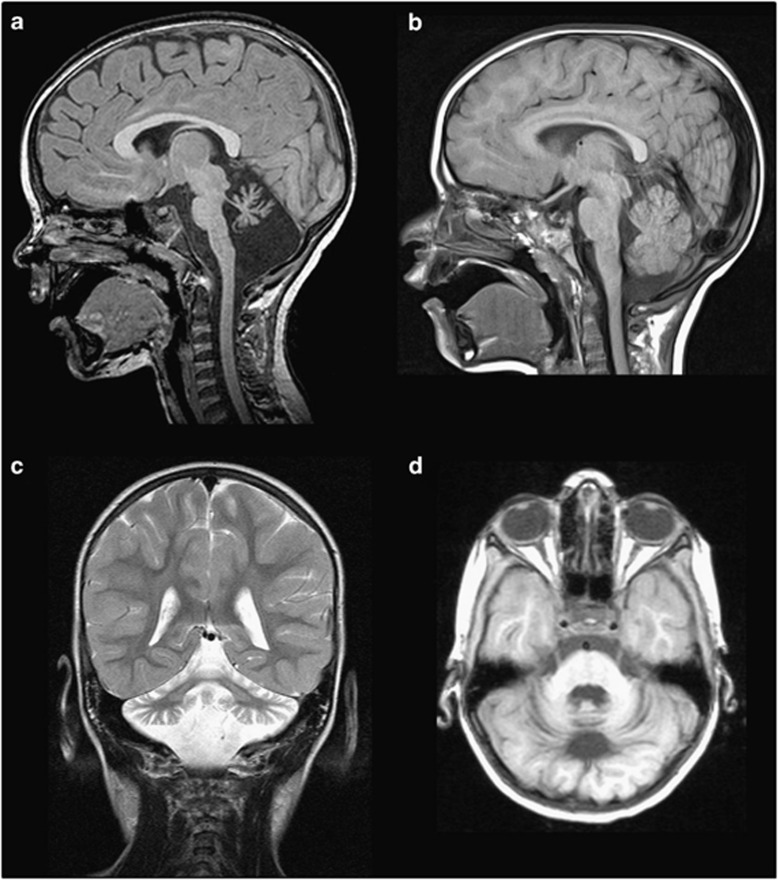
Figure 2.
The brain MRI of the patient with p.Q2202del showing pontocerebellar atrophy. (a) Sagittal T1 image at midline showing severe atrophy of vermis with hypoplastic brainstem. The corpus callosum is larger than normal for the age of the patient. (b) Sagittal T1 image of an age and gender matched control showing normal pons and normal cerebellum (c) Coronal T2 image of the patient at the level of the posterior fossa, showing severe atrophy of both cerebellar hemispheres with T2 hypersignal of both middle cerebellar pedoncule. (d) Axial T1 image of the patient at the level of the cerebellum showing diffuse cerebellar atrophy.
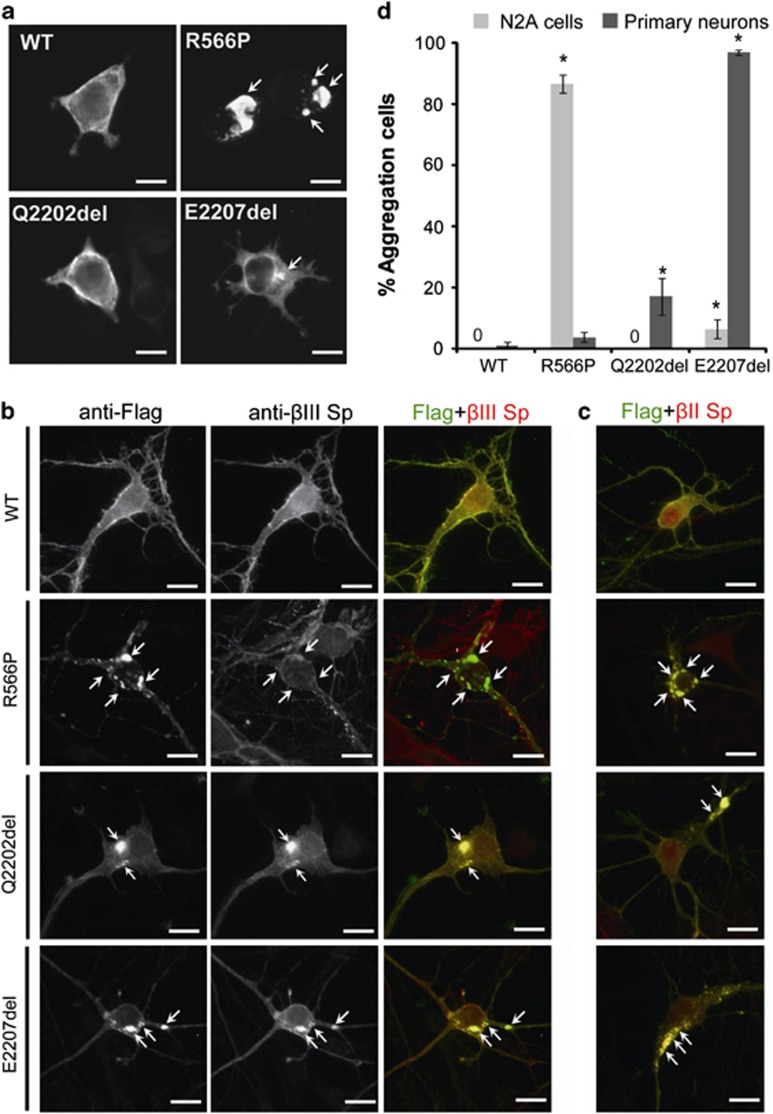
Expression of α-II spectrin containing p.E2207del or p.R2308_M2309dup induced subunit aggregates in primary neuronal cultures.4 We took advantage of this assay to compare the impact of p.R566P, p.Q2202del, and p.E2207del in mouse N2A cells and mouse cortical neurons (Figure 3). Expression of wild-type α-II spectrin in N2A cells resulted in its localization at cell periphery, without the formation of aggregates (Figures 3a and b). The p.Q2202del mutant showed a similar pattern of expression whereas the p.E2207del induced small aggregates in 6% of transfected cells. In contrast, the p.R566P mutant induced large aggregates in 86% of transfected N2A cells. These mutant α-II spectrin aggregates colocalized with endogenous β-II spectrin (Supplementary Figure S1). No endogenous β-III-spectrin subunits were detectable in N2A cells by immunofluorescence. In primary neuronal cultures, p.E2207del induced aggregates in most cells; these aggregates were composed of both β-II and β-III spectrin subunits (Figures 3b-d). The p.Q2202del mutant showed a similar aggregation profile, but in a lower proportion of cells (15–20%). In contrast, p.R566P induced aggregates in a much smaller number of cells (<5%). Moreover, double immunostaining revealed that these aggregates were composed of the β-II but not of the β-III spectrin subunit.
Figure 3.
Mutant α-II spectrin cause aggregation of α/β spectrin heterodimers. (a) The wildtype (WT) and the three mutant α-II spectrins were detected by immunofluorescence in transfected N2A cells. The WT α-II spectrin and p.Q2202del mutant were similarly expressed at cell periphery. However, the p.R566P and p.E2207del α-II spectrin mutants showed large and small aggregations (arrows), respectively. (b, c) Expression of the WT and the three mutant α-II spectrins at 7 days in vitro in primary cortical neurons. Flag tagged WT α-II spectrin was expressed at cell extensions and periphery, overlapping with the expression of β-II and β-III spectrins. Three mutant α-II spectrins (R566P, Q2202del, and E2207del) showed aggregation in cell bodies and neurites (arrows). Aggregations caused by the Q2202del and E2207del mutants were colocalized with both β-II and β-III spectrins (lower two panels). Aggregations caused by the R566P mutant were colocalized with β-II spectrin, but their colocalization with β-III spectrin was not evident. The scale bar represents 10 μm. (d) N2A cells and primary cortical neurons showing α-II spectrin aggregation were counted: Numbers of aggregated/total numbers of counted cells (expressing transfected α-II spectrin) in three experiments: N2A, WT: 0/194, R566P: 212/244, Q2202del: 0/241, E2207del: 9/180; primary neurons, WT: 3/300, R566P: 11/300, Q2202del: 51/300, E2207del: 291/300. Asterisks indicate that a significant difference (P<0.01) was observed compared with WT by Bonferroni's posttest analysis. The scale bar represents 10 μm.
Discussion
Saitsu et al4 recently showed that in-frame deletion (p.E2207del) or duplication (p.R2308_M2309dup) in the C-terminal region of SPTAN1 causes a specific syndrome characterized by severe ID, IS, hypomyelination, and brain atrophy.4 We describe here a novel in-frame deletion in the same domain (p.Q2202del). The phenotype of the patient with this mutation (patient-2) overlaps with that of the previously described patients with in-frame C-terminal SPTAN1 mutations, although being milder. For instance, all three patients showed severe ID, but patient-2 was less impaired than the others. Moreover, the previously described patients presented early in life with IS, and their course was characterized by intractable seizures. In contrast, patient-2 did not show IS and his seizures were well controlled with a single antiepileptic drug. Finally, MRI studies revealed pontocerebellar atrophy in all three patients with the in-frame mutations but patient-2 did not show atrophy of other structures or hypomyelination, like the others.
All three in-frame mutations induced the aggregation of the α-II with the β-II and β-III spectrin subunits in primary neuronal cells, suggesting that they have a dominant–negative effect. Although the spectrin aggregation profile associated with p.Q2202del was similar to that of p.E2207del, it was observed at a lower frequency correlating with the less severe phenotype observed in patient-2. Heterozygous mutations in β-III spectrin (SPTBN2) were previously shown to cause spinocerebellar ataxia type-5.3 It is tempting to speculate that cerebellar atrophy in patients with mutations in SPTAN1 is caused at least in part by the aggregation of β-III spectrin. This is consistent with the fact that β-III, unlike β-II spectrin, is abundantly expressed in the human cerebellum (Allen Brain Atlas; http://human.brain-map.org/). All together, these observations strengthen the causal relationship between in-frame mutations at the C-terminus of SPTAN1, severe ID, and pontocerebellar atrophy, while expanding the phenotypical spectrum associated with these mutations.
We also identified a de novo missense (p.R566P) in SPTAN1 in a patient with mild non-syndromic ID without epilepsy or any gross brain abnormalities. Interestingly, the aggregation profiles induced by p.R566P and by the in-frame mutations are different. First, p.R566P induced aggregates in a great proportion of N2A cells but only in a negligible proportion of primary neuronal cells. The reverse pattern was found with the in-frame mutations. Second, p.R566P only aggregated with β-II spectrin whereas the in-frame mutations showed aggregations with both β-II and β-III spectrin subunits in cortical neurons. The phenotypic differences between the patient with the missense and the patients with the in-frame mutations may thus be explained by the preferential aggregation of p.R566P α-II with β-II spectrin. It is important to underline that the sister of patient-1 was also diagnosed with non-syndromic ID but lacked p.R566P in her blood DNA. Although it is possible that she is a phenocopy, it appears more likely that p.R566P is not responsible for her brother's condition. The identification of other patients with deleterious mutations in the N-terminal region of SPTAN1 would help to distinguish these possibilities.
In summary, our work indicates that in-frame mutations in the C-terminus of SPTAN1 cause a core set of manifestations that include severe ID, generalized epilepsy, and pontocerebellar atrophy. Although it is unclear whether p.R566P is pathogenic, we found that this mutation induces a distinct pattern of spectrin aggregation. Additional studies are needed to determine whether this pattern of aggregation is associated with a specific phenotype. Our study thus provides a paradigm to validate candidate variants in SPTAN1 and to establish correlations between genotypes and phenotypes.
Acknowledgments
This study is supported by grants from the Canadian Institute of Health Research (CIHR) (J Michaud, G Rouleau, J-C Lacaille), Réseau de Génétique Médicale Appliquée (RMGA)/Fonds de la Recherche en Santé du Québec (FRSQ) (J Michaud), Genome Canada and Genome Quebec and co-funding by Université de Montréal for the Synapse to diseases (S2D) project (G Rouleau), the Ministry of Health, Labour and Welfare (H Saitsu, N Matsumoto), the Japan Science and Technology Agency (N Matsumoto), a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (N Matsumoto), and a Grant-in-Aid for Young Scientist from the Japan Society for the Promotion of Science (H Saitsu). J Michaud is the recipient of a Clinical Investigatorship Award of the CIHR (Institute of Genetics) and of a Senior Scientist Award from FRSQ. G Rouleau holds the Canada Research Chair and a Jeanne-et-J-Louis-Levesque Chair for the Genetics of Brain Diseases. We are grateful for the dedicated work of members of the S2D team (CHUM Notre-Dame Hospital Research Center, Montreal), including management (Claude Marineau and Ronald Lafrenière) and bioinformatics (Edouard Henrion and Ousmane Diallo). We are thankful for the efforts of the members of McGill University and Génome Québec Innovation Centre Sequencing and Bioinformatics groups.
The authors declare no conflict of interest
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Bennett V, Healy J. Organizing the fluid membrane bilayer: diseases linked to spectrin and ankyrin. Trends Mol Med. 2008;14:28–36. doi: 10.1016/j.molmed.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Baines AJ. Evolution of spectrin function in cytoskeletal and membrane networks. Biochem Soc Trans. 2009;37:796–803. doi: 10.1042/BST0370796. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Dick KA, Weatherspoon MR, et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat Genet. 2006;38:184–190. doi: 10.1038/ng1728. [DOI] [PubMed] [Google Scholar]
- Saitsu H, Tohyama J, Kumada T, et al. Dominant-negative mutations in alpha-II spectrin cause West syndrome with severe cerebral hypomyelination, spastic quadriplegia, and developmental delay. Am J Hum Genet. 2010;86:881–891. doi: 10.1016/j.ajhg.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher DW, Weglarz L, DeSilva TM. Properties of human red cell spectrin heterodimer (side-to-side) assembly and identification of an essential nucleation site. J Biol Chem. 1992;267:14775–14782. [PubMed] [Google Scholar]
- Conrad DF, Keebler JE, DePristo MA, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Araki Y, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–11012. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Saitsu H, Kato M, Mizuguchi T, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008;40:782–788. doi: 10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.