Abstract
Survivin is an inhibitor of apoptosis protein and has a crucial role in the development of cancer. The survivin −31G>C (rs9904341) promoter polymorphism influences survivin expression and has been implicated in cancer risk. However, conflicting results have been published from studies on the association between survivin −31G>C polymorphism and the risk of cancer. To clarify the role of this polymorphism in cancer, we performed a meta-analysis of all available and relevant published studies, involving a total of 3485 cancer patients and 3964 control subjects. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the strength of the associations. The overall results indicated that the variant genotypes were associated with a significantly increased cancer risk (CC vs GG: OR=1.58, 95% CI=1.20–2.10; CC/GC vs GG: OR=1.23, 95% CI=1.00–1.51; CC vs GG/GC: OR=1.51, 95% CI=1.23–1.85). In the stratified analyses, significantly increased risk was associated with the Asian populations (CC vs GG: OR=1.67, 95% CI=1.16–2.40; CC vs GG/GC: OR=1.50, 95% CI=1.17–1.91). We also performed the analyses by cancer type, and no statistical association was observed. The results suggest that the survivin −31G>C promoter polymorphism might be associated with an increased risk of cancer, especially in the Asian populations.
Keywords: survivin, genetic polymorphism, carcinoma, susceptibility, meta-analysis
Introduction
It is well known that the incidence of cancer, which is a multifactorial disease that resulted from complex interactions between environmental and genetic factors,1 has increased alarmingly. In addition, genetic variations may contribute to carcinogenesis. Signal transmissions resulting from these variations activate various pathways or mechanisms that disrupt the balance of normal cellular processes. Apoptosis is involved in the maintenance of these balances and has key roles in homeostasis.2, 3 Until now, two major apoptosis signaling pathways have been described: the extrinsic and intrinsic pathways.4 These two pathways have independent groups of initiator caspases that transmit the death signal downstream but share the same group of effector caspases to execute the final cell death program.4, 5 Inappropriate regulation of apoptosis may lead to a number of human disorders, including cancer.6
Survivin, a member of the inhibitor of apoptosis protein family, is involved in inhibition of apoptosis and regulation of cell division.7, 8, 9 Accumulating evidence has shown that increased expression of survivin favors the development and progression of malignancy by reducing tumor cell apoptosis.10 The human survivin gene (also known as BIRC5), located on chromosome 17q25, consists of four exons spanning 14.7 kb.11 In the promoter region of the survivin gene, the most widely studied polymorphisms are the G to C substitution at position −31 (survivin −31G>C, rs9904341, −31 from the first nucleotide of the ATG start codon). Xu et al12 first investigated the role of this polymorphism in cancer cell lines and found the presence of the mutation correlated with increased survivin expression at both the mRNA and protein levels. They also showed that this mutation altered cell cycle-dependent transcription by modifying the binding motif of the cell cycle-dependent element (CDE)/cell cycle genes homology region (CHR) repressor, which is located in the proximal region of the survivin promoter.12 Following this finding, researchers used in vitro promoter assays to demonstrate that the −31G allele had significantly lower transcriptional activity than the −31C allele, suggesting that the −31G/C polymorphism influences survivin expression, thus contributing to the genetic susceptibility to lung cancer.13
Recently, many studies have investigated the role of the survivin −31G>C polymorphism in the etiology of various type of cancers,13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 including lung, gastric, bladder, esophageal, colorectal, urothelial, and pancreatic cancer. However, the results of these studies remain inconclusive. To clarify the effect of variation in the role of survivin in cancer, we performed a meta-analysis of all eligible case–control and cohort studies to derive a more precise estimation of the overall cancer risk of the survivin −31G>C polymorphism and to quantify the potential for heterogeneity between studies.
Materials and methods
Identification and eligibility of relevant studies
Relevant publications were identified with a literature search using the keywords ‘survivin', ‘polymorphism', and ‘cancer' in the Medline database (the last search update was 23 February 2011), and the research was limited to English-language journals. Additional studies were identified by a manual search of the references of original studies. The following criteria were used for inclusion of the identified articles in our meta-analysis: (1) a case–control or cohort design was used and (2) studies contained available genotype frequencies. The major reasons for exclusion of studies were: no usable data were reported. Finally, the data for this analysis were available from 13 case–control studies and one cohort study, totaling 3485 cancer cases and 3964 controls for the survivin −31C/G promoter polymorphism.
Data extraction
Two investigators independently extracted data and jointly reached a consensus on all of the studies researched. The following information was sought from each article: the first author's name, year of publication, country of origin, ethnicity, number of cases and controls, genotype frequencies for cases and controls, and Hardy–Weinberg equilibrium (HWE) of controls.
Meta-analysis
The strength of the association between the survivin −31G/C promoter polymorphism and risk of cancer was measured by odds ratios (ORs) with 95% confidence intervals (CIs). We examined the association between allele C of the survivin −31G/C polymorphism and cancer risk, and made comparisons with homozygotes (CC vs GG), heterozygotes (GC vs GG), the dominant genetic model (GC/CC vs GG), and the recessive genetic model (CC vs GG/GC). Trend analysis was performed across the three genotypes. Stratified analyses were also carried out by ethnicity and cancer type (limited to gastric and esophageal cancer). Heterogeneity assumption was evaluated with a χ2-based Q-test. If the P-value was >0.05 of the Q-test, thus indicating a lack of heterogeneity among studies, then the effects model was used (the Mantel–Haenszel method).26 Otherwise, the random-effects model (the DerSimonian and Laird method)27 was performed. Funnel plots and Egger's linear regression tests were used to provide diagnosis of the potential publication bias. All statistical analyses were performed with the Stata software (version 9.2; StataCorp. LP, College Station, TX, USA), using two-sided P-values.
Results
Characteristics of eligible studies
A total of 30 articles relevant to the search keywords were identified. Twelve of these articles did not explore the survivin −31G/C polymorphism and were excluded.28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 Four of these articles did not explore cancer risk and were excluded.40, 41, 42, 43 Finally, 13 case–control studies and 1 cohort study44 that comprised a total of 3485 cancer cases and 3964 controls were included in our meta-analysis, and are presented in Table 1.
Table 1. Characteristics of studies included in the meta-analysis.
| ID | First author | Year | Country | Ethnic group | Cancer type | Case | Control | HWE |
|---|---|---|---|---|---|---|---|---|
| 1 | Borbely | 2006 | Hungary | European | Cervical cancer | 81 | 180 | 0.856 |
| 2 | Kawata | 2010 | Japan | Asia | Bladder cancer | 235 | 346 | 0.228 |
| 3 | Borges | 2010 | Brazilian | Mixed | Gastric cancer | 47 | 57 | 0.784 |
| 4 | Theodoropoulos | 2010 | Greece | European | Pancreatic cancer | 80 | 160 | 0.062 |
| 5 | Wang | 2009 | China | Asia | Urothelial cancer | 190 | 210 | 0.024 |
| 6 | Jang | 2008 | Korea | Asia | Lung cancer | 582 | 582 | 0.867 |
| 7 | Gazouli | 2009 | Greece | European | Colorectal cancer | 312 | 362 | 0.110 |
| 8 | Yang | 2009 | China | Asia | Esophageal cancer | 221 | 268 | 0.249 |
| 9 | Upadhyay | 2010 | India | European | Esophageal cancer | 250 | 250 | 0.094 |
| 10 | Cheng | 2008 | China | Asia | Gastric cancer | 96 | 67 | 0.667 |
| 11 | Yang | 2009 | China | Asia | Gastric cancer | 220 | 220 | 0.104 |
| 12 | Bayram | 2011 | Turkey | European | Hepatocellular cancer | 160 | 241 | 0.109 |
| 13 | Ma | 2011 | China | Asia | Nasopharyngeal cancer | 844 | 1021 | 0.357 |
| 14 | Han | 2009 | USA | European | Ovarian cancer | 167 | / | / |
Abbreviations: HWE, Hardy–Weinberg Equilibrium of Genotype of Control C, confirmed to HWE.
The 14 separate studies consisted of six Caucasians and seven Asian individuals. The genotype frequencies of the survivin −31G/C polymorphism were extracted from all eligible studies. The results of HWE test for the distribution of the genotypes in the control population are shown in Table 1. All the eligible studies were in HWE.
Quantitative synthesis
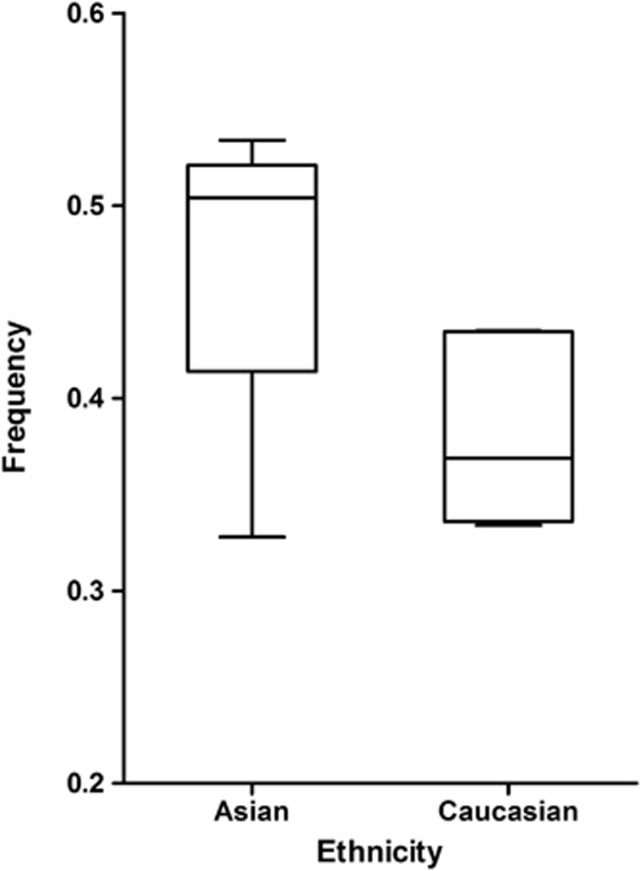
The frequency of C allele varied widely across the 14 studies, ranging from 0.33 to 0.53. As shown in Figure 1, the average frequency of the C allele in the Asian populations was 0.47, which was higher than in the Caucasian populations (0.38).
Figure 1.
Survivin −31C allele frequency among controls as stratified by ethnicity.
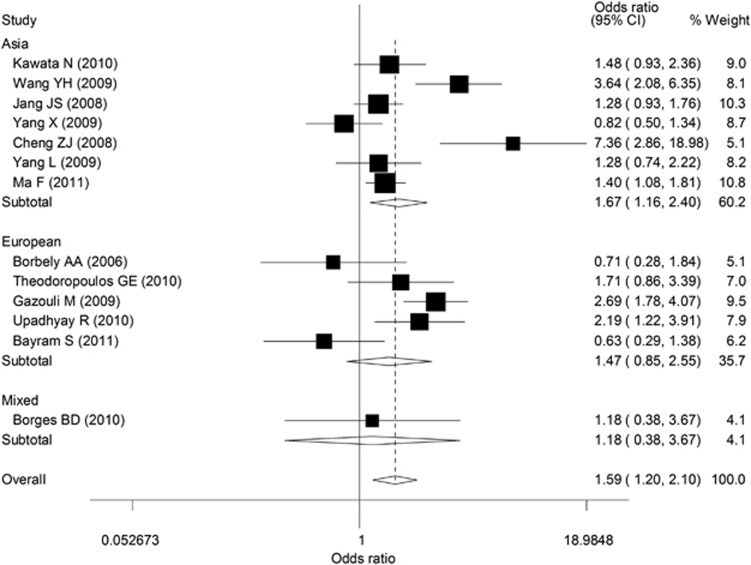
The Q-test of heterogeneity was always significant, and we conducted analyses using random effect models for the overall population. Overall, there was evidence for an association between increased cancer risk and the variant genotypes in different genetic models. As shown in Table 2 and Figure 2, the variant homozygote genotype CC was associated with significantly increased cancer risk (OR=1.58, 95% CI=1.20–2.10), compared with the wild-type homozygote genotype GG. In addition, increased cancer risks were also observed when we compared CC/GC vs GG (dominant model, OR=1.23, 95% CI=1.00–1.51) and CC vs GG/GC (recessive model, OR=1.51, 95% CI=1.23–1.85). Furthermore, the trend for the number of the C allele in the genotypes (the −31GG, −31GC, and −31CC genotypes) was statistically significant (P<0.001), indicating there was an evidence of a dose–response with increasing number of the variant allele. When stratified by ethnicity, increased cancer risk was found in the Asian populations (CC vs GG: OR=1.67, 95% CI=1.16–2.40; CC vs GG/GC: OR=1.50, 95% CI=1.17–1.91). We also performed the analysis stratified by gastric cancer and esophageal cancer, and no statistical association was observed.
Table 2. Meta-analysis of the Survivin −31C/G polymorphism and cancer risk association.
| Sample size | Test of association | Test of heterogeneity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphism | Study | Case | Control | Na | OR (95% CI) | Z | P-value | Modelb | χ2 | Pc | I2 (%) |
| CC vs GG | Overall | 1897 | 2018 | 14 | 1.59 (1.20–2.10) | 3.20 | 0.001 | R | 44.27 | <0.001 | 72.9 |
| Ethnicity | |||||||||||
| Asian | 1280 | 1353 | 7 | 1.67 (1.16–2.40) | 2.73 | 0.006 | R | 27.74 | <0.001 | 78.4 | |
| European | 578 | 636 | 6 | 1.47 (0.85–2.55) | 1.36 | 0.173 | R | 14.59 | 0.006 | 72.6 | |
| Cancer type | |||||||||||
| Gastric cancer | 197 | 166 | 3 | 2.20 (0.71–6.88) | 1.36 | 0.173 | R | 10.51 | 0.005 | 81.0 | |
| Esophageal cancer | 253 | 271 | 2 | 1.32 (0.51–3.46) | 0.57 | 0.568 | R | 6.34 | 0.012 | 84.2 | |
| GC vs GG | Overall | 2514 | 3134 | 14 | 1.08 (0.90–1.28) | 0.80 | 0.421 | R | 25.69 | 0.012 | 53.3 |
| Ethnicity | |||||||||||
| Asian | 1656 | 2072 | 7 | 1.32 (0.87–1.48) | 0.92 | 0.358 | R | 18.40 | 0.005 | 67.4 | |
| European | 820 | 1013 | 6 | 1.06 (0.87–1.29) | 0.60 | 0.548 | F | 6.24 | 0.182 | 35.9 | |
| Cancer type | |||||||||||
| Gastric cancer | 252 | 277 | 3 | 1.06 (0.74–1.53) | 0.34 | 0.736 | F | 4.66 | 0.097 | 57.1 | |
| Esophageal cancer | 369 | 415 | 2 | 0.99 (0.74–1.31) | 0.09 | 0.925 | F | 0.00 | 0.947 | 0.0 | |
| CC/GC vs GG | Overall | 3485 | 3964 | 14 | 1.23 (1.00–1.51) | 2.00 | 0.045 | R | 39.23 | <0.001 | 69.4 |
| Ethnicity | |||||||||||
| Asian | 2388 | 2714 | 7 | 1.32 (0.99–1.78) | 1.86 | 0.063 | R | 26.18 | <0.001 | 77.1 | |
| European | 1050 | 1193 | 6 | 1.18 (0.85–1.65) | 0.98 | 0.328 | R | 11.97 | 0.018 | 66.6 | |
| Cancer type | |||||||||||
| Gastric cancer | 363 | 344 | 3 | 1.38 (0.62–3.08) | 0.79 | 0.429 | R | 9.48 | 0.009 | 78.9 | |
| Esophageal cancer | 471 | 518 | 2 | 1.06 (0.81–1.38) | 0.39 | 0.696 | F | 0.65 | 0.421 | 0.0 | |
| CC vs GG/GC | Overall | 3485 | 3964 | 14 | 1.51 (1.23–1.85) | 3.90 | <0.001 | R | 33.29 | 0.001 | 64.0 |
| Ethnicity | |||||||||||
| Asian | 2388 | 2714 | 7 | 1.50 (1.17–1.91) | 3.24 | 0.001 | R | 18.96 | 0.004 | 68.4 | |
| European | 1050 | 1193 | 6 | 1.44 (0.91–2.29) | 1.55 | 0.120 | R | 12.45 | 0.014 | 67.9 | |
| Cancer type | |||||||||||
| Gastric cancer | 363 | 344 | 3 | 2.06 (0.91–4.66) | 1.74 | 0.083 | R | 7.06 | 0.029 | 71.7 | |
| Esophageal cancer | 419 | 518 | 2 | 1.32 (0.50–3.50) | 0.57 | 0.571 | R | 8.30 | 0.004 | 87.9 | |
Number of comparisons.
Random-effects model (R) was used when P-value for heterogeneity test <0.05; otherwise, fix-effects model (F) was used.
P-value of Q-test for heterogeneity test.
Bold values indicate significant difference.
Figure 2.
Forest plot showing the association between the survivin −31G>C promoter polymorphism and risk of malignancy (CC vs GG). The random effect model was used.
Sensitivity analyses
Overall comparisons showed significant heterogeneity between studies, which may be due to grouping all cancer types together. We performed sensitivity analyses to assess the source of heterogeneity, which indicated that four independent studies18, 20, 22, 24 were the main origin of heterogeneity. This heterogeneity was effectively removed by exclusion of these four studies (CC vs GG: P heterogeneity=0.099; CC versus GG/GC: P heterogeneity=0.151; GC/CC versus GG: P heterogeneity=0.315). In addition, no other single study influenced the pooled OR qualitatively, as indicated by sensitivity analyses, suggesting that the results of this meta-analysis are stable.
Publication bias
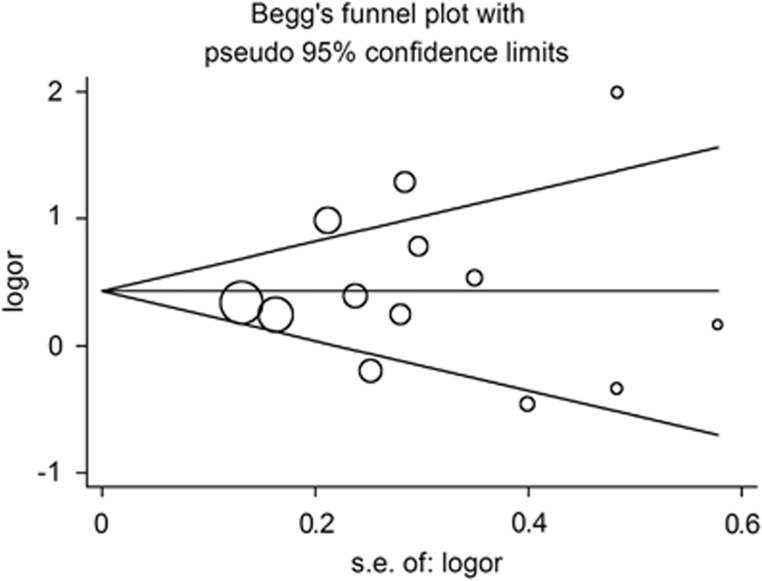
To assess the publication bias of the literature, Begger's funnel plot and Egger's test were performed. As shown in Figure 3, the shapes of the funnel plots did not indicate any evidence of obvious asymmetry in all comparison models. Thus, Egger's test was used to provide statistical evidence of funnel plot symmetry and also did not show any evidence of publication bias (t=0.36, P=0.729 for CC vs GG; t=0.25, P=0.806 for CC vs GG/GC; t=0.710, P=0.429 for GC/CC vs GG).
Figure 3.
Begg's funnel plot for publication bias test, CC vs GG; each point represents a separate study for the indicated association. Log (OR): natural logarithm of OR. Horizontal line represents size of effect.
Discussion
The regulation of programmed cell death is important for the prevention of tumorigenesis. Impairment of apoptosis facilitates the accumulation of genetic errors by prolonging the cell cycle, promoting resistance to immune-based cytotoxicity, and providing selective growth advantages for the altered cells, thus contributing to carcinogenesis.45, 46 Survivin is an important apoptosis inhibitor protein and has a key role in inhibiting apoptosis and facilitating cell proliferation.47 It has been observed that it is markedly overexpressed in almost all human malignancies, including lung, breast, brain, stomach, esophagus, and liver cancer, as well as ovarian and hematological cancers.7, 47 Therefore, it is biologically plausible that genetic variations of the survivin gene may modulate the risk of cancer.
The anti-apoptotic function of survivin has been identified both in vitro and in vivo.48, 49 The mechanisms through which survivin exerts this function are complicated, and includes binding to caspase-3 and caspase-7 to prevent their activation,50, 51 physically interacting with Smac/DIABLO to directly inhibit caspases,52 and inhibiting the AIF pathway to provide cytoprotection to cells against caspase-independent cell death.53 Regulation at the transcriptional level is an important mechanism for survivin expression. Survivin promoter activity is regulated by transcription factors such as β-catenin activated T-cell factor54 and hypoxia-inducible factor-1 α.55 In addition, survivin expression is also mediated by cell CDEs and cell CHRs located in the proximal region of the survivin promoter.56, 57 The −31G>C polymorphism of survivin is located at the CDE/CHR repressor binding site, and may influence the affinity of repressor binding to the CDE/CHR element.12 Functional studies on this polymorphism have shown that the −31C allele has significantly higher transcriptional activity than the −31G allele, and individuals with the −31CC genotype have upregulated survivin levels compared with those with the GC and GG genotypes.12, 13 Thus, the −31G>C polymorphism may influence an individual's susceptibility to cancer. To date, several epidemiological studies have investigated the association between the −31G>C polymorphism in survivin and risk of various types of cancer but have produced conflicting results. In order to resolve this conflict, we conducted a meta-analysis of 14 studies, comprising a total of 3485 cancer cases and 3964 controls, to evaluate the associations between the −31G>C polymorphism and cancer risk.
Consistent with the observations made in the above-mentioned functional studies, our results suggested that variant genotypes (CC and CC/GC) were associated with a significantly increased cancer risk in several genetic models. In the subgroup analysis by ethnicity, we found that Asian individuals with the −31CC genotype had an increased risk of cancer when compared with GG or GG/GC genotypes. However, no significant association was found among Europeans. Several reasons may lead to this ethnic difference. First, cancer is a multifactor disease with varying incidence in different populations. It has been suggested that this variation may depend on a combination of differences in polymorphism distributions with environmental factors.58 For instance, the average frequency of the C allele in the Asian populations was 0.47, which was higher than in the Caucasian populations (0.38). Second, a smaller sample size was enrolled from European than from Asia, and together with lower frequency of risk allele (C) in Europeans than in Asians, may contribute to the non-significant findings of Europeans. However, these observations should be interpreted with caution as the number of European studies enrolled and the sample size of each study are limited and may be underpowered to detect a significant association. Moreover, obvious differences in composition of cancer types between European and Asian may also contribute to the findings observed. Last, other factors such as selection bias and different matching criteria may also have a role. Additionally, although no statistical association was observed in the subgroup analysis by cancer type, due to the wide CIs of our data, a possibility of an effect between genotypes and gastric or esophageal cancer risk may still exist. Therefore, more studies may be needed to clarify the effect of this polymorphism on cancer in European and on the difference between cancer types.
Some other limitations of our meta-analysis should be addressed. First, only papers written in English were included; studies published in other languages were not included, which thus may bias the results. Second, our lack of access to the original data from the included studies limited further evaluation of the potential interactions, as gene–environment and gene–gene interactions, and even different polymorphic loci of the same gene, may also modulate cancer risk. Third, our results were based on unadjusted estimates, while a more precise analysis needs to be conducted if individual data such as age and sex are available. Thus, lack of the information for the data analysis may lead to serious confounding bias. Nevertheless, advantages in our meta-analysis should also be acknowledged. First, a systematic review of the association of survivin polymorphism with cancer risk is statistically more powerful than any single study. Second, the studies included in our meta-analysis strictly and satisfactorily met our selection criteria.
Studies of common polymorphisms in genetic variations, if large enough and unbiased, can provide insights into the in vivo associations between the risk of cancer and genetic variation. Such studies may explore empirical associations that indicate that a polymorphism in a gene of interest has an influence on cancer, independent of metabolic regulatory mechanisms and other genetic and environmental variability.59 Here, we performed a systematic literature review to evaluate the relationships between the survivin −31G>C promoter polymorphism and the risk of cancer. Individuals with variant genotypes of this polymorphism have an associated increased cancer risk, particularly those of Asian origin, which suggests that this increased risk may be ethno-specific. Additional larger studies are warranted to validate our findings. Future studies with larger numbers of standardized unbiased homogenous cancer patients and well-matched controls are required to examine associations between the survivin −31 G>C polymorphism and cancer risk and to draw more comprehensive conclusions. Moreover, investigations of the combined effects of gene and environment may lead to a better understanding of the role of the survivin −31 G>C polymorphism in cancers.
Acknowledgments
This work was supported by the China Natural Science Foundation (30872657 and 81072078), Natural Science Foundation of Jiangsu Province (2008475 and 2010580), Scientific Program of Ministry of Health (W2011BX009), Program for Development of Innovative Research Team in the First Affiliated Hospital of NJMU, and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
The authors declare no conflict of interest.
References
- Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev. 2004;4:850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- Mondello C, Scovassi AI. Apoptosis: a way to maintain healthy individuals. Subcell Biochem. 2010;50:307–323. doi: 10.1007/978-90-481-3471-7_16. [DOI] [PubMed] [Google Scholar]
- Raff M. Cell suicide for beginners. Nature. 1998;396:119–122. doi: 10.1038/24055. [DOI] [PubMed] [Google Scholar]
- Delhalle S, Duvoix A, Schnekenburger M, Morceau F, Dicato M, Diederich M. An introduction to the molecular mechanisms of apoptosis. Ann NY Acad Sci. 2003;1010:1–8. doi: 10.1196/annals.1299.001. [DOI] [PubMed] [Google Scholar]
- Degterev A, Boyce M, Yuan J. A decade of caspases. Oncogene. 2003;22:8543–8567. doi: 10.1038/sj.onc.1207107. [DOI] [PubMed] [Google Scholar]
- Hajra KM, Liu JR. Apoptosome dysfunction in human cancer. Apoptosis. 2004;9:691–704. doi: 10.1023/B:APPT.0000045786.98031.1d. [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- Altieri DC. The molecular basis and potential role of survivin in cancer diagnosis and therapy. Trends Mol Med. 2001;7:542–547. doi: 10.1016/s1471-4914(01)02243-2. [DOI] [PubMed] [Google Scholar]
- Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Sirugo G, Altieri DC. Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem. 1998;273:11177–11182. doi: 10.1074/jbc.273.18.11177. [DOI] [PubMed] [Google Scholar]
- Xu Y, Fang F, Ludewig G, Jones G, Jones D. A mutation found in the promoter region of the human survivin gene is correlated to overexpression of survivin in cancer cells. DNA Cell Biol. 2004;23:527–537. doi: 10.1089/dna.2004.23.527. [DOI] [PubMed] [Google Scholar]
- Jang JS, Kim KM, Kang KH, et al. Polymorphisms in the survivin gene and the risk of lung cancer. Lung Cancer. 2008;60:31–39. doi: 10.1016/j.lungcan.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Borbely AA, Murvai M, Szarka K, et al. Survivin promoter polymorphism and cervical carcinogenesis. J Clin Pathol. 2007;60:303–306. doi: 10.1136/jcp.2006.037804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata N, Tsuchiya N, Horikawa Y, et al. Two survivin polymorphisms are cooperatively associated with bladder cancer susceptibility. Int J Cancer. 2010;129:1872–1880. doi: 10.1002/ijc.25850. [DOI] [PubMed] [Google Scholar]
- Borges BD, Burbano RR, Harada ML. Survivin −31C/G polymorphism and gastric cancer risk in a Brazilian population. Clin Exp Med. 2011;11:189–193. doi: 10.1007/s10238-010-0122-5. [DOI] [PubMed] [Google Scholar]
- Theodoropoulos GE, Michalopoulos NV, Panoussopoulos SG, Taka S, Gazouli M. Effects of caspase-9 and survivin gene polymorphisms in pancreatic cancer risk and tumor characteristics. Pancreas. 2010;39:976–980. doi: 10.1097/MPA.0b013e3181d705d4. [DOI] [PubMed] [Google Scholar]
- Wang YH, Chiou HY, Lin CT, et al. Association between survivin gene promoter −31 C/G polymorphism and urothelial carcinoma risk in Taiwanese population. Urology. 2009;73:670–674. doi: 10.1016/j.urology.2008.09.048. [DOI] [PubMed] [Google Scholar]
- Gazouli M, Tzanakis N, Rallis G, et al. Survivin −31G/C promoter polymorphism and sporadic colorectal cancer. Int J Colorectal Dis. 2009;24:145–150. doi: 10.1007/s00384-008-0601-2. [DOI] [PubMed] [Google Scholar]
- Yang X, Xiong G, Chen X, et al. Polymorphisms of survivin promoter are associated with risk of esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2009;135:1341–1349. doi: 10.1007/s00432-009-0575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay R, Khurana R, Kumar S, Ghoshal UC, Mittal B. Role of survivin gene promoter polymorphism (−31G>C) in susceptibility and survival of esophageal cancer in Northern India. Ann Surg Oncol. 2011;18:880–887. doi: 10.1245/s10434-010-1371-y. [DOI] [PubMed] [Google Scholar]
- Cheng ZJ, Hu LH, Huang SJ. [Correlation of −31G/C polymorphisms of survivin promoter to tumorigenesis of gastric carcinoma] Ai Zheng. 2008;27:258–263. [PubMed] [Google Scholar]
- Yang L, Zhu H, Zhou B, et al. The association between the survivin C-31G polymorphism and gastric cancer risk in a Chinese population. Dig Dis Sci. 2009;54:1021–1028. doi: 10.1007/s10620-008-0441-5. [DOI] [PubMed] [Google Scholar]
- Bayram S, Akkiz H, Bekar A, Akgollu E. The association between the survivin −31G/C promoter polymorphism and hepatocellular carcinoma risk in a Turkish population. Cancer Epidemiol. 2011;35:555–559. doi: 10.1016/j.canep.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Ma F, Zhang H, Zhai Y, et al. Functional polymorphism −31C/G in the promoter of BIRC5 gene and risk of nasopharyngeal carcinoma among Chinese. PLoS One. 2011;6:e16748. doi: 10.1371/journal.pone.0016748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Wang YH, Chen DJ, Yi TN, Liu XH. The relationship among human papilloma virus infection, survivin, and p53 gene in lung squamous carcinoma tissue. Saudi Med J. 2010;31:1331–1336. [PubMed] [Google Scholar]
- Wang H, Holloway MP, Ma L, et al. Acetylation directs survivin nuclear localization to repress STAT3 oncogenic activity. J Biol Chem. 2010;285:36129–36137. doi: 10.1074/jbc.M110.152777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepela E, Dankova P, Moravcikova E, et al. Increased expression of inhibitor of apoptosis proteins, survivin and XIAP, in non-small cell lung carcinoma. Int J Oncol. 2009;35:1449–1462. doi: 10.3892/ijo_00000464. [DOI] [PubMed] [Google Scholar]
- Farnebo L, Jerhammar F, Vainikka L, Grenman R, Norberg-Spaak L, Roberg K. Number of negative points: a novel method for predicting radiosensitivity in head and neck tumor cell lines. Oncol Rep. 2008;20:453–461. [PubMed] [Google Scholar]
- Wagner M, Schmelz K, Dorken B, Tamm I. Epigenetic and genetic analysis of the survivin promoter in acute myeloid leukemia. Leuk Res. 2008;32:1054–1060. doi: 10.1016/j.leukres.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Boidot R, Vegran F, Jacob D, et al. The expression of BIRC5 is correlated with loss of specific chromosomal regions in breast carcinomas. Genes Chromosomes Cancer. 2008;47:299–308. doi: 10.1002/gcc.20533. [DOI] [PubMed] [Google Scholar]
- Vegran F, Boidot R, Oudin C, Defrain C, Rebucci M, Lizard-Nacol S. Association of p53 gene alterations with the expression of antiapoptotic survivin splice variants in breast cancer. Oncogene. 2007;26:290–297. doi: 10.1038/sj.onc.1209784. [DOI] [PubMed] [Google Scholar]
- Floyd HS, Jennings-Gee JE, Kock ND, Miller MS. Genetic and epigenetic alterations in lung tumors from bitransgenic Ki-rasG12C expressing mice. Mol Carcinog. 2006;45:506–517. doi: 10.1002/mc.20181. [DOI] [PubMed] [Google Scholar]
- Nakano J, Huang CL, Liu D, Ueno M, Sumitomo S, Yokomise H. Survivin gene expression is negatively regulated by the p53 tumor suppressor gene in non-small cell lung cancer. Int J Oncol. 2005;27:1215–1221. [PubMed] [Google Scholar]
- Teodoridis JM, Hall J, Marsh S, et al. CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res. 2005;65:8961–8967. doi: 10.1158/0008-5472.CAN-05-1187. [DOI] [PubMed] [Google Scholar]
- Kimura M, Okano Y. [Aurora kinases and cancer] Gan To Kagaku Ryoho. 2005;32:1–5. [PubMed] [Google Scholar]
- Saitoh Y, Yaginuma Y, Ishikawa M. Analysis of Bcl-2, Bax and Survivin genes in uterine cancer. Int J Oncol. 1999;15:137–141. doi: 10.3892/ijo.15.1.137. [DOI] [PubMed] [Google Scholar]
- Boidot R, Vegran F, Jacob D, et al. The transcription factor GATA-1 is overexpressed in breast carcinomas and contributes to survivin upregulation via a promoter polymorphism. Oncogene. 2010;29:2577–2584. doi: 10.1038/onc.2009.525. [DOI] [PubMed] [Google Scholar]
- Farnebo L, Jedlinski A, Ansell A, et al. Proteins and single nucleotide polymorphisms involved in apoptosis, growth control, and DNA repair predict cisplatin sensitivity in head and neck cancer cell lines. Int J Mol Med. 2009;24:549–556. doi: 10.3892/ijmm_00000264. [DOI] [PubMed] [Google Scholar]
- Loof J, Pfeifer D, Adell G, Sun XF. Significance of an exon 2 G4C14-to-A4T14 polymorphism in the p73 gene on survival in rectal cancer patients with or without preoperative radiotherapy. Radiother Oncol. 2009;92:215–220. doi: 10.1016/j.radonc.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Yang X, Xiong G, Chen X, et al. Survivin expression in esophageal cancer: correlation with p53 mutations and promoter polymorphism. Dis Esophagus. 2009;22:223–230. doi: 10.1111/j.1442-2050.2008.00885.x. [DOI] [PubMed] [Google Scholar]
- Han CH, Wei Q, Lu KK, Liu Z, Mills GB, Wang LE. Polymorphisms in the survivin promoter are associated with age of onset of ovarian cancer. Int J Clin Exp Med. 2009;2:289–299. [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, O'Donovan N, Brennan DJ, Gallagher WM, Ryan BM. Survivin: a promising tumor biomarker. Cancer Lett. 2007;249:49–60. doi: 10.1016/j.canlet.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Melet A, Song K, Bucur O, Jagani Z, Grassian AR, Khosravi-Far R. Apoptotic pathways in tumor progression and therapy. Adv Exp Med Biol. 2008;615:47–79. doi: 10.1007/978-1-4020-6554-5_4. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther. 2006;5:1087–1098. doi: 10.1158/1535-7163.MCT-05-0375. [DOI] [PubMed] [Google Scholar]
- Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- Reed JC. The Survivin saga goes in vivo. J Clin Invest. 2001;108:965–969. doi: 10.1172/JCI14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I, Wang Y, Sausville E, et al. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58:5315–5320. [PubMed] [Google Scholar]
- Shin S, Sung BJ, Cho YS, et al. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry. 2001;40:1117–1123. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- Sun C, Nettesheim D, Liu Z, Olejniczak ET. Solution structure of human survivin and its binding interface with Smac/Diablo. Biochemistry. 2005;44:11–17. doi: 10.1021/bi0485171. [DOI] [PubMed] [Google Scholar]
- Liu T, Brouha B, Grossman D. Rapid induction of mitochondrial events and caspase-independent apoptosis in Survivin-targeted melanoma cells. Oncogene. 2004;23:39–48. doi: 10.1038/sj.onc.1206978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PJ, Plescia J, Clevers H, Fearon ER, Altieri DC. Survivin and molecular pathogenesis of colorectal cancer. Lancet. 2003;362:205–209. doi: 10.1016/S0140-6736(03)13910-4. [DOI] [PubMed] [Google Scholar]
- Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, Yang L. Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J Biol Chem. 2006;281:25903–25914. doi: 10.1074/jbc.M603414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Altieri DC. The cancer antiapoptosis mouse survivin gene: characterization of locus and transcriptional requirements of basal and cell cycle-dependent expression. Cancer Res. 1999;59:3143–3151. [PubMed] [Google Scholar]
- Otaki M, Hatano M, Kobayashi K, Ogasawara T, Kuriyama T, Tokuhisa T. Cell cycle-dependent regulation of TIAP/m-survivin expression. Biochim Biophys Acta. 2000;1493:188–194. doi: 10.1016/s0167-4781(00)00142-1. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang H, Li H, et al. Association between X-ray repair cross complementing group 1 codon 399 and 194 polymorphisms and lung cancer risk: a meta-analysis. Cancer Lett. 2009;285:134–140. doi: 10.1016/j.canlet.2009.05.005. [DOI] [PubMed] [Google Scholar]