Abstract
The 17q21.31 microdeletion syndrome is characterised by intellectual disability, epilepsy, distinctive facial dysmorphism, and congenital anomalies. To date, all individuals reported with this syndrome have been simplex patients, resulting from de novo deletions. Here, we report sibling recurrence of the 17q21.31 microdeletion syndrome in two independent families. In both families, the mother was confirmed to be the parent-of-origin for the 17q21.31 deletion. Fluorescence in situ hybridisation analyses in buccal mucosa cells, of the mother of family 1, identified monosomy 17q21.31 in 4/50 nuclei (8%). In mother of family 2, the deletion was identified in 2/60 (3%) metaphase and in 3/100 (3%) interphase nuclei in peripheral lymphocytes, and in 7/100 (7%) interphase nuclei in buccal cells. A common 17q21.31 inversion polymorphism predisposes to non-allelic homologous recombination and hereby to the 17q21.31 microdeletion syndrome. On the basis of the 17q21.31 inversion status of the parents, we calculated that the probability of the second deletion occurring by chance alone was 1/14 438 and 1/4812, respectively. If the inversion status of the parents of a child with the 17q21.31 microdeletion syndrome is unknown, the overall risk of a second child with the 17q21.31 microdeletion is 1/9461. We conclude that the presence of low-level maternal somatic–gonadal mosaicism is associated with the microdeletion recurrence in these families. This suggests that the recurrence risk for parents with a child with a 17q21.31 microdeletion for future pregnancies is higher than by chance alone and testing for mosaicism in the parents might be considered as a helpful tool in the genetic counselling.
Keywords: 17q21.31, microdeletion, familial, germline mosaicism
Introduction
The 17q21.31 microdeletion syndrome (Online Mendelian Inheritance in Man, #610443) is a genomic disorder characterised by intellectual disability, hypotonia, epilepsy, congenital cardiac- and urogenital defects, and distinctive facial dysmorphism, including a tall or broad forehead, long face, upslanting palpebral fissures, epicanthal folds, and a tubular or pear-shaped nose with bulbous nasal tip.1, 2, 3, 4 The syndrome is caused by a recurrent 500–650 kb heterozygous deletion at chromosome 17q21.31,5, 6, 7 which is thought to result from non-allelic homologous recombination (NAHR),8 mediated by flanking low-copy repeats. The minimum critical region is 424 kb (41 046 729–41 470 954 Mb, hg18) in size1 and encompasses four known genes, CRHR1, IMP5, MAPT, and STH, in addition to three putative genes, FLJ25168, BC018035, and LOC284058.
In every parent–child trio tested to date, the transmitted deleted chromosome 17 carries an H2 chromosome, which is discernable from H1 by a 900-kb inversion polymorphism, encompassing the 17q21.31 genomic interval.1, 9 This suggests that the inversion may be a necessary pre-disposing factor for the deletion to occur, but it is unlikely to be sufficient in itself, given its relatively high frequency of up to 20% in the European populations.9 The latter is underscored by all individuals with the syndrome being simplex patients, resulting from de novo deletions.
Here, we report the sibling recurrence of a 17q21.31 microdeletion in two independent families. In both families, we identified low-grade mosaicism for monosomy 17q21.31 in blood and/or buccal cells from the mother, strengthening our hypothesis for the presence of low-level somatic–gonadal mosaicism. This is the first report of familial recurrence of the 17q21.31 microdeletion syndrome.
Patients and methods
Clinical reports
Family 1
Patients 1 and 2 (Figures 1a and b) were referred to the Sheffield Clinical Genetics Service, Sheffield Children's Hospital, UK, because of developmental delay and dysmorphism. They are two half-siblings, born to the same mother, but with different, unrelated fathers. Their mother is healthy and of normal intelligence. Conventional chromosome analyses in lymphocytes (GTG banding, 550 band level) demonstrated normal karyotypes for both siblings. However, their facial gestalt was consistent with the 17q21.31 microdeletion syndrome, thus this was studied further.
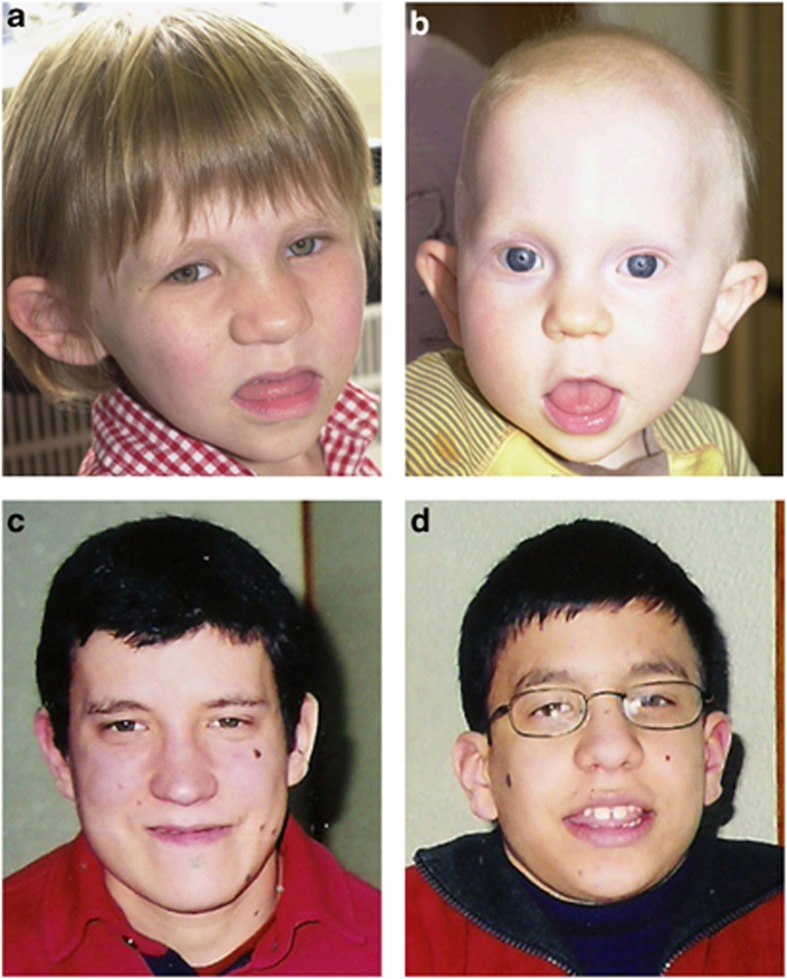
Figure 1.
Clinical photographs. (a) Patient 1, (b) patient 2, (c) patient 3, and (d) patient 4. Note in patients 1 and 2 the upslanting palpebral fissures, large protuberant ears, broad nasal bridge, bulbous nasal tip, prominent tongue, and a broad chin. In patients 3 and 4 facial dysmorphism, consistent with the 17q21.31 microdeletion syndrome, includes long face, prominent ears, broadnasal bridge, bulbous nasal tip, and everted lower lip. Written consent to publish these images has been obtained.
Patient 1
The first patient is an 8-year-old girl. She was born to a 28-year-old healthy mother and a 35-year-old healthy father, who are non-consanguineous. She was born after a normal vaginal delivery, at 41+5 weeks gestation, following a normal pregnancy. Her birth weight (BW) was 2980 g (10th centile), birth length (BL) 47 cm (<3rd centile), and head circumference (HC) 35 cm (50th centile). Apgar scores were 9 and 10, at 1 and 5 min, respectively. The neonatal history was complicated by feeding difficulties and hypotonia.
She had a ventricular septal defect that resolved spontaneously and a unilateral congenital hip dislocation that required surgery. She was hospitalised for pneumonia at 18 months of age. She has mild hearing loss associated with chronic serous otitis media with effusion. She is also prone to constipation. Facial dysmorphism includes upslanting palpebral fissures, large protuberant ears, a broad nasal bridge, bulbous nasal tip, prominent tongue, and a broad chin (Figure 1a). She also has a large sacral dimple.
Developmental milestones were delayed, with independent sitting achieved at 8 months (directly after the removal of the first hip spica), walking at 3 years and expressive speech at 3 years. She speaks with a stammer. Intellectual function was estimated to be at a level of approximately half her chronological age and she attends a special school for children with general learning difficulties.
Patient 2
The second patient is a 3-year-old boy. He was born to the same mother as patient 1, but a different, 35-year-old healthy father. This was also a non-consanguineous union and this father also has no family history of note. Patient 2 was born 5 years following the birth of Patient 1. Delivery occurred at 40 weeks gestation, by Caesarean section, due to breech presentation, following an uncomplicated pregnancy. BW was 3080 g (15th centile). Apgar scores were 8 and 9, at 1 and 5 min, respectively.
He fed well initially, but was investigated subsequently for failure to thrive. Hypotonia, dilatation of the renal collecting system, a mild degree of plagiocephaly, hip dysplasia, and similar facial dysmorphism to his sister were present (Figure 1b).
Developmental milestones were delayed, including independent sitting at 12 months and walking at 3 years. At the current age of 3 years, he has no expressive speech. He attends a mainstream school with additional support.
Family 2
Patients 3 and 4 (Figures 1c and d) were referred to the Departments of Genetics, Santa Maria University Hospital, Lisbon, Portugal, because of intellectual disability and dysmorphism. Their parents are healthy and of normal intelligence. Conventional chromosome analyses in lymphocytes (GTG banding, 550 band level) demonstrated normal karyotypes for both siblings.
Patient 3
The third patient is a 25-year-old man. He was born to a 28-year-old healthy mother and a 32-year-old healthy father. Parents are non-consanguineous, with no significant family history. Following a normal pregnancy, he was born at term, by Caesarean section because of a breech presentation. His BW was 3300 g (50–75th centile) and his height was 49 cm (25–50th centile). Apgar scores were 9 and 10, at 1 and 5 min, respectively. The neonatal period was uncomplicated, without hypotonia, feeding, or respiratory difficulties.
He has post-lingual, moderate, bilateral sensorineural hearing loss, and has used hearing aids since childhood. At the age of 18 years, he developed seizures and currently he is on antiepileptic drugs without crises. Facial dysmorphism includes a long face, prominent ears, a broad nasal bridge, bulbous nasal tip, macroglossia with a tongue groove, hypodontia (absence of the lateral inferior incisors), and everted lower lip. He also has multiple nevi, localised on the face and trunk, and macroorchidism. Magnetic resonance imaging (MRI) of the brain revealed colpocephaly, without other abnormalities.
Developmental milestones were delayed: smiling at 3 months, independent sitting at 8 months, walking at 19 months, and first words at 18 months. He had an expressive language disorder, requiring speech therapy during childhood. He has a mild cognitive impairment and a friendly personality. At the age of 25 years, his weight was 56 kg (5–10th centiles), his height was 163 cm (3rd centile), and his HC was 56 cm (50th centile). He finished secondary school with educational support. He is able to read and write, but has problems with mathematics. He lives with his parents and works at a catering company, packing food.
Patient 4
The fourth patient is an 18-year-old boy, the younger brother of patient 3. He was born 7 years following the birth of patient 3. The pregnancy was uneventful. He was born at term, by Caesarean section, again because of a breech presentation. His BW was 3530 g (75th centile), his BL was 49.5 cm (50th centile), and HC was 35.5 cm (75–90th centiles). Apgar scores were 8 and 10, at 1 and 5 min, respectively. At birth mild plagiocephaly, a sacral dimple, and bilateral hip dysplasia were noticed. The neonatal period was complicated with transitory, conjugated hyperbilirrubinemia, which required exchange transfusion. No hypotonia, feeding, or respiratory difficulties were noticed.
At 9 months of age, he developed seizures and currently he is on antiepileptic drugs without crises. He has facial features similar to his brother, including a long face, prominent ears, a broad nasal bridge, bulbous nasal tip, macroglossia with a tongue groove, hypodontia (absence of the lateral inferior incisors), and an everted lower lip. He also has multiple nevi localised on the face and trunk, and macroorchidism. He has myopia and astigmatism, but a hearing assessment was normal. MRI of the brain also revealed colpocephaly, without other abnormalities.
Developmental milestones were delayed: independent sitting was achieved at 7 months and walking at 20 months of age. He had marked speech delay, with his first words at 36 months, and only monosyllables at 4 years of age. He suffers from anxiety and stammering, and received speech therapy and psychological support. Similar to his brother, he has a mild cognitive impairment and a friendly personality. At the age of 18 years, his weight was 50 kg (<3rd centile), his height was 159 cm (<3rd centile), and his HC was 56.7 cm (95th centile). He finished secondary school with educational support and has begun a secretarial course.
Microarray studies
In family 1, 100 ng of genomic DNA from the patients and their mother were hybridised to Affymetrix Cytogenetics Whole-Genome 2.7 M arrays, according to the protocol provided by the manufacturer (Cytogenetics Assay Protocol P/N 702761 Rev.2; Affymetrix, Santa Clara, CA, USA). Signal intensities were calculated and analysed using Chromosome Analysis Suite v29 (ChAS; Affymetrix), using default settings. Suspected mosaicism for the 17q21.31 microdeletion in the mother led to an additional analysis being performed, with a minimum mosaicism confidence of 60% for the region involved.
In Family 2, 100 ng of genomic DNA from the patients and both parents were hybridised to Affymetrix GeneChip 250K SNP arrays, according to the standard protocols. Genome-wide copy number variants (CNVs) were identified using the Copy Number Analyzer for Affymetrix GeneChip v2.0 software.
FISH analyses
Standard fluorescence in situ hybridisation (FISH) assays were performed, on cultured peripheral-blood lymphocytes and buccal mucosa of the mothers with the use of the commercial 17q21.31 FISH probe, Vysis LSI MAPT 17q21 SG (Abbott Molecular, Des Plaines, IL, USA) and the Vysis SpectrumGreen CEP 17 probe as control. We performed control experiments to determine the false positive rate with FISH analysis on metaphase spreads and interphase nuclei with both the locus-specific identifier probe as well as a centromere probe. Slides were prepared from three control individuals and two observers examined the slides individually. From each control, at least 100 metaphases and at least 200 interphase nuclei were scored. Vysis LSI probes 22q11.2 and 22q13.3 were tested together with LSI MAPT 17q21.31 in combination with the centromere probe for chromosome 17.
Genotyping for H1 and H2, and parent-of-origin analysis
The presence of an intronic 238-bp deletion in intron 9 of the MAPT gene, characteristic for the H2 background, was used to genotype individuals with the 17q21.31 deletion and the accompanying parental DNA samples as described previously.9 The dinucleotide marker, DG17S142 in intron 9 of the MAPT gene, was used for independent validation of the H1/H2 genotyping. Two additional variable number tandem repeats (VNTR) inside the deletion interval (D17S27TG and D17S22TG), and one flanking VNTR (D17S26TG) and short tandem repeat (D17S810), were used to study the parental origin of the deletions as described before.1
Results
DNA- and cytogenetic analysis
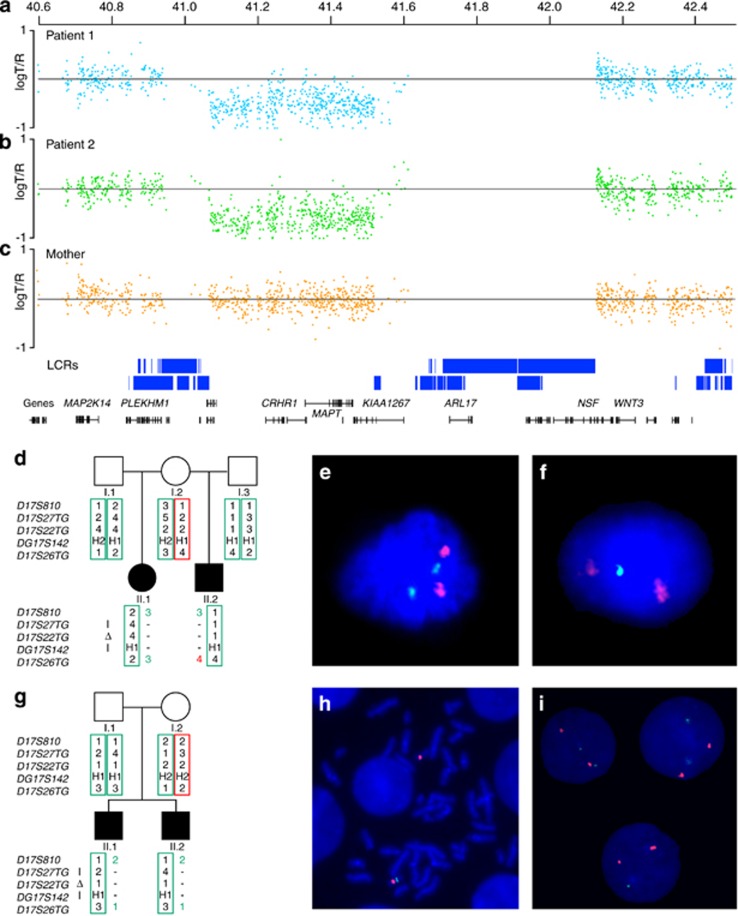
We report the recurrence of a 17q21.31 deletion, in two siblings, in two families. In the first family, the facial gestalt of patients 1 and 2 were consistent with a clinical diagnosis of the 17q21.31 microdeletion syndrome (Figures 1a and b), which was confirmed by Affymetrix Cytogenetics Whole-Genome 2.7 M SNP microarray analyses (Figures 2a and b). A 504-kb deletion (chr17: 41 062 760–41 567 167; hg18) was detected in patient 1; (Figure 2a) a 458-kb deletion (chr17: 41 062 760–41 520 813; hg18), followed by a 46-kb segment of normal ratio, and a 60-kb duplication (chr17: 41 572 115–41 635 816; hg18) were detected in patient 2 (Figure 2b). No additional disease causing CNVs were identified.
Figure 2.
Results of molecular and cytogenetic studies. (a–c) Panels (a, b) show the genomic microarray log T/R ratio plots of (a) patient 1, (b) patient 2, and (c) their mother. For patient 1, a 504-kb deletion (chr17: 41 062 760–41 567 167; hg18) was detected. For patient 2, this analysis revealed a 458-kb deletion (chr17: 41 062 760–41 520 813; hg18), followed by a 46-kb segment of normal ratio, and a 60-kb duplication (chr17: 41 572 115–41 635 816; hg18). Besides the 17q21.31 microdeletion, no other disease causing CNVs were identified. Microarray analysis of the mother, using DNA isolated from blood, did not show a CNV spanning the 17q21.31 microdeletion syndrome critical region. (d, g) Genotyping results. Circles represent female family members, squares male family members, open symbols unaffected family members, and the solid symbols the patients. The two columns of numbers next to each symbol indicate the alleles of each of the five markers on chromosome 17, in the order in which the markers are diagrammed below the pedigree. Marker segregation indicates that the mother is the parent-of-origin for the deletions observed in both families. (e, f) FISH testing of interphase spreads from buccal cells of the mother in family 1, using a FISH probe located within the critical region for the 17q21.31 microdeletion syndrome (green; Vysis LSI MAPT 17q21 SG FISH probe), and the control probe staining the chromosome 17 centromere (red; Vysis SpectrumGreen CEP 17 probe). Panel (e) shows two signals for both probes, whereas panel (f) shows only one green signal indicating monosomy 17q21.31. The percentage of interphase nuclei containing one MAPT probe was 8% (4/50). (h, i) FISH testing of peripheral blood cells from the mother of family 2. Panel (h) shows monosomy 17q21.31 in a metaphase spread and panel (i) shows the same in interphase nuclei. The percentage of metaphases/interphase nuclei in peripheral blood cells containing one MAPT probe was 3%.
In the first family, microarray analysis of the mother, using DNA isolated from peripheral blood, did not detect a CNV spanning the 17q21.31 microdeletion syndrome critical region (Figure 2c). Maternal array data were analysed subsequently for the presence of a mosaic deletion, but no such event was detected in peripheral lymphocytes. Consistent with these findings, FISH analyses from peripheral blood cells from the mother showed a normal signal pattern in metaphase (n=5) and in interphase nuclei (n=200, data not shown).
Haplotype analysis to determine the status for the H1 and H2 alleles showed that the mother is a H1/H2 heterozygote, the father of patient 1 an H1/H2 heterozygote, and the father of patient 2 homozygous for the H1 lineage. Segregation of these markers was consistent with a maternal origin of the 17q21.31 deletion on the H2 maternal haplotype in both patients (Figure 2d).
We hypothesised that maternal mosaicism for the 17q21.31 microdeletion could be present and so a second tissue type was examined. Interphase FISH, for 4 out of 50 nuclei (8%) from buccal mucosa, showed a single signal for the probe located in the 17q21.31 microdeletion syndrome critical region, consistent with maternal low-grade mosaicism for monosomy 17q21.31 (Figures 2e and f).
In the second family, the 17q21.31 microdeletion syndrome was not recognised, based on the clinical presentation. The diagnosis was made by Affymetrix GeneChip 250K SNP microarray analyses (data not shown). In patient 3, a 526-kb deletion (chr17: 41 049 321–41 575 332; hg18) was detected, and in patient 4 a 601-kb deletion (chr17: 41 119 024–41 719 833; hg18). Also, in these two brothers, no other clinical relevant CNVs were detected. Microarray analysis of both parents, using DNA isolated from peripheral blood, did not show a 17q21.31 microdeletion.
We also performed haplotype analysis in family 2. This showed that the mother is a H2 homozygote, and the father is an H1/H2 heterozygote. Moreover, the 17q21.31 microdeletion in patient 3 and 4 is of maternal origin, on the H2 maternal haplotype (Figure 2g).
Subsequently, peripheral blood cells from the mother and buccal mucosa were investigated for the 17q21.31 deletion using FISH. In peripheral blood cells we identified the 17q21.31 deletion in 2/60 metaphase spreads (3%, Figure 2h), in 3/100 interphase nuclei (3%, Figure 2i), and also in 7 out of 100 nuclei (7%) from buccal mucosa cells (data not shown).
FISH analysis of three negative control samples showed that the false positive thresholds for the LSI MAPT 17q21.31 probe is 1% for metaphase FISH and 2% for interphase FISH analysis (data not shown).
Discussion
This is the first report of sibling recurrence of the 17q21.31 microdeletion syndrome. To date, all individuals reported with the 17q21.31 microdeletion syndrome are simplex patients, resulting from de novo deletions. Most de novo chromosome rearrangements are sporadic and reliable data on the recurrence risks are not available. Röthlisberger and Kotzot10 reviewed the medical literature to determine such recurrence risks and found 29 patient reports of siblings with identical chromosomal aberrations, born to karyotypically normal parents, including 13 patients of trisomy 21 due to an i(21q). In eight of these, low-level mosaicism in one of the parents was found in the fibroblasts, blood, or in both. In their review, only six familial recurrences of an interstitial (micro) deletion resulting from NAHR are described, including Williams–Beuren syndrome, Prader–Willi/Angelman syndrome, and velocardiofacial syndrome. Interestingly, the transmitting parent of the patients with Williams–Beuren syndrome also showed a predisposing inversion.10
We describe the recurrence of the 17q21.31 microdeletion syndrome in two independent families. In family 1, the two affected children are born to the same mother, but different, unrelated fathers. We hypothesised that it was therefore likely that the mother was the parent-of-origin of the 17q21.31 deletion for both patients and this was confirmed by genotype analysis (Figure 2d). In family 2, we showed that the mother was also the parent-of-origin (Figure 2g).
We speculated that the recurrence of the 17q21.31 microdeletion in these two families could be due to one of four possible reasons: (i) chance recurrence; (ii) trans regulators of meiotic recombination hot spots; (iii) a balanced chromosomal rearrangement involving 17q21.31; or (iv) low-level maternal somatic–gonadal mosaicism.
The probability that the recurrence is based on chance alone cannot be excluded. The estimated frequency of the 17q21.31 microdeletion syndrome is ∼1 in 16 000 newborns.1 A distinct MAPT haplotype, H2, in one of the parents has been suggested to be a prerequisite to facilitate the microdeletion.5, 6, 7, 11 The H2 haplotype is detected in 20% of the independent chromosomes, in the Utah HapMap project sample of individuals of the Northern and Western European ancestry, and is linked to an inversion polymorphism of ∼900 kb.9 In family 1, the mother of the siblings is a H1/H2 heterozygote. On the basis of this information, the chance that the second deletion occurred by chance alone is 1/14 438 (for calculation see Supplementary File 1). In family 2, the mother is a H2 homozygote and the father H1/H2 heterozygote, resulting in a chance of 1/4812. If the inversion status of the parents of a child with the 17q21.31 microdeletion syndrome is unknown, the overall risk of a second child with the 17q21.31 microdeletion is 1/9461 (Supplementary File 1).
The 900-kb H2 inversion may be a necessary risk factor for the deletion to occur, but it is unlikely to be sufficient, given the relatively high frequency of the inversion in the European populations,9 compared with the lower frequency of the 17q21.31 microdeletion syndrome. Therefore, other genetic factors might increase the chance of NAHR. For instance, variation in the PR domain-containing 9 (PRDM9) was recently reported to strongly influence recombination hot-spot activity and meiotic instability in humans.12 Variation in this gene could therefore promote recombination errors such as needed in NAHR.
A third possibility is the involvement of a maternal balanced chromosomal rearrangement involving 17q21.31. Numerous examples have indicated the importance of fully unravelling the nature of rearrangements, as this may have major impact on the recurrence risk.13 The rearrangements mostly involve balanced translocations, but in addition, Carelle-Calmels et al14 reported on a complex scenario in which the father of a child with a 22q11.2 deletion carried a 22q11.2 deletion on one copy of chromosome 22 and a reciprocal 22q11.2 duplication on the other copy of chromosome 22, compensating for a genomic disorder. To our knowledge, parents with balanced rearrangements, only involving 17q21.31, have not yet been reported. In our families, extensive FISH analyses on peripheral blood cells from the parents-of-origin did not reveal the presence of such a balanced rearrangement.
We also hypothesised that the recurrence of the 17q21.31 deletion in the children was due to germline, or somatic–gonadal mosaicism in the parent-of-origin (the mother in both families). Germline mosaicism is well known for autosomal dominant and X-linked inherited disorders. In this study, further investigations to demonstrate gonadal mosaicism in the mothers were not feasible, and therefore it neither can be demonstrated, nor excluded. Initial investigations in the mother of family 1 failed to detect 17q21.31 somatic mosaicism in blood. A second cell type (buccal mucosa) was then tested for such mosaicism. Interestingly, this analysis showed one signal for the probe in the 17q21.31 microdeletion syndrome critical region in 4 out of 50 cells, consistent with low-grade mosaicism for monosomy 17q21.31. In family 2, FISH analyses showed 17q21.31 somatic mosaicism both in blood and buccal cells of the mother. In both mothers, the mosaic losses were significantly higher than the thresholds that were set at 1% for metaphase FISH and 2% for interphase FISH analysis. On the basis of these findings, and given the recurrence of this microdeletion in both families, it would be expected that the maternal germline contains this deletion in mosaic form.
Concluding remarks
We conclude that low-level somatic–gonadal mosaicism in both mothers of these siblings predisposed to a recurrent microdeletion in both families. Although the recurrence risk for parents with a child with a 17q21.31 microdeletion for future pregnancies is low, these two families suggest that it is higher than that in the general population, that is, by chance alone. Testing for mosaicism may be offered to unaffected parents, who have had a child with the 17q21.31 microdeletion syndrome. In case of somatic mosaicism, prenatal testing should be discussed due to the recurrence risk associated with possible germline mosaicism. However, as the risk of fetal loss after amniocentesis or chorionic villous sampling is 0.5–1%,15 invasive prenatal investigation should only be performed after genetic counselling.
Acknowledgments
We are grateful to these two families, for agreeing to this publication. We acknowledge Rolph Pfundt, Leo Derksen, and Guillaume van de Zande for technical assistance and Tony Roscioli for critical reading of the manuscript. This work was funded in part by grants from the Netherlands Organisation for Health Research and Development (ZonMW grants 916-86-016 to LELMV and 917-86-319 to BBAdV), and the AnEUploidy project (LSHG-CT-2006-37627 to BBAdV).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Koolen DA, Sharp AJ, Hurst JA, et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet. 2008;45:710–720. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TY, Aftimos S, Worgan L, et al. Phenotypic expansion and further characterisation of the 17q21.31 microdeletion syndrome. J Med Genet. 2009;46:480–489. doi: 10.1136/jmg.2008.065391. [DOI] [PubMed] [Google Scholar]
- Sharkey FH, Morrison N, Murray R, et al. 17q21.31 microdeletion syndrome: further expanding the clinical phenotype. Cytogenet Genome Res. 2009;127:61–66. doi: 10.1159/000279260. [DOI] [PubMed] [Google Scholar]
- Dubourg C, Sanlaville D, Doco-Fenzy M, et al. Clinical and molecular characterization of 17q21.31 microdeletion syndrome in 14 French patients with mental retardation. Eur J Med Genet. 2011;54:144–151. doi: 10.1016/j.ejmg.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Vissers LE, Pfundt R, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C, Pittman AM, Willatt L, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Helgason A, Thorleifsson G, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- Rothlisberger B, Kotzot D. Recurrence risk in de novo structural chromosomal rearrangements. Am J Med Genet A. 2007;143A:1708–1714. doi: 10.1002/ajmg.a.31826. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genome structural variation and sporadic disease traits. Nat Genet. 2006;38:974–976. doi: 10.1038/ng0906-974. [DOI] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam KW, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel A.Catalogue of Unbalanced Chromosome Aberrations in Man2nd edn,New York: Walter de Gruyter; 2001 [Google Scholar]
- Carelle-Calmels N, Saugier-Veber P, Girard-Lemaire F, et al. Genetic compensation in a human genomic disorder. N Engl J Med. 2009;360:1211–1216. doi: 10.1056/NEJMoa0806544. [DOI] [PubMed] [Google Scholar]
- Tabor A, Vestergaard CH, Lidegaard O. Fetal loss rate after chorionic villus sampling and amniocentesis: an 11-year national registry study. Ultrasound Obstet Gynecol. 2009;34:19–24. doi: 10.1002/uog.6377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.