Abstract
Chromosomal band 1q21.1 can be divided into two distinct regions, proximal and distal, based on segmental duplications that mediate recurrent rearrangements. Microdeletions and microduplications of the distal region within 1q21.1, which are susceptibility factors for a variety of neurodevelopmental phenotypes, have been more extensively studied than proximal microdeletions and microduplications. Proximal microdeletions are known as a susceptibility factor for thrombocytopenia-absent radius (TAR) syndrome, but it is unclear if these proximal microdeletions have other phenotypic consequences. Therefore, to elucidate the clinical significance of rearrangements of the proximal 1q21.1 region, we evaluated the phenotypes in patients identified with 1q21.1 rearrangements after referral for clinical microarray testing. We report clinical information for 55 probands with copy number variations (CNVs) involving proximal 1q21.1: 22 microdeletions and 20 reciprocal microduplications limited to proximal 1q21.1 and 13 microdeletions that include both the proximal and distal regions. Six individuals with proximal microdeletions have TAR syndrome. Three individuals with proximal microdeletions and two individuals with larger microdeletions of proximal and distal 1q21.1 have a ‘partial' TAR phenotype. Furthermore, one subject with TAR syndrome has a smaller, atypical deletion, narrowing the critical deletion region for the syndrome. Otherwise, phenotypic features varied among individuals with these microdeletions and microduplications. The recurrent, proximal 1q21.1 microduplications are enriched in our population undergoing genetic testing compared with control populations. Therefore, CNVs in proximal 1q21.1 can be a contributing factor for the development of abnormal phenotypes in some carriers.
Keywords: 1q21.1, microdeletion, microduplication, TAR, segmental duplication
Introduction
Chromosomal region 1q21.1 is structurally complex with many segmental duplications (SDs) that make it prone to non-allelic homologous recombination (NAHR).1 The 1q21.1 region contains multiple sequence gaps based on the UCSC March 2006 build (hg18); of the ∼5.6 Mb of sequence comprising 1q21.1, only 25% is composed of unique sequence.2 Band 1q21.1 can be divided into two distinct regions based on SD-mediated recurrent microdeletions/microduplications: a proximal and a distal region (Figure 1). The proximal region extends from ∼144.1 to ∼144.5 Mb from the 1p telomere, contains 16 genes in its unique sequence, and is flanked by SDs that mediate recurrent breakpoints (BPs) BP2 and BP3. The distal region is located at ∼145 to ∼146.35 Mb, contains 13 genes, and is flanked by SDs that mediate BP3 and BP4.2
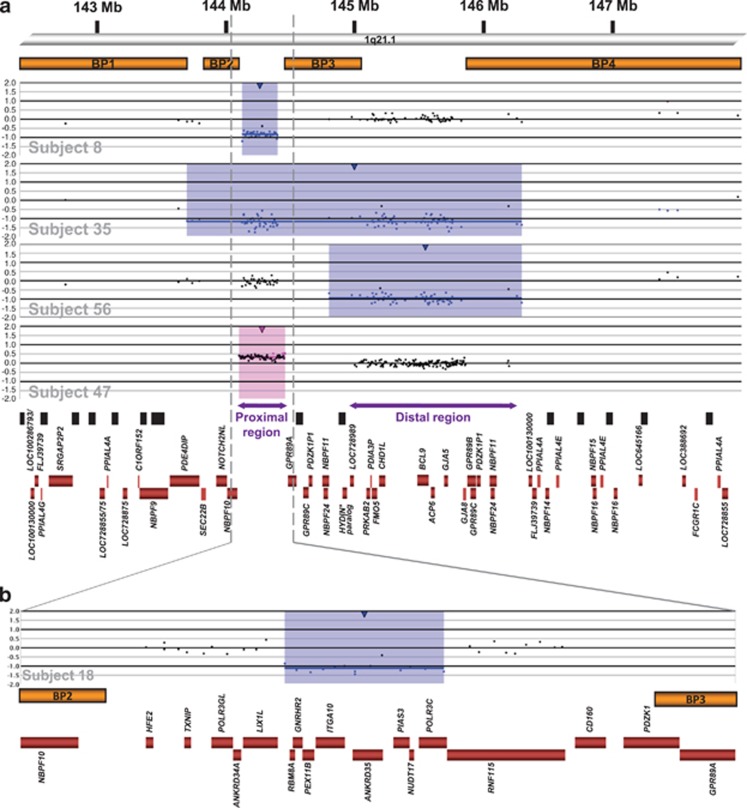
Figure 1.
(a) Genomic architecture of 1q21.1 and representative aCGH results showing microdeletions and microduplications. At the top of the figure is a partial ideogram showing chromosome band 1q21.1 with genomic coordinates corresponding to the hg18 build of the human genome. Orange bars represent clusters of SD blocks BP1, BP2, BP3, and BP42 that contain the BPs of the recurrent rearrangements. Plots show the representative recurrent 1q21.1 microdeletions (blue shading) and microduplications (pink shading) found in subjects reported here. Values along the y axis represent log2 ratios of patient:control signal intensities. The first three plots showing deletions are from a 105K-feature oligonucleotide microarray (SignatureChipOS, version 1), and the plot showing the duplication is from a 135K-feature oligonucleotide microarray (SignatureChipOS, version 2). Black boxes represent sequence gaps in the hg18 human genome build, and red boxes represent all known genes annotated in the Reference Sequence database in 1q21.1, except genes between BP2 and BP3. *The HYDIN paralog is localized within the distal sequence gap in BP3.6 (b) Zoomed-in view of the BP2–BP3 interval and aCGH results from subject 18, who had an atypical, small deletion within the proximal 1q21.1 region and clinically diagnosed TAR syndrome (chr1:144 215 938–144 320 529). His deletion minimally extends from RBM8A to POLR3C and may extend proximally into LIX1L and distally into RNF115. Red boxes represent all known genes annotated in the Reference Sequence database in the region.
Current literature demonstrates the phenotypic variability associated with copy number variations (CNVs) in chromosome band 1q21.1. Microdeletions and microduplications of the distal 1q21.1 region have been linked with a variety of morbidities, including intellectual disability (ID),2, 3, 4, 5, 6 autism,2, 6, 7, 8, 9 schizophrenia,10, 11, 12, 13, 14, 15, 16 microcephaly/macrocephaly,2, 6 congenital heart defects,17, 18, 19, 20 and renal and urinary tract anomalies21 (Supplementary Table 1). Proximal 1q21.1 microdeletions are hypothesized to be a prerequisite for thrombocytopenia-absent radius (TAR) syndrome,22 but otherwise proximal microdeletions and microduplications have been rarely reported, in cohorts of individuals with Mayer–Rokitansky–Küster–Hauser syndrome,23 congenital heart defects,24 and autism25 (Supplementary Table 1). Both proximal and distal CNVs of 1q21.1 are reported in phenotypically affected individuals and apparently normal carriers.2, 6, 22 Although microdeletions and microduplications of distal 1q21.1 have been shown to be enriched in populations with abnormal phenotypes,2, 13, 17 some conflicting data exists in case-control comparisons for proximal 1q21.1 CNVs,26, 27 and it remains unclear if these proximal CNVs have phenotypic consequences other than TAR syndrome. Therefore, to clarify the clinical significance of rearrangements of the proximal 1q21.1 region, we performed molecular and clinical characterization of 55 probands with microdeletions or microduplications encompassing the proximal 1q21.1 region in a patient population undergoing clinical microarray-based comparative genomic hybridization (aCGH) testing and compared the frequencies of these rearrangements with reported control groups.
Materials and methods
Subject ascertainment
Individuals with 1q21.1 CNVs reported here were identified after referral for molecular cytogenetic testing to clinical laboratories, including Signature Genomics, University of Nebraska, Nationwide Children's Hospital, Emory Genetics Laboratory, GeneDx, Hospital for Sick Children, and University of North Carolina. Common indications for study included developmental delay (DD), ID, dysmorphic features, and congenital anomalies. To compare the phenotypes resulting from rearrangements of the proximal and/or distal regions of 1q21.1, we included 18 subjects with deletions limited to distal 1q21.1, in addition to 55 subjects with proximal 1q21.1 rearrangements, in the clinical analysis. Informed consent was provided for the publication of photographs.
Molecular cytogenetic testing
Microdeletions and microduplications were identified at Signature Genomics using bacterial artificial chromosome (BAC)-based microarrays or oligonucleotide-based microarrays. Versions 1–4 of the BAC-based SignatureChip (Signature Genomics, Spokane, WA, USA) only covered proximal 1q21.1, whereas all other BAC-based and oligo-based arrays covered the whole genome and, therefore, covered proximal and distal 1q21.1. Available samples tested on versions 1–4 of the BAC arrays and found to have proximal 1q21.1 rearrangements were reanalyzed using arrays that covered the proximal and distal regions on 1q21.1. Reanalysis was performed with either a BAC-based array (SignatureChipWG, Signature Genomics) or an oligo-based array (SignatureChipOS versions 1 (105K-feature, Agilent Technologies, Santa Clara, CA, USA) or 2 (135K-feature, Roche NimbleGen, Madison, WI, USA), both custom designed by Signature Genomics), according to previously described methods.28, 29, 30, 31 Some samples were analyzed at other laboratories either by BAC array (SignatureSelect v1.0 (Signature Genomics; subject 13) or Spectral Chip 2600 (PerkinElmer, Waltham, MA, USA; subject 67)), oligonucleotide array (Agilent 44K-feature custom array (Agilent Technologies; subjects 10, 28–31, 33, 49, and 68–69),32 44K-feature EmArray Cyto6000 (Emory Genetics Laboratory, Decatur, Georgia; subject 9), or 105K-feature GenomeDx (GeneDx, Gaithersburg, MD, USA), subjects 27, 34)), or single nucleotide polymorphism (SNP) array (Genome-wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA; subject 12)).
Fluorescence in situ hybridization (FISH)
All abnormal microarray results obtained at Signature Genomics were visualized by metaphase or interphase FISH using one or more BAC clones located within the abnormal regions.33, 34
Results
We identified 55 probands with abnormalities involving the proximal region of 1q21.1 (Figure 2). Of this group, 22 subjects (#1–22) had the smaller proximal microdeletion flanked by BP2 and BP3, and 20 subjects (#36–55) had the reciprocal microduplication. The remaining 13 subjects (#23–35) had a larger microdeletion flanked by BP1 or BP2 and BP4 involving the proximal and distal regions (Figure 1). Also, clinical information was available for 18 subjects (#56–73) with distal microdeletions flanked by BP3 and BP4 (Supplementary Tables 3 and 4). Subject 19 had a complex rearrangement with a distal, BP3–BP4 duplication as well as the proximal deletion. Subject 53 had a complex rearrangement with a proximal, BP2–BP3 duplication and a duplication of the distal portion of the distal region, with normal copy number intervening sequence. Subjects 20–22, 35, 54–55, and 71–73 had additional clinically significant abnormalities found by aCGH and were excluded from phenotypic analysis (Supplementary Table 4). After these exclusions, clinical information for the remaining 49 probands with abnormalities involving proximal 1q21.1 is presented in Supplementary Table 2, and comparisons of clinical features among individuals with 1q21.1 deletions and duplications are shown in Tables 1 and 2, respectively.
Figure 2.
(a, b) Subject 6 (with proximal microdeletion) at 20 months. Note slight epicanthal folds, mild left ptosis, broad nasal root with round tip, and normal ears. (c–e) Subject 13 (with proximal microdeletion and clinically diagnosed TAR syndrome) at 17 months. Note shortened forearms and adducted thumb. (f) Radiograph of subject 13, who had radial aplasia, hooking clavicles, shortened ulna, and normal hands and humeri. (g, h) Subject 27 (with deletion of proximal and distal 1q21.1) at 17 months. Note brachycephaly, full cheeks, flat philtrum, and posteriorly rotated ears. (i) Subject 33 (with deletion of proximal and distal 1q21.1) at 8 months. Note radial hypoplasia. (j) Subject 40 (with proximal microduplication) at 4 months. Note square forehead, prominence of the eyes, low nasal bridge, and flame nevus. (k) Subject 41 (with proximal microduplication) at 6 years. Note strabismus, broad nasal bridge, prominent ears, and myopathic facies with open mouth. (l, m) Subject 47 (with proximal microduplication) at 8 years. Note epicanthal folds, cleft lip repair, toenail dystrophy, and wide feet. (n) Subject 70 (with distal microdeletion) at 3.5 years. Note slight midfacial flattening and triangular nose.
Table 1. Frequencies of phenotypic features in probands with 1q21.1 microdeletions.
| Feature | Proximal (BP2–BP3) deletions without TAR syndromea | Large (BP2–BP4) deletionsb | Distal (BP3–BP4) deletionsc |
|---|---|---|---|
| Short stature | 3/10 | 7/17 | 17/47 |
| Failure to thrive/feeding problems | 6/11 | 13/16 | 21/48d |
| Microcephaly | 3/11 | 13/16 | 28/50 |
| Macrocephaly | 2/11 | 0/16 | 0/50 |
| Developmental delay/intellectual disability | 11/12 | 18/22e,f | 45/70d,e,f,g,h |
| Hypotonia | 1/12 | 2/22 | 11/59h |
| Seizures | 4/12 | 2/23f | 9/60g,h |
| Autistic features | 4/13i | 0/22 | 6/57f,h,j |
| Other behavioral problems | 4/12 | 1/22 | 15/58h |
| Hearing loss | 2/12 | 4/22 | 3/58 |
| Brain abnormalities | 4/9 | 2/4 | 6/11 |
| Dysmorphic features | 8/12 | 14/17 | 43/57 |
| Cataracts | 1/12 | 0/22 | 2/58 |
| Other ophthalmologic abnormalities | 4/12 | 4/22 | 11/58 |
| Craniosynostosis | 0/12 | 1/22 | 1/58 |
| Skeletal limb abnormalities | 1/12 | 2/22 | 3/58 |
| Polydactyly | 0/12 | 1/22 | 8/58 |
| Other skeletal anomalies | 3/12 | 1/22 | 2/58 |
| Ligamentous laxity | 0/12 | 2/22 | 5/58 |
| Cardiac anomalies | 2/12 | 5/22 | 15/58 |
| Lung abnormalities | 0/12 | 2/22 | 0/58 |
| Renal anomalies | 1/12 | 5/22 | 4/58 |
| Genital anomalies | 1/12 | 3/22 | 1/58 |
| Blood disorders | 2/12 | 0/22 | 0/58 |
Subjects 1–12 from this report, who do not meet clinical criteria for TAR syndrome.
Also includes a previously reported proband in a schizophrenia cohort.15
Also includes previously reported probands in a schizophrenia cohort.11
Also includes previously reported probands in a schizophrenia cohort.12
Also includes a previously reported proband in a schizophrenia cohort.10
Also includes a previously reported proband in an epilepsy cohort.53
Also includes a previously reported proband in an autism cohort.25
Also includes a previously reported proband in an autism cohort.8
Table 2. Frequencies of phenotypic features in probands with 1q21.1 microduplications.
| Feature | Proximal (BP2–BP3) duplicationsa | Large (BP2–BP4) duplicationsb | Distal (BP3–BP4) duplicationsc |
|---|---|---|---|
| Short stature | 3/13 | NS | 2/21 |
| Failure to thrive/feeding problems | 5/10 | NS | 4/20 |
| Microcephaly | 2/12 | NS | 2/23 |
| Macrocephaly | 1/12 | NS | 10/23 |
| Developmental delay/intellectual disability | 12/16 | 0/2 | 21/27 |
| Hypotonia | 5/16 | 0/2 | 4/27 |
| Seizures | 2/16 | 0/2 | 5/28 |
| Autistic features | 4/14 | 0/2 | 14/34d |
| Other behavioral problems | 6/14 | 0/2 | 1/25 |
| Hearing loss | 1/16 | 0/2 | 0/27 |
| Brain abnormalities | 5/7 | NS | 3/4 |
| Dysmorphic features | 9/16 | 0/2 | 14/27 |
| Cataracts | 0/16 | 0/2 | 0/27 |
| Other ophthalmologic abnormalities | 5/16 | 0/2 | 4/27 |
| Craniosynostosis | 1/16 | 0/2 | 0/27 |
| Skeletal limb abnormalities | 4/17 | 0/2 | 0/27 |
| Other skeletal anomalies | 3/17 | 0/2 | 2/27 |
| Clinodactyly | 6/17 | 0/2 | 1/27 |
| Ligamentous laxity | 1/16 | 0/2 | 1/27 |
| Cardiac anomalies | 3/17 | 1/2 | 8/28e |
| Lung abnormalities | 2/17 | 0/2 | 0/27 |
| Renal anomalies | 1/17 | 0/2 | 1/27 |
| Genital anomalies | 3/17 | 1/2 | 3/27 |
| Blood disorders | 0/16 | 0/2 | 0/27 |
Four of the probands with deletions limited to the proximal 1q21.1 region had clinically diagnosed TAR syndrome, and two additional prenatal cases had skeletal findings consistent with TAR syndrome. One of these probands, subject 18, had an atypical partial deletion of the proximal 1q21.1 region (chr1:144 215 938–144 320 529, UCSC hg18 coordinates; Figure 1). The phenotypic features of TAR syndrome and other individuals with proximal 1q21.1 microdeletions not meeting full clinical criteria for TAR syndrome are summarized in Table 3.
Table 3. Comparison of features in TAR syndrome and probands reported with proximal 1q21.1 deletions.
| Feature | TAR syndrome (%) | TAR syndrome and proximal 1q21.1 deletionsa | Proximal 1q21.1 deletions without TAR syndromeb |
|---|---|---|---|
| Bilateral radial aplasia | 99c,d | 35/35 | 0/12 |
| Thrombocytopenia | 98c,d | 32/32 | 1/12 |
| Cow milk intolerance/feeding problems | 47c | 10/26 | 6/11 |
| Ulnar abnormalities | 74d | 28/29 | 1/12 |
| Humerus abnormalities | 41d | 19/29 | 0/12 |
| Hand abnormalities | 85d | 24/28 | 1/12 |
| Lower limb abnormalities | 46c,d | 17/28 | 0/12 |
| Foot abnormalities | 40e | 6/28 | 0/12 |
| Spine/rib abnormalities | 7c,e | 1/28 | 3/12 |
| Cardiac anomalies | 19c,d | 5/27 | 2/12 |
| Renal anomalies | 23c | 4/26 | 1/12 |
| Uterine or genital anomalies | 5c,e | 4/26 | 1/12 |
| Cleft palate | 1c,d | 2/27 | 0/12 |
| Micrognathia | 14d | 1/2 | 1/12 |
| Hemangioma | 24c | 2/26 | 0/12 |
| Dysmorphic features | NA | 2/26 | 8/12 |
| Developmental delay/intellectual disability | 2c,e | 1/28 | 11/12 |
| Behavior problems | NA | 0/2 | 7/13f |
| Seizures | 6c | 2/26 | 4/12 |
| Brain abnormalities | NA | 1/3 | 4/9 |
| Hearing loss | 3c | 2/26 | 2/12 |
| Ophthalmologic abnormalities | NA | 0/2 | 4/12 |
Abbreviation: NA, not available.
Previously reported probands with TAR syndrome and deletions limited to the proximal region of 1q21.1,22, 23, 56 and subjects 13–18 in this report, who had clinical diagnoses or prenatally suspected diagnosis of TAR syndrome.
Subjects 1–12 from this report, who do not meet clinical criteria for TAR syndrome.
Based on review by Hedberg et al.36
Based on cohort published by Greenhalgh et al.55
Based on review by Hall et al.35
Also includes individual reported by Shen et al.25
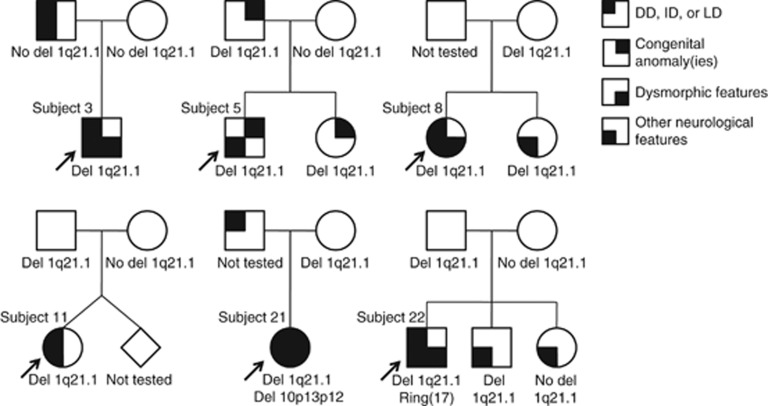
Among the 55 probands with proximal 1q21.1 abnormalities, inheritance was known for 22 deletions and 7 duplications. Microdeletions flanked by BP2 and BP3 were apparently de novo in 6 subjects and inherited in 11 subjects, including 1 from a mildly affected father and at least 4 from apparently normal mothers. Pedigrees of selected families with proximal deletions are shown in Figure 3. Microdeletions flanked by BP1/2 and BP4 were apparently de novo in four subjects and inherited in one, from a mildly affected father. Microduplications flanked by BP2 and BP3 were inherited in seven subjects, including one from a mother with mild learning delays and short stature, one from a father with learning disabilities and cleft lip, and at least one from an apparently normal parent.
Figure 3.
Pedigrees of families with individuals carrying the proximal, BP2–BP3 1q21.1 microdeletion. Individuals shown to have ‘other neurological features' had seizures, behavioral abnormalities, abnormal head size, and/or other brain abnormalities detected by imaging. LD, learning disability.
To enable a frequency comparison of individuals with proximal 1q21.1 abnormalities in ‘normal' and ‘abnormal' populations, data from 45744 probands undergoing aCGH testing at Signature Genomics were analyzed. Analysis for distal 1q21.1 was limited to 30215 probands tested on arrays with coverage of the distal 1q21.1 region (Table 4). The frequency of microduplications limited to the proximal, BP2–BP3 segment of 1q21.1 in our patient population who underwent aCGH testing compared with a sum of control individuals reported in the literature (Supplementary Table 1) showed an enrichment in our patient population (72/45744 vs 9/11057; one-tailed P=0.033, Fisher's exact test). The microdeletions limited to the proximal, BP2–BP3 region in our cases did not show a significant difference in frequencies compared with controls (26/45744 vs 2/11757; one-tailed P=0.055, Fisher's exact test).
Table 4. Inheritance of 1q21.1 abnormalities detected by Signature Genomics.
| BP2–BP3 deletions | BP2–BP4 deletions | BP3–BP4 deletions | BP2–BP3 duplications | BP2–BP4 duplications | BP3–BP4 duplications | |
|---|---|---|---|---|---|---|
| Frequency (%) | 26/45744 (0.057)a,b | 11/45744 (0.024) | 86/30215 (0.28)c | 72/45744 (0.16)d,e | 12/45744 (0.026)d | 57/30215 (0.19)b |
| de novo | 5 | 2 | 6 | 0 | 2 | 6 |
| Maternally inherited | 9 | 0 | 15 | 6 | 1 | 17 |
| Paternally inherited | 5 | 1 | 12 | 6 | 1 | 8 |
| Unknown (parents not tested) | 7 | 8 | 53 | 60 | 8 | 26 |
Five of these individuals had clinical diagnoses of TAR syndrome or demonstrated the skeletal manifestations of TAR syndrome prenatally.
Includes one individual with a complex, proximal deletion/distal duplication.
Includes two individuals with a complex, proximal duplication/distal deletion.
Two individuals with proximal duplications and unknown involvement of the distal region are not included in either group.
Includes six individuals with a complex, proximal duplication/distal deletion, and two individuals with a proximal duplication and a duplication of a distal portion of the BP3–BP4 region.
Discussion
Individuals reported here with 1q21.1 microdeletions and microduplications show variability in phenotypes. The features present in a majority or near majority of individuals with microdeletions or microduplications limited to proximal, BP2–BP3 1q21.1 were a history of failure to thrive or feeding problems, DD/ID, dysmorphic features, and behavior problems. Also, clinodactyly was common among individuals with proximal microduplications (Tables 1 and 2). Some of these trends may reflect an ascertainment bias based on common indications for study among individuals undergoing aCGH testing. To determine if these proximal microdeletions and microduplications are benign or a susceptibility locus for the development of an abnormal phenotype, we compared the frequencies of the proximal microdeletion and microduplication in our patient population with published control populations. Proximal BP2–BP3 microduplications were enriched among individuals referred for aCGH testing, suggesting that this microduplication can contribute to abnormal phenotypes in some carriers. Lack of enrichment for the proximal BP2–BP3 microdeletion cannot support a similar conclusion for the reciprocal microdeletion. This conclusion opposes that reached in another recent study showing significant enrichment of proximal 1q21.1 microdeletions and lack of enrichment for the reciprocal microduplication among a population of individuals undergoing clinical aCGH testing.27 This is likely because of slight differences in control frequencies for proximal microdeletions. Furthermore, proximal microduplications had a significantly lower frequency in that patient population than in ours; this may be because of underreporting by some laboratories, which considered these microduplications to be likely benign (Dr Christa Lese Martin, personal communication). Given the rarity of the proximal microdeletions, larger control populations may be required to reach definite conclusions about the pathogenicity of these deletions. However, previous studies show this microdeletion to be significantly enriched in cases of TAR syndrome,22 so this microdeletion at least contributes to the development of that syndrome.
Subject 18, who has TAR syndrome, had an atypical, smaller deletion within the BP2–BP3 1q21.1 segment, which may allow refinement of a critical 1q21.1 region for the syndrome. Previously proposed TAR candidate genes PIAS3, a regulator of hematopoietic growth factor signaling, and LIX1L, which shares homology with a gene involved in limb development,22 are at least partially within this subject's deletion region (Figure 1). Of note, 3 subjects not meeting the clinical criteria for TAR syndrome but with proximal 1q21.1 microdeletions presented with features related to the hallmark features of the syndrome: subject 9 had radioulnar synostosis, subject 7 had a history of autoimmune neutropenia, and subject 22, who also carried a mosaic 1.4-Mb supernumerary ring chromosome 17, had chronic thrombocytopenia. Also, subject 33, who had a larger BP1/2–BP4 microdeletion, had bilateral radial hypoplasia (Figure 2), and subject 25, who had a large deletion, had dysplasia of the radial head and lateral humeral epicondyle. Other features occasionally associated with TAR syndrome were also seen in our cohort, including cow milk intolerance and/or feeding problems, spinal abnormalities, heart defects, renal and genital anomalies, seizures, and hearing loss (Table 3). Some differences between phenotypic incidences in TAR syndrome and individuals with proximal microdeletions not meeting TAR syndrome criteria, including dysmorphic features, ID/DD, seizures, and behavioral problems, may reflect our ascertainment bias of individuals undergoing genetic testing. Cardiac and renal anomalies occurred in similar frequencies among all individuals with proximal microdeletions, which may be more representative of the incidence of these anomalies with this CNV. These results suggest that TAR syndrome may be at the severe end of a spectrum of phenotypic abnormalities caused by proximal 1q21.1 microdeletions, and some individuals may only develop part of the syndrome's phenotype. This is consistent with the previously proposed model for TAR syndrome, as genetic changes at one or more other, unidentified modifier loci (mTAR) likely determine the ultimate phenotypic expression in these individuals.22
Early reports on individuals with TAR syndrome noted a 7% incidence of significant DD. The original presumptions were that the delays were caused by micro-infarcts secondary to the thrombocytopenia.35, 36 Several lines of evidence, however, have suggested otherwise. First, patients with other causes of thrombocytopenia did not demonstrate the same level of delays. Second, MRI data has suggested an etiology that is primarily dysgenesis, not injury.37 The understanding of the nature of the 1q21.1 deletions now offers a better explanation. Similar to individuals with TAR syndrome, the brain anomalies in some individuals in this cohort with proximal 1q21.1 microdeletions, including corpus callosum thinning and cysts identified by MRI (Supplementary Table 2), were not suggestive of injury. The proximal 1q21.1 region likely contains genes that, when deleted, can contribute to TAR syndrome in addition to genes that impact cerebral development, which would explain why some individuals with TAR syndrome and some with proximal 1q21.1 microdeletions not meeting clinical criteria for TAR syndrome show DD.
Like the individuals with the proximal microdeletions, individuals with the reciprocal BP2–BP3 microduplication had variable phenotypic features, with the most common features – ID/DD, dysmorphic features, and behavioral problems (Table 2) – possibly because of an ascertainment bias. Additionally, individuals in our cohort had other environmental and genetic factors that could contribute to the abnormal phenotypes, including documented prenatal exposures, poor social situations, and consanguinity. Despite the presence of these complicating factors, enrichment of the microduplication in our patient population suggests that the proximal, BP2–BP3 1q21.1 microduplication has a causative role in abnormal phenotypes.
Some rarer, specific phenotypes were seen in several individuals with 1q21.1 abnormalities. These may represent a rare causative association with these features, or the association may be owing to chance. Congenital cystic adenomatoid malformation (CCAM) type II of the lung has been reported in two individuals with large, BP1/2-BP4 deletions (subject 32 here and patient 17 in Brunetti-Pierri et al4). As CCAM is likely because of the focal arrest in lung maturation during the fetal period,38 candidate genes for this malformation include genes with roles in developmental pathways, including PIAS3 and RBM8A, which regulate STAT3,39, 40 involved in lung branching morphogenesis;41 BCL9, which is involved in the β-catenin/WNT pathway,42 also active in the lung development;43 and PEX11B, which encodes a peroxisome biogenesis factor hypothesized to be important in developing airways and during alveolarization.44 There have been multiple reports of polydactyly associated with the distal, BP3–BP4 microdeletion (Table 1). A candidate gene for this malformation is BCL9 (homolog of Drosophila gene legless), which is essential in vertebrate body axis determination.45 Finally, it has been hypothesized that the head size variation observed with distal 1q21.1 rearrangements is associated with copy number changes of genes within the SDs in the region, either the HYDIN paralog within BP36 or the NPBF family genes within all of the BPs (Figure 1).46 If micro- and macrocephaly are associated with copy number alteration of genes within the SDs, then proximal rearrangements could also alter the copy number of these genes. However, head sizes are not significantly different between individuals with microdeletions and microduplications limited to BP2–BP3 (P=0.85, unpaired t-test; mean z-score −0.64 and 95% confidence interval −2.72 to 1.43 for microdeletions, and mean z-score −0.45 and 95% confidence interval −1.74 to 0.84 for microduplications; Tables 1 and 2; Supplementary Table 2). This lack of consistent head size abnormalities with proximal CNVs could be attributed to the involvement of different genes within the SDs in proximal and distal rearrangements, or additional genes within the BP3–BP4 region may contribute to alterations in head size. However, our sample sizes are still small and many factors, including parental head size, contribute to patient head size, so data from more individuals and fine mapping of the common BPs within the SDs will help clarify these conclusions.
Despite the lack of enrichment for the proximal microdeletions in our patient population, some evidence is present for increased selective pressure against the proximal microdeletion compared with the reciprocal microduplication. In our patient population, de novo microdeletions have been observed, whereas de novo microduplications have not (Table 4); this indicates a greater proportion of parents with the microduplications are fit enough to reproduce. Also, proximal microdeletions are significantly less frequent than the reciprocal microduplications in our population of individuals undergoing aCGH testing (two-tailed P=3.6 × 10−6, Fisher's exact test) and control populations (two-tailed P=0.034, Fisher's exact test), which may indicate increased selective pressure against the proximal microdeletions, especially given that it has been predicted that, absent selection or other factors, NAHR-mediated deletions should be more common than duplications.47 This is similar to the effect seen to a greater extent with small, reciprocal, NAHR-mediated CHRNA7 deletions and duplications in 15q13.3; presumably benign duplications are much more common than the harmful deletions (55/8800 vs 3/8800 in one population).48, 49 Although the relative frequencies of proximal 1q21.1 microdeletions and microduplications fit a pattern associated with more significant phenotypic consequences with microdeletions, we have also established that proximal 1q21.1 microduplications contribute to abnormal phenotypes. The reason that there is increased selective pressure against the microdeletion may be solely because of selection against microdeletion carriers with TAR syndrome.
We show that the recurrent microdeletion of the proximal 1q21.1 region, which had previously only been significantly associated with TAR syndrome,22 may also cause a partial TAR syndrome phenotype in some carriers. The reciprocal duplications are enriched in our patient population undergoing clinical aCGH testing, which indicates that these duplications can also contribute to abnormal phenotypes. Clinically normal individuals may also carry these 1q21.1 rearrangements; abnormal phenotypes segregate with the microdeletion in some but not all families, and the same CNV may contribute to different phenotypes within the same family (Figure 3). Because of the phenotypic variability and lack of consistent dysmorphic features, with the exception of individuals with TAR syndrome, proximal 1q21.1 microdeletions and microduplications are unlikely to be recognized clinically and diagnosis will likely continue to follow molecular cytogenetic testing. Similar to what is observed with TAR syndrome, these proximal 1q21.1 microdeletions and microduplications are likely one factor in the development of abnormal phenotypes, and additional unidentified factors, both genetic and environmental, will determine what, if any, phenotypic manifestations will be present in an individual.50
Acknowledgments
We thank Aaron Theisen (Signature Genomics, Spokane, WA, USA) for his critical editing of this manuscript and Evan Eichler, Bradley Coe, and Gregory Cooper (University of Washington, Seattle, WA, USA) for sharing the control data.
Appendix
1q21.1 Study Group Contributors
The 1q21.1 Study Group includes the following: Dianne Abuelo6, Ilse Anderson7, Brad Angle8, Holly Ardinger9, Alexander Asamoah10, Joan F Atkin11, Judith Axelrod10, Patricia Bader12, Carrie Blout11, Campbell Brasington13, Lauren C Briere14, Pamela L Brock10, Barbara Burton8, David Chitayat15, Lisa J Cushman16, Dawn L Earl17, Dima El-Khechen18, Luis F Escobar18, Aline Hamati16, David J Harris19, Gail Herman11, Jacqueline Hoover20, Kelly E Jackson10, Kathleen A Kaiser-Rogers5, Lawrence C Kaplan21, Abigail Klemsz16, Yves Lacassie22, Roger Ladda23, Allen N Lamb1, Molly M Lund9, Helen Lyon19, Gregory P MacDonald24, Suneeta Madan-Khetarpal20, Michael Marble22, Paul R Mark16, Laura S Martin25, Nicole Martin15, Juliann S McConnell20, Elizabeth McCracken20, Marie McDonald26, Marianne McGuire20, Roberto Mendoza-Londono15, Amanda N Miller16, John B Moeschler21, Amanda G Noyes7, Kathryn Platky10, Cynthia M Powell5, Abbey Putnam7, Victoria Roberts8, Warren G Sanger27, Roger A Schultz1, Susan Sell23, Yasmin Senturias10, Vandana Shashi28, Natasha Shur6, Komudi Siriwardena15, Annemarie Sommer11, J Edward Spence13, James Stavropoulos15, Cathy A Stevens29, Meghan E Strenk9, Mark Tarnopolsky30, Ellen Thomas31, Matthew J Thomas32, Beth S Torchia1, Amy Venter33, David D Weaver16, William G Wilson32
6Rhode Island Hospital/Hasbro Children′s Hospital, Providence, RI, USA; 7University of Tennessee Genetics Center, Knoxville, TN, USA; 8Children's Memorial Hospital, Chicago, IL, USA; 9Children′s Mercy Hospitals and Clinics, Kansas City, MO, USA; 10Department of Pediatrics, Weisskopf Child Evaluation Center, University of Louisville, Louisville, KY, USA; 11Nationwide Children's Hospital, Columbus, OH, USA; 12Northeast Indiana Genetic Counseling Center, Ft. Wayne, IN, USA; 13Carolinas Medical Center, Charlotte, NC, USA; 14Beth Israel Deaconess Medical Center, Boston, MA, USA; 15Mount Sinai Hospital, The Prenatal Diagnosis and Medical Genetics Program and The Hospital for Sick Children, Division Of Clinical and Metabolic Genetics, University of Toronto, Toronto, Ontario, Canada; 16Indiana University School of Medicine, Indianapolis, IN, USA; 17Seattle Children's Hospital, Seattle, WA, USA; 18St Vincent Hospital, Indianapolis, IN, USA; 19Children's Hospital Boston, Boston, MA, USA; 20Children's Hospital Pittsburgh, Pittsburgh, PA, USA; 21Dartmouth-Hitchcock Medical Center, Lebanon, NH, USA; 22Department of Pediatrics, LSUHSC, and Children′s Hospital, New Orleans, LA, USA; 23Hershey Medical Center, Hershey, PA, USA; 24Providence Sacred Heart Children′s Hospital, Spokane, WA, USA; 25Nemours Children′s Clinic, Jacksonville, FL, USA; 26Duke University Medical Center, Durham, NC, USA; 27University of Nebraska Medical Center, Omaha, NE, USA; 28Wake Forest University Baptist Medical Center, Winston-Salem, NC, USA; 29TC Thompson Children's Hospital, Chattanooga, TN, USA; 30McMaster University Medical Centre, Hamilton, Ontario, Canada; 31Southern Illinois University School of Medicine, Springfield, IL, USA; 32University of Virginia Health System, Department of Pediatrics, Division of Genetics, Charlottesville, VA, USA; 33The Children's Hospital, Aurora, CO, USA
Jill A Rosenfeld, Ryan N Traylor, Blake C Ballif, Lisa G Shaffer, Allen N Lamb, Roger A Schultz, and Beth S Torchia are employees of Signature Genomic Laboratories, PerkinElmer Inc. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Sharp AJ, Locke DP, McGrath SD, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- de Vries BB, Pfundt R, Leisink M, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillard S, Drunat S, Bendavid C, et al. Identification of gene copy number variations in patients with mental retardation using array-CGH: novel syndromes in a large French series. Eur J Med Genet. 2010;53:66–75. doi: 10.1016/j.ejmg.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Aleksic B, Kirov G, et al. Copy number variation in schizophrenia in the Japanese population. Biol Psychiatry. 2010;67:283–286. doi: 10.1016/j.biopsych.2009.08.034. [DOI] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium: Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Ge D, Weale ME, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Duan J, Oh S, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway SC, Pereira AC, Lin JC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan F, Larsen LA, Zhang L, et al. High frequency of submicroscopic genomic aberrations detected by tiling path array comparative genome hybridisation in patients with isolated congenital heart disease. J Med Genet. 2008;45:704–709. doi: 10.1136/jmg.2008.058776. [DOI] [PubMed] [Google Scholar]
- Christiansen J, Dyck JD, Elyas BG, et al. Chromosome 1q21.1 contiguous gene deletion is associated with congenital heart disease. Circ Res. 2004;94:1429–1435. doi: 10.1161/01.RES.0000130528.72330.5c. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S, Landwehr C, Renkert M, et al. Mapping candidate regions and genes for congenital anomalies of the kidneys and urinary tract (CAKUT) by array-based comparative genomic hybridization. Nephrol Dial Transplant. 2010;26:136–143. doi: 10.1093/ndt/gfq400. [DOI] [PubMed] [Google Scholar]
- Klopocki E, Schulze H, Strauss G, et al. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am J Hum Genet. 2007;80:232–240. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledig S, Schippert C, Strick R, Beckmann MW, Oppelt PG, Wieacker P. Recurrent aberrations identified by array-CGH in patients with Mayer-Rokitansky-Kuster-Hauser syndrome. Fertil Steril. 2010;95:1589–1594. doi: 10.1016/j.fertnstert.2010.07.1062. [DOI] [PubMed] [Google Scholar]
- Brunet A, Armengol L, Heine D, et al. BAC array CGH in patients with velocardiofacial syndrome-like features reveals genomic aberrations on chromosome region 1q21.1. BMC Med Genet. 2009;10:144. doi: 10.1186/1471-2350-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Dies KA, Holm IA, et al. Clinical genetic testing for patients with autsim spectrum disorders. Pediatrics. 2010;125:e727–e735. doi: 10.1542/peds.2009-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky EB, Kaul V, Paschall J, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Hornor SA, Jenkins E, et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Coppinger J, et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, McDonald-McGinn DM, et al. Identification of a previously unrecognized microdeletion syndrome of 16q11.2q12.2. Clin Genet. 2008;74:469–475. doi: 10.1111/j.1399-0004.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- Duker AL, Ballif BC, Bawle EV, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010;18:1196–1201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin EL, Lee JY, Blake DM, et al. Enhanced detection of clinically relevant genomic imbalances using a targeted plus whole genome oligonucleotide microarray. Genet Med. 2008;10:415–429. doi: 10.1097/GIM.0b013e318177015c. [DOI] [PubMed] [Google Scholar]
- Traylor RN, Fan Z, Hudson B, et al. Microdeletion of 6q16.1 encompassing EPHA7 in a child with mild neurological abnormalities and dysmorphic features: case report. Mol Cytogenet. 2009;2:17. doi: 10.1186/1755-8166-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, McCaskill C, Han JY, et al. Molecular characterization of de novo secondary trisomy 13. Am J Hum Genet. 1994;55:968–974. [PMC free article] [PubMed] [Google Scholar]
- Hall JG, Levin J, Kuhn JP, Ottenheimer EJ, van Berkum KA, McKusick VA. Thrombocytopenia with absent radius (TAR) Medicine (Baltimore) 1969;48:411–439. doi: 10.1097/00005792-196948060-00001. [DOI] [PubMed] [Google Scholar]
- Hedberg VA, Lipton JM. Thrombocytopenia with absent radii. A review of 100 cases. Am J Pediatr Hematol Oncol. 1988;10:51–64. doi: 10.1097/00043426-198821000-00010. [DOI] [PubMed] [Google Scholar]
- MacDonald MR, Schaefer GB, Olney AH, Patton DF. Hypoplasia of the cerebellar vermis and corpus callosum in thrombocytopenia with absent radius syndrome on MRI studies. Am J Med Genet. 1994;50:46–50. doi: 10.1002/ajmg.1320500111. [DOI] [PubMed] [Google Scholar]
- Correia-Pinto J, Gonzaga S, Huang Y, Rottier R. Congenital lung lesions—underlying molecular mechanisms. Semin Pediatr Surg. 2010;19:171–179. doi: 10.1053/j.sempedsurg.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Muromoto R, Taira N, Ikeda O, et al. The exon-junction complex proteins, Y14 and MAGOH regulate STAT3 activation. Biochem Biophys Res Commun. 2009;382:63–68. doi: 10.1016/j.bbrc.2009.02.127. [DOI] [PubMed] [Google Scholar]
- Dabir S, Kluge A, Aziz MA, Houghton JA, Dowlati A. Identification of STAT3-independent regulatory effects for protein inhibitor of activated STAT3 by binding to novel transcription factors. Cancer Biol Ther. 2011;12:139–151. doi: 10.4161/cbt.12.2.15732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro G, El-Hashash A, Guidolin D, et al. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol. 2009;333:238–250. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, et al. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109:47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang F, Ornitz DM. Mesothelial- and epithelial-derived FGF9 have distinct functions in the regulation of lung development. Development. 2011;138:3169–3177. doi: 10.1242/dev.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnati S, Baumgart-Vogt E. Peroxisomes in airway epithelia and future prospects of these organelles for pulmonary cell biology. Histochem Cell Biol. 2009;131:447–454. doi: 10.1007/s00418-009-0566-4. [DOI] [PubMed] [Google Scholar]
- Kennedy MW, Cha SW, Tadjuidje E, Andrews PG, Heasman J, Kao KR. A co-dependent requirement of xBcl9 and Pygopus for embryonic body axis development in Xenopus. Dev Dyn. 2010;239:271–283. doi: 10.1002/dvdy.22133. [DOI] [PubMed] [Google Scholar]
- Dumas L, Sikela JM. DUF1220 domains, cognitive disease, and human brain evolution. Cold Spring Harb Symp Quant Biol. 2009;74:375–382. doi: 10.1101/sqb.2009.74.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinawi M, Schaaf CP, Bhatt SS, et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet. 2009;41:1269–1271. doi: 10.1038/ng.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski P, Schaaf CP, Person RE, et al. Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: benign or pathological. Hum Mutat. 2010;31:840–850. doi: 10.1002/humu.21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:R176–R187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velinov M, Dolzhanskaya N. Clavicular pseudoarthrosis, anomalous coronary artery and extra crease of the fifth finger-previously unreported features in individuals with class II 1q21.1 microdeletions. Eur J Med Genet. 2010;53:213–216. doi: 10.1016/j.ejmg.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Goldberg-Stern H, Mimouni-Bloch A, Shkalim V, Bohm D, Kohlhase J. An emerging 1q21.1 deletion-associated neurodevelopmental phenotype. J Child Neurol. 2011;26:113–116. doi: 10.1177/0883073810377658. [DOI] [PubMed] [Google Scholar]
- de Kovel CG, Trucks H, Helbig I, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheroki C, Krepischi-Santos AC, Szuhai K, et al. Genomic imbalances associated with mullerian aplasia. J Med Genet. 2008;45:228–232. doi: 10.1136/jmg.2007.051839. [DOI] [PubMed] [Google Scholar]
- Greenhalgh KL, Howell RT, Bottani A, et al. Thrombocytopenia-absent radius syndrome: a clinical genetic study. J Med Genet. 2002;39:876–881. doi: 10.1136/jmg.39.12.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig S, Schlembach D, Waldispuehl-Geigl J, et al. Impact of array comparative genomic hybridization-derived information on genetic counseling demonstrated by prenatal diagnosis of the TAR (thrombocytopenia-absent-radius) syndrome-associated microdeletion 1q21.1. Am J Hum Genet. 2007;81:866–868. doi: 10.1086/521338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.