If you can look into the seeds of time, and say which grain will grow and which will not, speak then to me … —William Shakespeare (Macbeth, Act I, Scene 3)
In the mustard family (Brassicaceae), pollen grains delivered to the stigma of a flower follow one of two possible fates: One, in which germination and growth of the pollen tube lead to fertilization and seed production; the other, in which activation, pollen tube development, and invasion of the stigma epidermis are aborted (Fig. 1) because of the operation of a genetic barrier to self-fertilization, termed self-incompatibility (SI). SI is based on the ability of cells of the stigma epidermis to discern the presence of self-pollen and to inhibit the germination or subsequent development of self-related, but not genetically unrelated, pollen. In the mustard family, SI occurs in nearly half of the species, and these must then rely on outcrossing via crosspollination by insect vectors to complete their life cycle in the wild. The genetics of SI in this family were deciphered in the early 1950s by Bateman (1) who described control by a single Mendelian locus, the S (Sterility) locus, which exists as multiple alleles or variants, each of which encodes a distinct mating specificity. In the self-incompatible plants of this family, pollen will not develop on a stigma that expresses the same alleles as the pollen parent. As expected for a system in which new alleles have a reproductive advantage and therefore will increase in frequency toward equilibrium, the number of S-locus alleles is usually large, being estimated at 22 in Iberis (1), 34 in Raphanus (2), 50 in Brassica oleracea (3) and 30 in B. campestris (4). Through the efforts of groups in the United Kingdom and Japan, the Brassica S-locus variants have been placed in accessible collections where they are maintained as homozygotes by forced self-pollination in immature young buds before the stigma acquires the ability to reject self-pollen.
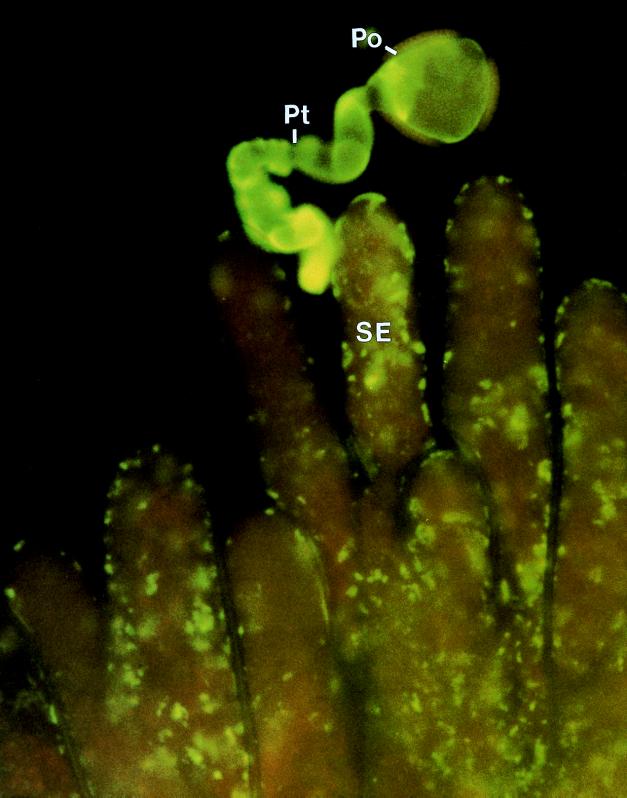
Figure 1.
An incompatible pollination in Brassica showing the inhibition of self-pollen (Po) and the inability of the emerging pollen tube (Pt) to invade the wall of a stigma epidermal cell (SE).
Several predictions for variation at genes controlling the SI response can be made (5). Because of self-sterility, homozygotes rarely are produced, resulting in strong heterozygote advantage. Thus, it is expected that polymorphisms will persist for long periods of time, and that there will be a great deal of ancestral polymorphism. Because alleles are maintained for such long periods of time, substantial differentiation is expected to accumulate between alleles. In addition, because alleles are present essentially only as heterozygotes, it is expected that intragenic recombination and/or gene conversion would have occurred, potentially leading to new self-incompatibility alleles.
The molecular cloning of genes from the Brassica S locus (6–8) has provided an opportunity to test these predictions. Molecular analysis of the S-locus region has shown that this Mendelian locus is a complex locus spanning several hundred kilobases and containing several physically linked transcriptional units that cosegregate perfectly with SI phenotype (Fig. 2; refs. 8–10). A subset of genes within the S-locus complex (or “S haplotype”) is highly polymorphic as expected for genes involved in recognition, and specific combinations of allelic forms of each of these genes are thought to define different SI specificities. Thus, the S locus may be viewed as a master recognition locus that encodes the function(s) required for the stigma to distinguish self-related from self-unrelated pollen as well as the molecules borne by pollen that identify the pollen grain as being self or non-self. An implication of this statement is that each S haplotype encodes distinct gene products adapted for structural compatibility, and that the proper functioning of the SI system depends on the maintenance of the gene complex in a tightly linked genetic unit. An understanding of the evolution of S-locus specificities therefore becomes a question of investigating not only the evolutionary forces that have led to the diversification of alleles of one gene, but also those that allow for the coevolution of genes required for self-recognition and the maintenance of their genetic linkage.
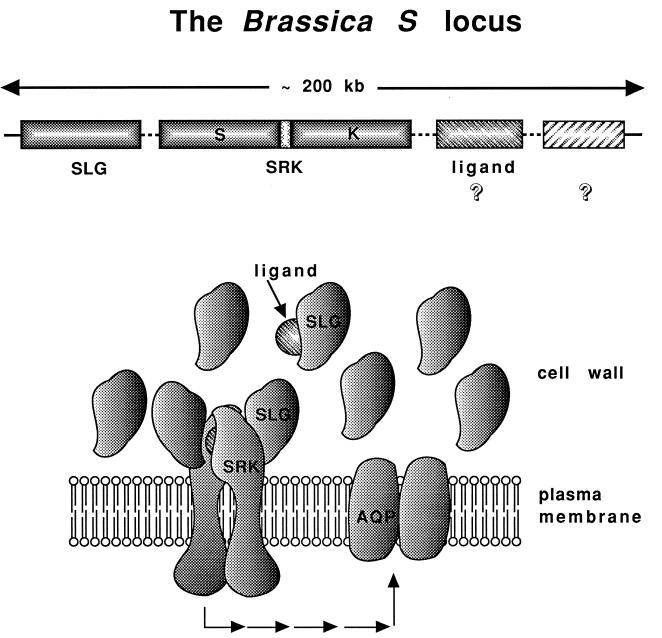
Figure 2.
The S-locus complex and a model of self-incompatibility in Brassica. (Upper) Diagram shows the genes that code for the stigmatic receptors SLG and SRK as well as a hypothetical pollen ligand-encoding gene. The unlabeled box to the right represents one of several genes that map to the S-locus complex but whose role in the self-incompatibility response is unclear. The locus is shown as spanning approximately 200 kb, but its size as well as the order of its genes may vary in different S haplotypes. For simplicity, the SRK gene is depicted without introns; the three domains of its encoded protein, the extracellular (S) domain linked to the kinase (K) domain via a transmembrane domain are shown. (Lower) A model depicting the hypothetical interactions between SLG, SRK, and pollen ligand at the surface of a stigma epidermal cell. The series of arrows represents a signal transduction pathway initiated by activation of SRK and acting on its putative target, a membrane protein related to water-transporting aquaporins (AQP) (24).
Two highly polymorphic S-locus genes are required for the ability of the stigma to inhibit self-pollen (Fig. 2; refs. 11–13): the S-locus glycoprotein (SLG) gene, which encodes a soluble cell wall-localized glycoprotein (6, 7), and the S-locus receptor kinase (SRK) gene, which encodes a receptor-like plasma membrane-spanning kinase (8, 14) that belongs to a family of serine/threonine receptor kinases, the first member of which was isolated from Caenorhabditis elegans in 1990 (15). SLG shares extensive sequence similarity with the extracellular (S) domain of SRK and apparently arose from SRK by a gene duplication event early in the evolution of S haplotypes (16). The structural features of SRK make the compelling argument that the S-locus encoded pollen determinant of SI is a ligand for the receptor. A current model of self-recognition views SLG and SRK as stigmatic cell surface receptors that function in concert, presumably through haplotype-specific binding to this as-yet-unidentified polymorphic pollen-borne ligand, thus precipitating an intracellular phoshorylation cascade that leads to the arrest of self-pollen (Fig. 2). It is not known which part of the SLG protein or the extracellular S domain of SRK acts as the primary specificity-determining region, which is the ligand-binding domain, and which (if any) functions in possible interactions between the soluble and membrane-bound receptors. Ever since the isolation of the first SLG alleles in the mid-80s, it has been hoped that allelic sequence comparisons of S-locus genes not only would suggest mechanisms for sequence diversification at the locus, but also might uncover those regions of the encoded proteins on which natural selection has acted most strongly and thus possibly identify regions that serve as specificity determinants in the SI recognition system.
Based on a comparison of DNA sequences for three SLG alleles, the SLG protein was described as consisting of alternating relatively well conserved domains and hypervariable regions (7). The report of Kusaba and coworkers (17) now extends these data by reporting the most extensive comparative sequence analysis of SLG alleles to date. They compared 24 B. oleracea alleles and 18 B. campestris alleles. Two conclusions consistent with population genetic predictions are evident from this and previous studies. First, SLG alleles exhibit an extraordinarily high degree of DNA and protein sequence variability, with the majority (but not all) of nonsynonymous substitutions clustered in three “hypervariable” regions. Second, SLG polymorphisms are ancient, and allele diversification has predated speciation in the genus Brassica. The great age of the alleles is evident from the observation of interspecific shared polymorphism, not only in SLG DNA and protein sequence (17, 18) but also in the structural heteromorphism of the S locus (9). Although the exact age of the alleles is debatable, analyses of the level of synonymous site substitution and estimates of divergence rates have suggested that S-locus polymorphisms are at least 20–40 million years old (19, 20). This SI system thus represents a more extreme example of the selective maintenance of ancestral polymorphism than that reported for the major histocompatibility loci in mammals where alleles have persisted for 3–10 million years (21, 22).
The expectation for the occurrence of intragenic recombination is borne out by Kusaba et al.’s analysis (17) of the number of substitutions in each of the hypervariable segments of the SLG gene. Homogeneous levels of divergence (particularly at synonymous sites) across the gene are expected if one assumes a clocklike accumulation of divergence between alleles because they diverged from a common ancestral allele. However, intragenic recombination or gene conversion between alleles will obscure this simple allele phylogeny, because alleles now will be composed of segments with potentially very different evolutionary histories. Kusaba et al.’s analysis indicates that the hypervariable regions have been shuffled between alleles, thus suggesting that intragenic recombination, as well as base pair substitutions and insertions/deletions, has been a factor in the DNA sequence diversification of SLG alleles.
Although it is likely that recombination also has contributed to the generation of at least some new self-incompatibility specificities, this cannot be confirmed without knowledge of the amino acid changes that determine allele specificity. And in this regard, sequence comparisons of SLG alleles have not provided the anticipated insight. An examination of SLG hydropathy plots indicates that the three hypervariable regions described by Kusaba et al. (17) do occur in mostly hydrophilic regions of the protein as would be expected if they are on the exterior of the folded molecule and involved in recognition phenomena related to SI. However, many amino acid substitutions occur outside these regions. Further, other regions at least equally hydrophilic or more antigenic and with a higher surface probability are scattered throughout the length of the protein. Indeed, the sequence identity of the hypervariable regions observed between the SLG8 and SLG46 alleles of B. campestris (assuming that the plants from which these sequences were amplified were tested for crosscompatibility by reciprocal pollination) would seem to suggest that these regions are not the sought-after specificity determinants; they may simply be relatively free to diverge because they represent segments of the protein that are subject to fewer selective constraints than other functionally more important regions. Thus, a determination of which regions of the protein are involved in S-allele specific recognition (assuming that specificity is a function of a particular domain of the protein rather than of the protein as a whole) requires in vitro mutagenesis or reconstruction experiments in which regions are altered or switched between alleles having different specificities. Unfortunately, transformation experiments designed to address these issues have been unsuccessful, largely due to the phenomenon of sense suppression whereby the introduction of an SLG or SRK transgene results in the silencing of the endogenous S-locus genes (23).
If recombination occurred frequently during evolution of the S locus, what then accounts for the apparent stability of present-day haplotypes? Frequent recombination in the S-locus region, including unequal crossing over events between SLG and the extracellular domain of SRK, is expected not only to shuffle domains between allelic forms of SLG and SRK but also to disrupt the linkage of stigma and pollen functions that together determine S specificity. A comparison of the long-range maps of different S haplotypes (9) has revealed the occurrence of sequence rearrangements, haplotype-specific sequences, and repetitive sequences. In addition, embedded within the locus are highly conserved single-copy sequences that are expressed in vegetative tissues as well as in reproductive tissues and may perform an essential function in the plant. These various features all may contribute to reduced rates of recombination or recovery of recombinant types in the S-locus region. However, whether recombination frequency at the locus is in fact reduced relative to other regions of the Brassica genome remains to be determined.
Also unresolved is the functional significance of the sequence similarity between SLG and the extracellular domain of SRK. Kusaba et al. (17) analyzed seven SLG/SRK gene pairs from sequences deposited in databases. In three pairs (those derived from the B. campestris S12 and the B. oleracea S3 and S29 haplotypes), the two genes were found to share less sequence similarity with each other than with genes from other haplotypes, prompting the suggestion that SLG and SRK might bind different sites of the same pollen ligand. Clearly what is required to address this question fully is the amplification of SRK alleles from the same genotypes used for the SLG study: analysis of a large number of SLG/SRK gene pairs might identify protein domains that are conserved within an SLG/SRK gene pair but differ between haplotypes. However, a definitive resolution of the various issues raised by the tremendous polymorphisms of S-locus genes must await the identification of the still elusive pollen determinant of self-recognition.
References
- 1.Bateman A J. Heredity. 1955;9:52–68. [Google Scholar]
- 2.Sampson D R. J Heredity. 1957;48:26–29. [Google Scholar]
- 3.Brace J, King G J, Ockendon D J. Sex Plant Reprod. 1994;7:203–208. [Google Scholar]
- 4.Nou I S, Watanabe M, Isogai A, Hinata K. Sex Plant Reprod. 1993;6:79–86. [Google Scholar]
- 5.Wright S. Genetics. 1939;24:538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nasrallah J B, Kao T-H, Goldberg M L, Nasrallah M E. Nature (London) 1985;318:263–267. [Google Scholar]
- 7.Nasrallah J B, Kao T-H, Chen C-H, Goldberg M L, Nasrallah M E. Nature (London) 1987;326:617–619. [Google Scholar]
- 8.Stein J C, Howlett B, Boyes D C, Nasrallah M E, Nasrallah J B. Proc Natl Acad Sci USA. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyes D C, Nasrallah M E, Vrebalov J, Nasrallah J B. Plant Cell. 1997;9:237–247. doi: 10.1105/tpc.9.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu K, Schafer U, Glavin T L, Goring D R, Rothstein S J. Plant Cell. 1996;8:2369–2380. doi: 10.1105/tpc.8.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasrallah M E, Kandasamy M K, Nasrallah J B. Plant J. 1992;2:497–506. [Google Scholar]
- 12.Goring D R, Glavin T L, Schafer U, Rothstein S J. Plant Cell. 1993;5:531–539. doi: 10.1105/tpc.5.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasrallah J B, Rundle S J, Nasrallah M E. Plant J. 1994;5:373–384. [Google Scholar]
- 14.Stein J C, Dixit R, Nasrallah M E, Nasrallah J B. Plant Cell. 1996;8:429–445. doi: 10.1105/tpc.8.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgi L L, Albert S S, Riddle D L. Cell. 1990;61:635–645. doi: 10.1016/0092-8674(90)90475-t. [DOI] [PubMed] [Google Scholar]
- 16.Tantikanjana T, Nasrallah M E, Stein J C, Chen C-H, Nasrallah J B. Plant Cell. 1993;5:657–666. doi: 10.1105/tpc.5.6.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusaba M, Nishio T, Satta Y, Hinata K, Ockendon D. Proc Natl Acad Sci USA. 1997;94:7673–7678. doi: 10.1073/pnas.94.14.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dwyer K G, Balent M A, Nasrallah J B, Nasrallah M E. Plant Mol Biol. 1991;16:481–486. doi: 10.1007/BF00024000. [DOI] [PubMed] [Google Scholar]
- 19.Uyenoyama M. Genetics. 1995;139:975–992. doi: 10.1093/genetics/139.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasrallah J B, Nasrallah M E. Plant Cell. 1993;5:1325–1335. doi: 10.1105/tpc.5.10.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueroa F, Gunther E, Klein J. Nature (London) 1988;335:265–267. doi: 10.1038/335265a0. [DOI] [PubMed] [Google Scholar]
- 22.Lawlor D A, Ward F E, Ennis P D, Jackson A P, Parham P. Nature (London) 1988;335:268–271. doi: 10.1038/335268a0. [DOI] [PubMed] [Google Scholar]
- 23.Conner J A, Tantikanjana T, Stein J C, Kandasamy M K, Nasrallah J B, Nasrallah M E. Plant J. 1997;11:809–823. [Google Scholar]
- 24.Ikeda S, Nasrallah J B, Dixit R, Preiss S, Nasrallah M E. Science. 1997;276:1564–1566. doi: 10.1126/science.276.5318.1564. [DOI] [PubMed] [Google Scholar]