SUMMARY
Thousands of long intervening non-coding RNAs (lincRNAs) have been identified in mammals. To better understand the evolution and functions of these enigmatic RNAs, we used chromatin marks, poly(A)-site mapping and RNA-Seq data, to identify more than 550 distinct lincRNAs in zebrafish. Although these shared many characteristics with mammalian lincRNAs, only 29 had detectable sequence similarity with putative mammalian orthologs, typically restricted to a single short region of high conservation. Other lincRNAs had conserved genomic locations without detectable sequence conservation. Antisense reagents targeting conserved regions of two zebrafish lincRNAs caused developmental defects. Reagents targeting splice sites caused the same defects and were rescued by adding either the mature lincRNA or its human or mouse ortholog. Our study provides a roadmap for identification and analysis of lincRNAs in model organisms and shows that lincRNAs play crucial biological roles during embryonic development with functionality conserved despite limited sequence conservation.
INTRODUCTION
The availability of sequenced genomes for many species has shifted the focus from determining the genetic makeup of organisms to the delineation of the functional elements they encode. These analyses have revealed that, in addition to loci generating known genes, many other loci are transcribed, often in a regulated and tissue-specific fashion (Bertone et al., 2004; Carninci et al., 2005; Dinger et al., 2008; Mercer et al., 2008; De Lucia and Dean, 2011; Jouannet and Crespi, 2011). In the human and mouse genomes, thousands of loci produce RNA molecules longer than 200 nucleotides (nt) that are capped, polyadenylated and often spliced, yet do not overlap protein-coding genes or previously characterized classes of noncoding RNAs (ncRNAs); these have been called long intervening ncRNAs (lincRNAs) (Guttman et al., 2009; Khalil et al., 2009). Although a few dozen mammalian lincRNAs have been characterized to some extent and reported to function in important cellular processes such as X-chromosome inactivation, imprinting, pluripotency maintenance, and transcriptional regulation (Rinn et al., 2007; Mercer et al., 2009; Gupta et al., 2010; Huarte et al., 2010; Orom et al., 2010; Guttman et al., 2011; Hung et al., 2011), the functions of most annotated lincRNAs are unknown.
Comparative sequence analysis and functional studies in non-mammalian species have greatly advanced the understanding of protein-coding genes as well as microRNAs and other ncRNAs. However, these approaches were not immediately applied to lincRNAs because of their more limited sequence conservation (Ponjavic et al., 2007; Marques and Ponting, 2009). Thousands of lincRNAs have been reported in human and mouse, some of which have recognizable sequence homology in the other species (Ponjavic et al., 2007; Khalil et al., 2009; Marques and Ponting, 2009; Guttman et al., 2010). However, there have been only a few hints that orthologs of mammalian lincRNAs exist outside of mammals (Chodroff et al., 2010; Stadler, 2010; Wang et al., 2011). Therefore, the promise of model organisms for providing insight into mammalian lincRNA genomics, evolution and function has awaited accurate experimental identification of lincRNAs in a non-mammalian model organism.
Although high-throughput RNA sequencing (RNA-Seq) provides information useful for lincRNA identification (Guttman et al., 2010), the short reads of current technologies limit the ability to accurately delineate full-length transcriptional units, especially those of lincRNAs, which typically are expressed at low levels (Guttman et al., 2009; Khalil et al., 2009; Guttman et al., 2010). Therefore, to build a robust pipeline for lincRNA discovery, complementary datasets augmenting RNA-Seq-based reconstruction must be acquired and integrated. Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) generates genome-wide chromatin-state maps (Barski et al., 2007; Mikkelsen et al., 2007), which enabled the preliminary annotation of many mammalian lincRNAs (Guttman et al., 2009; Khalil et al., 2009). Particularly informative have been the maps of histone H3 lysine 4 trimethylation (H3K4me3), which marks promoters of genes actively transcribed by RNA polymerase II, and maps of histone H3 lysine 36 trimethylation (H3K36me3), which marks the bodies of these genes (Schubeler et al., 2004; Marson et al., 2008). Another approach that provides important information for defining boundaries of transcriptional units is poly(A)-position profiling by sequencing (3P-Seq) (Jan et al., 2011). This method isolates distal segments of polyadenylated transcripts and identifies them by high-throughput sequencing. Although the initial description and application of 3P-Seq focused on protein-coding genes (Jan et al., 2011), the method defines 3′ termini of all polyadenylated transcripts, including lincRNAs.
To identify lincRNAs of zebrafish (Danio rerio), we acquired chromatin maps and poly(A) sites from three developmental stages and developed a framework for lincRNA discovery that integrates these new datasets with transcriptome datasets, which included RNA-Seq reads, annotated ESTs and full-length cDNAs. We report more than 550 distinct lincRNAs in zebrafish and analyze their sequence and genomic properties, temporal and spatial expression, and conservation. For functional studies, we examined two lincRNAs with short stretches of deep conservation across vertebrates and found that these lincRNAs play important roles in brain morphogenesis and eye development, and that these functions are retained in their mammalian orthologs.
RESULTS
Identification of lincRNAs in Zebrafish
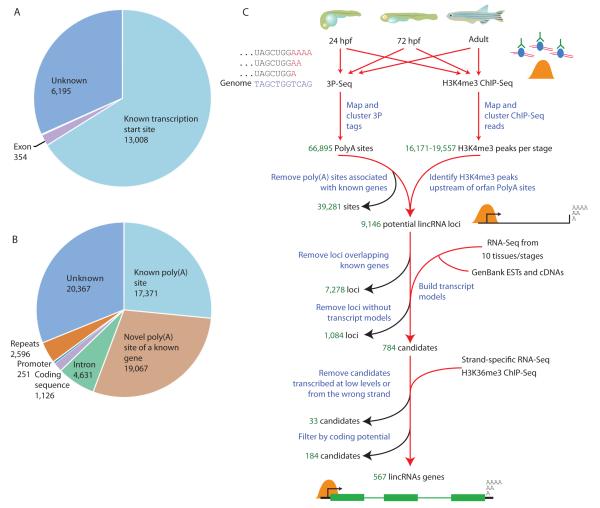
To allow a systematic overview of actively transcribed regions, we generated genome-wide chromatin maps of histone H3 modifications with ChIP-Seq, focusing on zebrafish embryos at 24 and 72 hours post-fertilization (hpf) and mixed-gender adults (Table S1). As in other species (Barski et al., 2007; Marson et al., 2008; Kolasinska-Zwierz et al., 2009), H3K4me3 marks were strongly enriched around transcription start sites, H3K36me3 levels were higher in gene bodies, and the amplitudes of both marks reflected gene expression levels (Figure S1A-B). At each stage, between 16,171 and 19,557 H3K4me3 peaks were identified (Figure 1A and Table S2), most of which were present in all three stages (Figure S1C). We also applied 3P-Seq to poly(A)-selected RNA from the same three stages and identified 66,895 poly(A) sites (Figure 1B and Tables S1 and S2).
Figure 1. Identification of Zebrafish lincRNA Genes.
(A) Positions of H3K4me3 peaks from 24-hpf embryos with respect to annotations of known protein-coding genes and genes of small ncRNAs (<200 nt) annotated in Ensembl or RefSeq.
(B) Positions of poly(A) sites with respect to annotated protein-coding and small ncRNA genes.
(C) Pipeline for identification of lincRNAs. See text and extended experimental procedures for description.
Although the majority of H3K4me3 peaks and poly(A) sites could be assigned to known genes, a significant fraction occurred in regions without transcript annotation (Figure 1A-B) Poly(A) sites that could not be assigned to protein-coding or microRNA genes were used as seeds for identification of lincRNAs (Figure 1C), because sites identified by 3P-Seq unambiguously determine the strand of the transcript and one of its termini, which substantially constrained the subsequent search space. For each of these poly(A) sites, the closest upstream H3K4me3 peaks were identified, and putative lincRNA domains were defined as regions spanning from an H3K4me3 peak to a poly(A) site. After filtering out domains overlapping exons of protein-coding genes or small RNAs in the sense orientation, or coding exons in the antisense orientation, the remaining domains were significantly enriched for H3K36me3, indicating that they were enriched in bona fide transcriptional units (Figure S1D). Using publicly available RNA-Seq data (SRA accession ERP000016) and cDNAs and ESTs deposited in GenBank, transcript models of long RNA molecules were generated, which were then extensively filtered to exclude those with predicted coding potential or insufficient transcription from the predicted strand. This procedure yielded 567 lincRNA gene annotations giving rise to 691 isoforms (Table S2). Of those, 27 genes (4.8%) were contained within introns of protein-coding genes (14 in the sense and 13 the anti-sense orientation). Hand curation of a subset of the 567 genes confirmed the specificity of our pipeline, uncovering only a few false-positives resulting from either gaps in the genome assembly or short unannotated coding regions (Table S2). As in mammals (Guttman et al., 2009; Khalil et al., 2009), lincRNA genes were assigned provisional names based on the closest annotated protein-coding gene (Table S2), except for cases in which vertebrate synteny that involved another nearby gene suggested a more suitable name (e.g., linc-plcb2).
For comparison to the zebrafish lincRNAs, we filtered human and mouse lincRNA annotations to remove those overlapping protein-coding genes, pseudogenes or small ncRNA genes, such as microRNA genes (Table S3). Despite this filtering, our curated sets of mammalian lincRNA genes, numbering 2,458 in human and 3,345 in mouse, were larger than our set of zebrafish lincRNA genes, in part because our pipeline for lincRNA discovery was more stringent than those used previously (Ponjavic et al., 2007; Guttman et al., 2009; Guttman et al., 2010; Jia et al., 2010; Orom et al., 2010), in that it required independent experimental evidence for transcriptional initiation, elongation and termination at each locus. In analyzing whole animals our analysis also might have missed many lincRNAs with very restricted expression patterns.
Genomics of Zebrafish lincRNAs
Our set of zebrafish lincRNAs shared many characteristics with mammalian lincRNAs. Most (61.7%) were spliced. They averaged 1,951 nt spanning an average of 2.3 exons, were more A/U-rich than coding sequences and 5′UTRs, but less so than 3′UTRs, and resembled 5′UTRs in prevalence of short homopolymers (Figure S1E-H). lincRNAs from zebrafish, mouse and human are more likely than protein-coding genes to overlap with repetitive elements, but compared with mammalian lincRNAs, a smaller portion of zebrafish lincRNA sequence was repetitive (Figure S1I).
Mammalian lincRNA genes tend to be within <10 kb of protein-coding genes (Bertone et al., 2004; Ponjavic et al., 2007; Jia et al., 2010). Although zebrafish lincRNA genes also tended to reside near protein-coding genes (empirical P <0.001 compared to random intergenic regions; Figure S1J), the distances between lincRNA genes and the closest protein-coding genes were similar to the distances between adjacent protein-coding genes (Figure S1J). The closest neighboring protein-coding gene was most likely to appear in a divergent orientation with respect to the lincRNA (Figure S1K).
Mammalian lincRNAs have also been reported to be enriched near genes encoding transcription factors and genes involved in nervous system development (Mercer et al., 2008; Guttman et al., 2009; Ponjavic et al., 2009), although these trends are potentially confounded by larger intergenic regions surrounding these genes (Taher and Ovcharenko, 2009). We tested for GO enrichments in the set of protein-coding genes flanking zebrafish lincRNA loci (Figure S1L). The closest neighbors of zebrafish lincRNAs were significantly more likely to function in transcription-related processes [2.85-fold enrichment, hypergeometric test false-discovery rate (FDR) <0.05], an enrichment that could not be explained by the larger intergenic regions flanking those genes (empirical P <0.001). Enrichment for developmental genes was not significant after correction for multiple hypothesis testing or for the sizes of the intergenic regions, with similar trends observed in mouse and human (Figure S1L).
Tissue-Specific Expression of Zebrafish lincRNAs
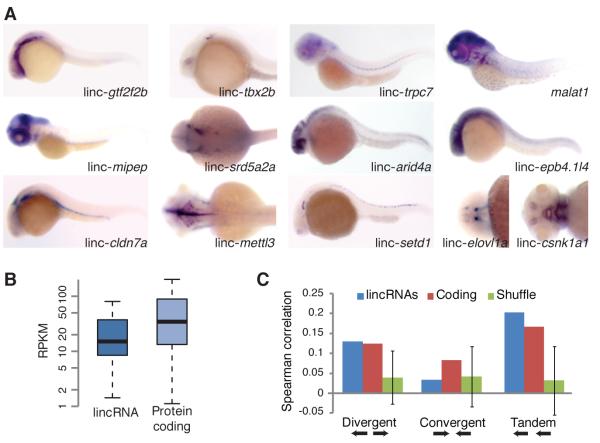
We selected a subset of lincRNAs with relatively high expression or conservation and determined their spatial-temporal expression by in situ hybridization at two developmental stages in zebrafish embryos (Table S4). Most tested lincRNAs were expressed in a highly tissue-specific manner (Figure 2A and S2), predominantly in different parts of the central nervous system, although some were expressed in non-neural tissues and cell types, such as the pronephros (linc-cldn7a) and notochord (linc-trpc7). Although our pipeline was expected to miss lincRNAs expressed in very few cells (because their ChIP-Seq signal from entire embryos might not have exceeded background), it did identify many lincRNAs with tissue-specific expression, suggesting diverse and specific roles for these non-coding RNAs.
Figure 2. Expression of Zebrafish lincRNAs.
(A) Whole-mount in situ hybridizations of selected lincRNAs. Control experiments using sense probes for selected lincRNAs were also performed (Figure S2).
(B) Expression levels of lincRNA and protein-coding genes evaluated using RNA-Seq results from ten stages/tissues. Plots indicate the median, quartiles, and 10th and 90th percentiles. RPKM is reads per kilobase per million reads.
(C) Correlations between levels of neighboring transcripts. For each gene, the Spearman correlation between its expression profile (across ten stages/tissues) and that of the closest protein-coding gene was determined, and the average is plotted for the lincRNA and coding genes. Error bars are 95% confidence intervals based on 1000 random shuffles of lincRNA positions.
We used RNA-Seq data from ten developmental stages and tissues (SRA study ERP000016) to characterize lincRNA expression across zebrafish development. Akin to mammalian lincRNAs (Ponjavic et al., 2009; Guttman et al., 2010), expression levels of zebrafish lincRNAs were generally lower than those of protein-coding genes (Kolmogorov-Smirnov test P <10−15, Figure 2B). Across the ten developmental stages/tissues, expression of lincRNAs tended to correlate with that of their nearest protein-coding neighbors (average Spearman correlation r2=0.14, P <0.001). Correlation of a similar magnitude was observed for adjacent protein-coding genes (r2=0.136, which dropped to r2=0.121 after excluding homologous pairs). The correlation between lincRNAs and their neighbors was significantly high for divergent and tandem orientations, but not for pairs in convergent orientation (Figure 2C). Thus, based on expression similarity and relative distances, lincRNAs and their adjacent protein-coding genes are no more likely to share the same primary transcript than two adjacent protein-coding genes. Instead, the significant co-expression and proximity between lincRNAs and their adjacent protein-coding genes presumably stems from common cis-regulatory modules (especially in the case of divergent transcripts) or shared chromatin domains.
Our lincRNA discovery was performed in late embryonic stages (24 hpf and 72 hpf), but 270 lincRNAs were also expressed (RPKM>1) during early development [oocyte to 5.5 hpf, using data from Aanes et al. (2011)], including 43 with transcripts up-regulated more than 4-fold in the poly(A)+ fraction during the transition from the 1-cell to the 16- or 32-cell stage. As transcription is not thought to occur during these stages (Aanes et al., 2011), our results suggest that some lincRNAs undergo cytoplasmic polyadenylation.
More Positional Conservation than Sequence Conservation Across Vertebrates
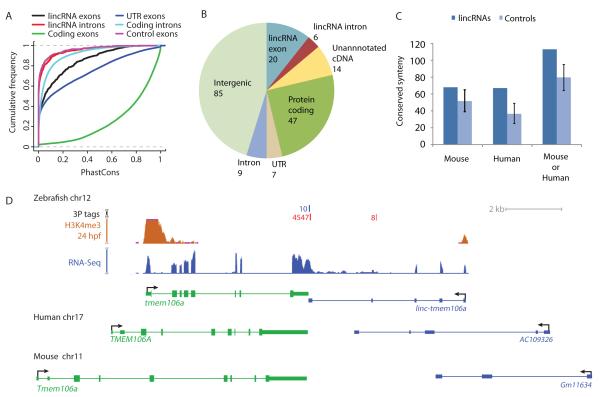
To examine the sequence conservation of lincRNAs, we used the phastCons scores (Siepel et al., 2005) calculated from the UCSC 8-way vertebrate genome alignment seeded with the zebrafish genome (Blanchette et al., 2004). By this measure, lincRNA exons were less well conserved than mRNA coding regions or UTRs but better conserved than introns and random size-matched intergenic regions (control exons) (Figure 3A). This intermediate conservation trend resembled that observed in whole-genome alignments for mammalian lincRNAs for which the observation of conservation above background has provided a main argument for lincRNA functionality (Guttman et al., 2009; Khalil et al., 2009; Marques and Ponting, 2009; Guttman et al., 2010; Orom et al., 2010). Our annotation of zebrafish lincRNAs provided the impetus to take the analyses a step further to look not just at raw conservation but also at the annotations of the homologous regions.
Figure 3. lincRNA Conservation in Vertebrates.
(A) Conservation levels of lincRNA and protein-coding introns and exons, computed using phastCons (Siepel et al., 2005) applied to the 8-way whole-genome alignment. For each lincRNA locus, a computational control was generated by random sampling of a length-matched region from intergenic space of the same chromosome. Within this control region, exons were assigned to the same relative positions as in the authentic lincRNA locus (control exons).
(B) Annotation of human or mouse genomic regions aligned to 188 zebrafish lincRNA genes in the 8-way whole-genome alignment. Regions aligned with zebrafish lincRNAs were tested for overlap with (i) lincRNAs (Table S3), (ii) protein-coding sequence, (iii) 5′UTR or 3′UTR, (iv) introns, or (v) GenBank mRNAs (unannotated cDNAs), in this order, and assigned to the first category for which overlap was observed.
(C) Conserved orientation of protein-coding genes with adjacent lincRNAs. Orthologous protein-coding genes adjacent to zebrafish and mammalian lincRNAs were identified, and the corresponding lincRNAs were considered to have conserved positions, regardless of their sequence conservation. Plotted is the number of those with orientations conserved with respect to their anchoring proteins. The 95% confidence intervals were estimated using 200 cohorts of computational controls, generated as in (A).
(D) linc-tmem106a and its positionally conserved lincRNAs in the human and mouse genomes. The number of 3P tags mapping to the plus and minus strands are indicated (red and blue, respectively).
Of the annotated mouse and human lincRNAs, only 250 and 420 (9.0% and 16.1%), respectively, were aligned to any zebrafish sequence in the whole-genome alignments. Of those, only seven mouse and nine human lincRNAs mapped to zebrafish lincRNAs identified in this study (Table S2). In contrast, 100 mouse and 286 human lincRNAs mapped to at least one zebrafish coding exon. Thus, about half of mammalian lincRNA genes with recognizable sequence homology in the zebrafish genome are either misannotated protein-coding loci or actual lincRNA genes that derived from ancestral protein-coding genes.
An analogous picture emerged when starting from the zebrafish lincRNAs using the 8-genome alignment to the zebrafish genome (Figure 3B): 188 (33.2%) distinct zebrafish lincRNAs were aligned to human or mouse genomes (compared to 76.5% of protein-coding genes), and of these, 20 mapped to exons of mouse or human lincRNAs, which was a small but significantly enriched fraction (Figure 3B, empirical P <0.005 when compared to random intergenic regions). Another six of the 188 mapped to introns of a mammalian lincRNAs (Figure 3B), and for four of those six, further inspection revealed evidence for mapping to unannotated exons of the respective mammalian lincRNAs. An additional 14 were aligned to sequences overlapping GenBank cDNAs, indicating that they might correspond to lincRNAs that were not yet annotated in mammalian genomes. Another 47 (25% of the lincRNAs with recognizable mammalian homology) were aligned to the transcribed strand of protein-coding exons in human or mouse. Overall, 55% of zebrafish lincRNAs that were aligned to human or mouse in the 8-way alignment mapped to annotated transcribed units in those genomes (Figure 3B), compared to 14% for random regions (empirical P <0.005).
Direct comparison of mammalian and zebrafish lincRNAs using BLASTN found another three zebrafish lincRNAs with annotated mammalian orthologs, bringing the total to 29, with 90% of these cases supported by synteny extending to at least one protein-coding neighbor (Table S2). Although in most cases the mammalian ortholog was annotated as a lincRNA in only one of the two mammals, further inspection usually indicated that similar transcribed loci were present in both mammals. For these 29 lincRNAs, detectable homology with the proposed mammalian orthologs spanned a small portion of the transcript, averaging 308 nt (range 31–1,206 nt) and was typically restricted to a single exon. For nine of the 11 cases in which both the zebrafish lincRNA and its mammalian ortholog were spliced, the relative position of the exon with the conserved region was also conserved.
The zebrafish and mammalian genomes were extensively rearranged during >400 million years of independent evolution, which included a whole-genome duplication in the teleost fish lineage (Hoegg et al., 2004; Jaillon et al., 2004; Semon and Wolfe, 2007). As a result, only 14.7% of protein-coding gene pairs that both have mouse orthologs and are adjacent in the zebrafish genome also have adjacent orthologs in the mouse genome. Despite this extensive rearrangement, we found that adjacency to a lincRNA (limited to distance ≤ 100 kb) was conserved: Out of the 935 protein-coding genes flanking lincRNAs in zebrafish and conserved in human or mouse, 317 had an ortholog adjacent to a lincRNA annotated in either the human or the mouse genome (hypergeometric P = 0.0028). Of the lincRNAs adjacent to those 317 genes, 113 had conserved orientation with respect to at least one conserved protein-coding neighbor (empirical P <0.005, Figure 3C). The synteny blocks around these lincRNAs contained 2.7 protein-coding genes on average, and were larger than those around random intergenic regions (empirical P = 0.046). In most of these cases, including linc-tmem106a (Figure 3D), sequence similarity between the zebrafish lincRNA and its syntenic mammalian counterpart was not detected, and the number of lincRNAs with conserved position and orientation was significant even when we excluded the 29 lincRNAs with detected sequence conservation (P = 0.01), which suggested that some lincRNAs have conserved functional features, such as secondary structure or genomic position, that were retained without detectable primary-sequence conservation.
lincRNA cyrano Is Required for Proper Embryonic Development
To investigate the roles of lincRNAs during development, we analyzed the effects of lincRNA loss of function in zebrafish embryos using morpholino antisense oligos (MOs). One strategy was to inject MOs designed to target lincRNA splice sites in an attempt to disrupt maturation. The other strategy was to inject MOs designed to target highly conserved sites presumed to be important for interactions with other cellular factors. This approach follows successful use of MOs for blocking specific sites in mRNAs (Heasman et al., 2000; Draper et al., 2001; Choi et al., 2007), and an analogous approach using locked nucleic acid (LNA) antisense oligos to disrupt Xist lincRNA function in cultured mammalian cells (Sarma et al., 2010).
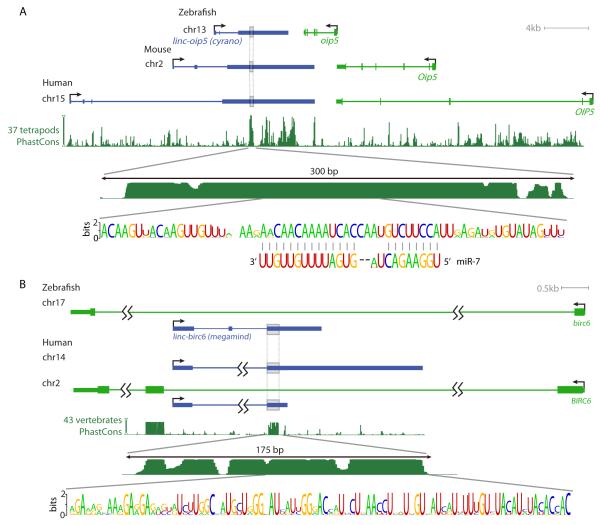
For functional studies, we selected two lincRNAs based on their tissue-specific expression and synteny with mammalian lincRNAs. One was a 4.5 kb transcript with three exons convergent with oip5 (Figures 4A and S3A). Although none of the lincRNA locus was aligned between mammals and zebrafish in any of the whole-genome alignments examined, our BLASTN search identified a conserved 67 nt match with human and mouse lincRNAs (Figures 4A and S3B-D). Both human and mouse orthologs shared a similar gene structure (2-3 short exons followed by a long terminal exon of 4-8 kb, Figure 4A) and were part of a synteny block containing not only the oip5 ortholog but also orthologs of three other zebrafish protein-coding genes (nusap1, ndufaf1 and rtf1). Ribosome profiling in HeLa cells (Guo et al., 2010), which express the human homolog (LOC729082), showed that it was not translated. The PhastCons plot from the UCSC whole-genome alignments to human (which did not include alignment to any fish genomes) showed several conserved regions within the terminal exon, including a ~300 nt region highly conserved among the tetrapods (Figure 4A). Within this region was the segment conserved to fish, which was identified in our BLASTN search (Figure S3D). This segment included a 26 nt site in which all but one nucleotide was perfectly conserved in all 52 homologous segments that we recovered from genomic sequences and ESTs of 52 vertebrate species (Figures 4A).
Figure 4. lincRNAs with Short Conserved Segments.
(A) Genomic context and sequence conservation of the linc-oip5 (cyrano) lincRNA gene. Gray boxes include the deeply conserved region. The conservation plot is relative to the human locus, and is based on aligned regions of 37 genomes, which do not include any fish genomes, as those do not contain any regions that are aligned with this human locus in the whole-genome alignments. The top consensus logo highlights the RNA sequence of the most conserved segment, which we identified in 45 vertebrate genomes, including fish genomes. Shown are the 67 aligned positions present in zebrafish, with a score of 2 bits indicating residues perfectly conserved in all 45 genomes. The bottom consensus logo shows conservation of vertebrate miR-7 sequences annotated in miRBase 18, with vertical lines indicating Watson-Crick base pairs.
(B) Genomic context and sequence conservation of the linc-birc6 (megamind) lincRNA gene. As in (A), except the region is aligned to fish genomes in the whole-genome alignments, and the consensus logo is for the RNA sequence inferred from 75 sequences from 47 vertebrate genomes. An alternative isoform of the zebrafish RNA retains the first intron (Figure S3E).
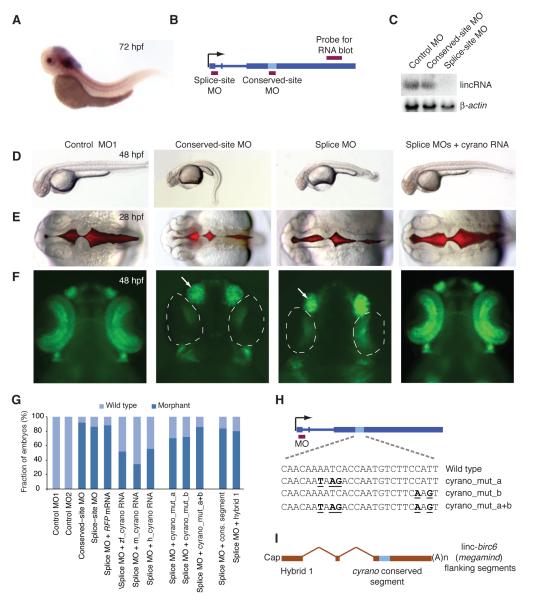
In zebrafish embryos, this lincRNA is expressed in the nervous system and notochord (Figure 5A and S4A). To characterize its role during development, MOs targeting either the first exon-intron splice junction or the most conserved site were injected at the one-cell stage (Figure 5B and Table S6). RNA-blot, qRT-PCR and in situ hybridization analyses showed that the splice-site MO reduced transcript accumulation, whereas targeting the conserved site did not affect either transcript levels or size (Figure 5C and S4B-C). Embryos injected with either splice- or conserved-site MOs exhibited similar developmental defects. These morphants had small heads and eyes, and short, curly tails, perhaps because of the reduced levels of this lincRNA in the notochord (Figure 5D). They also had defects in neural tube opening (Figure 5E), loss of NeuroD-positive neurons in the retina and tectum, and enlarged nasal placodes, as visualized by GFP expression under the control of the neurod promoter (Figure 5F and S4D). Embryos injected with either a conserved-site MO with five mismatches (control MO1) or an MO complementary to a non-conserved region (control MO2) lacked morphant phenotypes (Figures 5D-F and S4D). Because of the prominent nose phenotype in these morphants, we named this lincRNA cyrano (designating the gene as cyrano).
Figure 5. The Importance of linc-oip5 (cyrano) for Proper Embryonic Development.
(A) In situ hybridization showing cyrano expression in the CNS and notochord of zebrafish embryos at 72 hpf.
(B) Gene architecture of cyrano, showing the hybridization sites of the RNA-blot probe and MOs (red boxes).
(C) RNA blot monitoring cyrano accumulation in wild-type embryos (48 hpf) that had been injected with the indicated MOs. To control for loading, the blot was re-probed for β-actin mRNA.
(D) Embryos at 48 hpf that had been either injected with the indicated MO or co-injected with the splice-site MO and mature mouse cyrano RNA.
(E) Brain ventricles after injection with the indicated reagents, visualized using a red fluorescent dye injected into the ventricle space at 28 hpf.
(F) Embryos at 48 hpf that had been injected with the indicated reagents. NeuroD-positive neurons in the retina and nasal placode were marked with GFP expressed from the neurod promoter (Obholzer et al., 2008). Near absence of NeuroD-positive neurons in the retina (dotted line) and enlargement of the nasal placode (arrow) are indicated.
(G) Frequency of morphant phenotypes in injected embryos (Table S5).
(H) Schematic of DNA point substitutions in the cyrano conserved site.
(I) Architecture of a hybrid transcript containing the cyrano conserved segment in the context of linc-birc6 (megamind) flanking sequences.
As an additional control for specificity, we tested whether the observed developmental defects could be rescued by co-injection of spliced RNA, which would not be affected by the splice-site MO (Bill et al., 2009). Co-injection of the splice-site MO with spliced cyrano RNA reduced the fraction of embryos showing morphant phenotypes by over 40% compared to embryos injected either with only the splice-site MO or with the splice-site MO and RFP mRNA (Figures 5G and S4E).
Encouraged by the rescue experiment showing the function of mature zebrafish cyrano RNA during embryonic development, we tested whether the mammalian orthologs might also function in zebrafish. Remarkably, over 60% and over 35% of embryos injected with splice-site MO were rescued by co-injection of the mature mouse or human RNAs, respectively (Figure 5G). Rescued embryos had normal neural tube openings, restored neurogenesis, and normal sized brain, eyes and nasal placodes (Figure 5D-F and Figure S4D). To further investigate the functional importance of the conserved site, we introduced point substitutions (Figure 5H) and tested the potency of the mutated RNAs in rescuing the morphants. The rescuing potential of the mutated RNAs was diminished (cyrano_mut_a and cyrano_mut_b) or abolished (cyrano_mut_a+b) compared to that of the wild type (Figure 5G). Sufficiency for rescue was tested using two different constructs. One was a short RNA containing the 67 nt conserved segment and 32 flanking bases and the other was a long hybrid RNA in which this 99 nt segment replaced the conserved region of an unrelated lincRNA, linc-birc6 (hybrid 1, Figure 5I). In both cases, co-injection of these in vitro transcribed, capped and polyadenylated RNAs with the splice-site MO did not rescue the morphant phenotype, which indicated that the conserved segment was not sufficient for cyrano function (Figure 5G). Taken together, our results show that cyrano, acting in part through its conserved site, plays an important role during embryogenesis, and that the mammalian orthologs retain this function.
After completing these functional studies, we recognized that the conserved site of cyrano pairs perfectly to all but two central nucleotides of the miR-7 microRNA and that this extensive pairing is conserved in all vertebrates examined (Figure 4A). Experiments are underway to determine the reason that this pairing has been conserved. Possibilities include: (i) miR-7 regulates cyrano, (ii) cyrano regulates miR-7, and (iii) cyrano and miR-7 collaborate in a downstream function. Although we suspect that association with miR-7 confers some cyrano destabilization, our observation that disrupting pairing to miR-7 abrogates cyrano function (Figure 5G) suggests that the pairing has not been conserved merely for the repression of cyrano, thereby disfavoring possibility (i) and favoring possibilities (ii) or (iii).
lincRNA megamind Regulates Brain Morphogenesis and Eye Development
Another conserved lincRNA was a 2.4 kb transcript composed of 3 exons (with alternative splicing sometimes skipping the middle exon) overlapping intronic sequence of the protein-coding gene birc6 in an antisense orientation (Figures 4B and S3E). The expression and exon structure of this lincRNA was supported by both RNA-Seq and EST data. A region of about 340 bp near the 5′-end of the third exon was aligned to mammalian genomes in the 8-way whole-genome alignment to zebrafish. In fact, this lincRNA was aligned to two different regions of the human genome — one overlapping a BIRC6 intron in the antisense orientation (part of a large synteny block containing another five protein-coding genes) and another overlapping an annotated lincRNA in a gene desert upstream of BDKRB1/2 (Figure 4B). Using the zebrafish and human sequences, whole-genome alignments, and HMMER (http://hmmer.org/), we identified 75 homologous sequences in 47 vertebrate species, appearing in three distinct contexts: (i) in introns of birc6 homologs, (ii) in gene deserts upstream of bdkrb1/2 homologs, and (iii) near hhipl1 homologs (found only in fish genomes). In zebrafish, homologs with evidence of transcription were found at all three loci (Figure S3E-G), although only one of the two additional loci could be detected using BLASTN. In genomes with sufficient data, the homologous transcripts contained two exons (with an alternative cassette exon present in some species) and a broadly conserved region appearing in the 5′ end of the last exon. The core segment of this region spanned 93 nt and was depleted of both insertions and deletions. Forty positions in this segment had over 90% sequence identity across the 75 homologs, and 19 bases were perfectly conserved (Figures 4B and S3H). Interestingly, 15 of the conserved positions were Ts that occurred with perfect 3 nt periodicity.
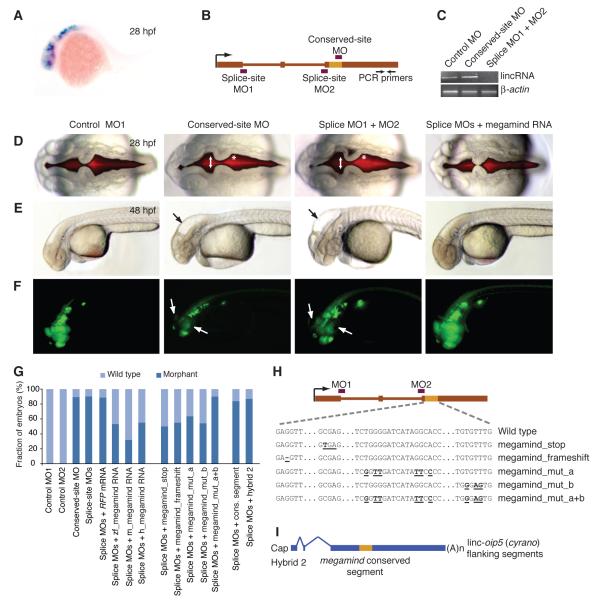
Of the three homologous transcripts in zebrafish, the lincRNA at the birc6 locus, which in embryos was expressed about 12-fold higher than the others, was selected for experimental interrogation. In zebrafish embryos, this lincRNA was expressed in the eyes and brain (Figure 6A and S5A), consistent with EST evidence indicating brain expression of its mammalian homologs. We perturbed this lincRNA using MOs targeting either two splice sites or the conserved segment (Figure 6B and Table S6). Splice-site morpholinos reduced the lincRNA accumulation (Figure 6C and S5B-C). Injections of the splice-site MOs and the conserved-site MO resulted in embryos with indistinguishable brain and eye defects (Figures 6D-F), as did injection of the two splice-site MOs individually. Morphants had defects in brain-ventricle morphology 28 hpf, including an unusual expansion of the midbrain ventricle, loss of the midbrain hingepoint, and contraction of the forebrain ventricle (Figure 6D). By 48 hpf, the morphants had smaller heads and eyes, enlarged brain ventricles (a hydrocephalic phenotype), and loss of NeuroD-positive neurons in the retina and tectum (Figures 6E-F). Embryos injected with MOs complementary to the conserved site with five mismatches (control MO1) or to a non-conserved region of the lincRNA (control MO2) did not show any mutant phenotypes (Figures 6D-F). Based on the head shape, we named this lincRNA megamind.
Figure 6. The Importance of linc-birc6 (megamind) for Proper Brain Development.
(A) In situ hybridization showing megamind expression in the brain and eyes of zebrafish embryos at 28 hpf.
(B) Gene architecture of megamind, showing the hybridization sites of the MOs (red boxes) and RT-PCR primers (arrows).
(C) Semi-quantitative RT-PCR of mature megamind in embryos at 72 hpf that had been injected with the indicated MOs. β-actin mRNA was used as a control.
(D) Brain ventricles after injection with either the indicated MOs or co-injected with the splice-site MO and mature mouse megamind RNA, visualized using a red fluorescent dye injected into the ventricle space at 28 hpf. An expanded midbrain ventricle (arrow) and abnormal hindbrain hinge point (asterisk) are indicated.
(E) Embryos at 48 hpf that had been injected with the indicated reagents. Abnormal head shape and enlarged brain ventricles are indicated (arrow).
(F) Embryos at 48 hpf that had been injected with the indicated reagents. NeuroD-positive neurons in the retina and nasal placode were marked with GFP expressed from the neurod promoter (Obholzer et al., 2008). Near absence of NeuroD-positive neurons in the retina and tectum (arrows) is indicated.
(G) Frequency of morphant phenotypes in injected embryos (Table S5).
(H) Schematic of DNA point substitutions in the megamind conserved segment.
(I) Architecture of a hybrid transcript containing the megamind conserved segment in the context of cyrano flanking sequences.
To test for rescue and function of the orthologous lincRNAs, we co-injected splice-site MOs with in vitro transcribed zebrafish megamind, or the orthologous mouse or human lincRNAs. Mature megamind RNA from each of the three species rescued the morphant phenotypes, despite the limited overall sequence conservation (Figures 6D-G). Embryos co-injected with the splice-site MO and RFP mRNA control showed no improvement (Figures 6G and S5D).
Although the inferred polypeptide of the most plausible open reading frame was poorly conserved and only 49 amino acids long, the conserved 3 nt periodicity in much of this region of megamind, essentially without insertions or deletions, suggested the possibility of a coding region. To test this possibility we introduced either a stop codon to disrupt the most plausible coding frame or a frameshift-inducing single-nucleotide deletion at the beginning of the highly conserved segment (Figure 6H). Co-injecting the splice-site MO with these mutated lincRNAs (megamind_stop and megamind_frameshift) rescued as well as co-injecting the wild-type RNA (Figure 6G), confirming that the conserved segment is very unlikely to act as part of a coding sequence. To test its functional importance, the conserved segment was mutated (Figure 6H). Point substitutions at six conserved nucleotides slightly reduced rescue (megamind_mut_a), and combining these with three point substitutions that had no detectable effect on their own (megamind_mut_b) completely abolished rescue (megamind_mut_a+b), thereby indicating that an intact conserved segment is required for megamind function (Figures 6G and 6H). The conserved segment was not sufficient on its own for function, as splice-site morphants were not rescued by co-injection of either a short RNA containing only the 93 nt conserved segment or a hybrid RNA (hybrid 2, Figure 6I) in which the megamind conserved segment replaced with that of cyrano (Figures 6G).
DISCUSSION
Origins and Evolution of Vertebrate lincRNAs
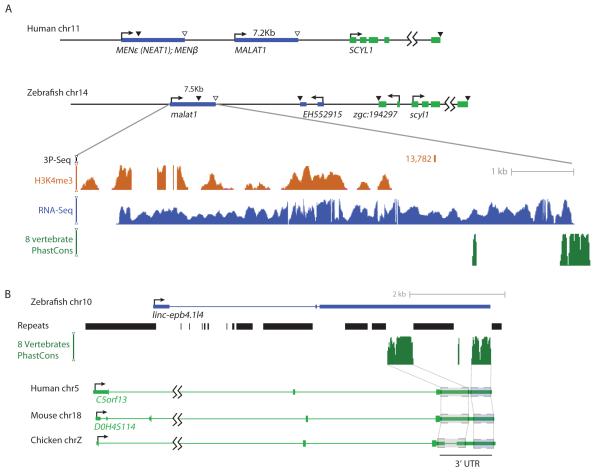
Our mapping of lincRNAs in a non-mammalian vertebrate genome indicates that such genomes encode hundreds of lincRNAs, of which only a few can be traced to potential mammalian homologs. In those cases, the homology spanned a small portion of the transcript, typically restricted to a single exon. Similar short regions of conservation nested in rapidly evolving sequence have been described for lincRNAs conserved only within mammals (Guttman et al., 2010; He et al., 2011). For both cyrano and megamind, the conserved regions were not extensively complementary to other conserved regions in the genome and did not show enrichment for known binding motifs of RNA-binding factors (data not shown), apart from the microRNA complementarity noted for cyrano. In addition to a short region of sequence conservation, cyrano and megamind also preserve genomic architecture, with respect to the sizes and arrangement of exons. Similar patterns were observed in several other conserved lincRNAs, one of which was MALAT1, a lincRNA characterized in mammalian cells (Tripathi et al., 2010), an ortholog of which we identified in zebrafish (malat1, Figure 7A). Sequence similarity between zebrafish and mammalian MALAT1 is restricted to the 3′ end, likely due to a conserved mechanism for 3′ end formation (Wilusz et al., 2008). Despite this limited sequence conservation, the length of MALAT1 (~7 kb) along with the lack of any efficiently spliced introns appeared to be roughly fixed in all vertebrates. These observations suggest that conserved functionality of some lincRNAs requires relatively small amount of specific sequence, supported by long flanking regions deprived of deeply conserved sequence elements.
Figure 7. lincRNA Conservation Patterns.
(A) The human MALAT1 locus and the orthologous locus in zebrafish. Protein-coding genes are in green and lincRNA genes are in blue. Arrows indicate direction of transcription, black triangles indicate canonical poly(A) sites and white triangles indicate 3′ termini obtained by RNAse P cleavage (Wilusz et al., 2008). The conservation plot is relative to the zebrafish locus and based on the 8-genome alignment.
(B) The zebrafish linc-epb4.1l4 gene showing homology to the 3′UTR of an mRNA expressed in human, mouse, chicken and other amniotes. Gray boxes indicate two deeply conserved regions. The repeats track indicates all the repetitive elements predicted by RepeatMasker, taken from the UCSC genome browser. The conservation plot is as in (A).
Although we found mammalian orthologs for only 29 (5.1%) of the zebrafish lincRNA genes, analysis of synteny suggested that a greater fraction might have orthologous function. Perhaps additional lincRNA sequence constraints are present but not detectable above background when carrying out whole-genome alignments or BLASTN comparisons. We used a relatively non-stringent BLASTN E-value threshold of 10−5. Reducing the stringency even further to a threshold of 10−4 recovered another nine zebrafish-mammal lincRNAs pairs, but none of these had conserved synteny, suggesting that such pairs with less significant sequence similarity were less likely to be truly orthologous. Regardless of whether undetectable homology exists, lincRNA genes clearly evolve more rapidly than do those of mRNAs, and they appear to be more rapidly gained and lost during evolution. Indeed, some lincRNAs identified here and elsewhere might be transcribed from newly emergent genes that have not yet acquired a biological function and might be lost before they do acquire one.
New lincRNAs can emerge from one of three mechanisms: (i) de novo formation from previously untranscribed genomic sequence, (ii) duplication of another lincRNA, or (iii) transformation of a protein-coding gene. Model (i) appears to have been the most frequent. A small portion (6.7%) of zebrafish lincRNAs showed similarity to another zebrafish lincRNA (compared to 44.2% of protein-coding genes showing similarity to another protein-coding gene within the genome). Although this comparison is confounded by weaker sequence constraints leading to rapid loss of recognizable sequence similarity following duplication, it provides little evidence for high prevalence of model (ii). In addition, 47 zebrafish lincRNAs (8.6%) showed significant sequence similarity to zebrafish protein-coding genes. These comprised about a quarter of the zebrafish lincRNAs that were aligned to mammalian genomes, and as mentioned above, about half of the mammalian lincRNAs that were aligned to fish mapped to coding exons. Although some of these might be mRNAs misannotated as lincRNAs, many are probably authentic lincRNAs, as indicated by the lack of long open reading frames or sequences predicted to encode conserved polypeptides. These findings suggest that some lincRNAs originated from genes that formerly coded for proteins (model iii), as has been proposed for XIST, a well-characterized lincRNA, which functions in X chromosome inactivation (Duret et al., 2006). We note that the conservation observed for these lincRNAs might arise from purifying selection in only the lineages of their mRNA cousins and not speak to lincRNA function, thereby illustrating a caveat of implying biological function from sequence conservation.
In model (iii), a functional lincRNA might arise from a pseudogene, which has already lost its protein-coding function, or lincRNA function might emerge while the transcript still codes for a functional protein, with subsequent evolutionary loss of the protein-coding function (sometimes after gene duplication and subfunctionalization) to produce a new lincRNA. This second scenario raises the possibility that some mRNAs might currently carry out important non-coding functions, thereby significantly contributing to the number of transcripts with conserved lincRNA-like functions. The same mature transcript carrying out both types of functions would extend the paradigm represented by the SRA1 gene, in which different transcript isoforms from the same gene carry out either coding or noncoding functions (Chooniedass-Kothari et al., 2004; Hube et al., 2006). Although moonlighting mRNAs are difficult to distinguish from normal mRNAs, our annotation of zebrafish lincRNAs enabled identification of bifunctional transcripts that emerged from the reciprocal evolutionary scenario, i.e., the acquisition of protein-coding potential by a lincRNA. For example, a highly expressed, brain-enriched lincRNA in zebrafish (linc-epb4.1l4) showed synteny and sequence conservation with the 3′UTR of a gene encoding neuronal protein 3.1 (P311) in human and mouse (Figures 2A and 7B). Although P311 is only 68 amino acids long, its translation was supported by ribosome footprinting in HeLa cells (Guo et al., 2010). However, despite the clear presence of a homologous transcript in bony and cartilaginous fish, no protein homologs of P311 were detected more basal to tetrapods, and only the 3′UTR of P311 was highly conserved throughout vertebrate genomes. These results suggest that the transcript originally was a lincRNA that performed a function that might still be retained throughout extant vertebrates and began moonlighting as a coding transcript in tetrapods.
A System for Studying lincRNA Functions
In addition to providing insights into lincRNA origins and evolution, the identification of lincRNAs in zebrafish unlocks the toolbox of zebrafish molecular genetics for the study of lincRNA function. With the exceptions of studies of Xist and its associated transcripts (Payer and Lee, 2008), and more recent work done with Neat1 (Nakagawa et al., 2011), lincRNA functions have been studied exclusively in cell lines. Moreover, for the vast majority of lincRNAs, biological functions remain unknown, as do answers to some basic questions: Do any lincRNAs employ common mechanisms of action? Do they function mostly in cis or in trans? For how many does the RNA itself have any importance over the mere act of its transcription? Finding answers to these questions has been hampered by a lack of a model system amenable to quick genetic manipulations and in which phenotypes can be scored on an organismal level. Using zebrafish, we identified biological functions of two lincRNAs in vertebrate development and began to unravel their sequence-function relationships, as well as the functional equivalence of lincRNAs from different species. We anticipate that this system and approach can be used to rapidly reveal biological functions of other lincRNAs and to identify additional lincRNAs with conserved roles across vertebrates, which can be prioritized for functional studies in mammals.
EXPERIMENTAL PROCEDURES
High-Throughput Datasets
Zebrafish were maintained and staged using standard procedures (Kimmel et al., 1995). ChIP-Seq, 3P-Seq and RNA-Seq were performed as described (Guenther et al., 2008; Guo et al., 2010; Jan et al., 2011), and reads were mapped to the genome using Bowtie (Langmead et al., 2009). H3K4me3 enrichment peaks meeting FDR <0.1 were determined using MACS (Zhang et al., 2008). Raw 3P-Seq data was processed as described (Jan et al., 2011). Additional RNA-Seq reads were obtained from SRA (accession ERP000016), and processed using TopHat (Trapnell et al., 2009) and Cufflinks (Trapnell et al., 2010). For additional details, see extended experimental procedures.
Identification of Zebrafish lincRNAs
Our lincRNA prediction pipeline (Figure 1C) is explained in the extended experimental procedures.
Human and Mouse lincRNA Collections
Sets of 2,458 human and 3,345 mouse lincRNAs (Table S3) were obtained by combining long (>200 bp) noncoding transcripts from Ensembl, RefSeq, UCSC genes and Guttman et al. (2010) and filtering for overlap with protein-coding genes, coding transcripts from other species mapped to the human genome in the UCSC genome browser, pseudogenes and “other RNAs” annotated in Ensembl. For additional details, see Extended Experimental Procedures.
Control Regions for lincRNA Genes
For each lincRNA locus, a computational control was generated by random sampling of a length-matched region from intergenic space of the same chromosome. To estimate confidence intervals, 200-1000 cohorts of computational controls were used.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Auyeung, O. Rissland, I. Drinnenberg, R. Shalgi, S. Hong, and D. Garcia for comments on the manuscript, T. DiCesare for illustration, M. Guenther for help with the ChIP-Seq protocol, and W. Johnston and G. Lafkas for technical support. The Tg(neurod:egfp) transgenic line was kindly provided by the Nicolson lab. RNA-Seq data was kindly made available by the Wellcome Trust Sanger Institute. This work was supported by a grant (GM067031) from the NIH (D.P.B.), an EMBO long-term fellowship (I.U.), a Human Frontiers Science Program long-term fellowship (A.S.), and an NSF predoctoral fellowship (C.H.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
ChIP-Seq, 3P-Seq and RNA-Seq sequencing data have been deposited into the Gene Expression Omnibus (GEO, accession number GSE32880).
REFERENCES
- Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, et al. Zebrafish mRNA sequencing decipher novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21:1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnar Z, Ponting CP. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, Hube F, Mutawe M, Watson PH, Leygue E. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–47. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- De Lucia F, Dean C. Long non-coding RNAs and chromatin regulation. Curr Opin Plant Biol. 2011;14:168–173. doi: 10.1016/j.pbi.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol Biol. 2011;11:102. doi: 10.1186/1471-2148-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube F, Guo J, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA, Blanchard AA, Wang X, Deng G, Myal Y, et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–428. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannet V, Crespi M. Long Nonprotein-Coding RNAs in Plants. Prog Mol Subcell Biol. 2011;51:179–200. doi: 10.1007/978-3-642-16502-3_9. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obholzer N, Wolfson S, Trapani JG, Mo W, Nechiporuk A, Busch-Nentwich E, Seiler C, Sidi S, Sollner C, Duncan RN, et al. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J Neurosci. 2008;28:2110–2118. doi: 10.1523/JNEUROSCI.5230-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A. 2010;107:22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semon M, Wolfe KH. Rearrangement rate following the whole-genome duplication in teleosts. Mol Biol Evol. 2007;24:860–867. doi: 10.1093/molbev/msm003. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler PF. Evolution of the Long Non-coding RNAs MALAT1 and MEN{it eta}{it psilon} In: Ferreira CE, Miyano S, Stadler PF, editors. Advances in Bioinformatics and Computational Biology. Springer; Rio de Janeiro, Brazil: 2010. [Google Scholar]
- Taher L, Ovcharenko I. Variable locus length in the human genome leads to ascertainment bias in functional inference for non-coding elements. Bioinformatics. 2009;25:578–584. doi: 10.1093/bioinformatics/btp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.