Abstract
Gut microbial communities represent one source of human genetic and metabolic diversity. To examine how gut microbiomes differ between human populations when viewed from the perspective of component microbial lineages, encoded metabolic functions, stage of postnatal development, and environmental exposures, we characterized bacterial species present in fecal samples obtained from 531 individuals representing healthy Amerindians from the Amazonas of Venezuela, residents of rural Malawian communities, and inhabitants of USA metropolitan areas, as well as the gene content of 110 of their microbiomes. This cohort encompassed infants, children, teenagers and adults, parents and offspring, and included mono- and dizygotic twins. Shared features of the functional maturation of the gut microbiome were identified during the first three years of life in all three populations, including age-associated changes in the representation of genes involved in vitamin biosynthesis and metabolism. Pronounced differences in bacterial species assemblages and functional gene repertoires were noted between individuals residing in the USA compared to the other two countries. These distinctive features are evident in early infancy as well as adulthood. In addition, the similarity of fecal microbiomes among family members extends across cultures. These findings underscore the need to consider the microbiome when evaluating human development, nutritional needs, physiological variations, and the impact of Westernization.
Genetic variation between human populations is typically viewed as differences in the allele frequencies of shared Homo sapiens genes. Another source of genetic and metabolic diversity resides in differences in the representation of the millions of genes and myriad gene functions within our gut microbial communities1–3. Sampling a broad population of healthy humans representing different ages and cultural traditions offers an opportunity to discover how our gut microbiomes evolve within a lifespan, vary between populations, and respond to our changing lifestyles4–9. Therefore, we conducted a demonstration project to address the question of whether there are discernible patterns of functional maturation of the gut communities of healthy infants and children living in geographically and culturally distinct settings.
Fecal samples were obtained from individuals in families of Guahibo Amerindians residing in two villages (Platanillal and Coromoto), separated by 10 miles, located near Puerto Ayacucho in the Amazonas State of Venezuela (see Table S1a,b for information about their diets). Fecal samples were also procured from members of families living in four rural communities of Malawi located within 10–70 miles of one another (Chamba, Makwhira, Mayaka, Mbiza). Lifestyles in these villages are very similar, and diets are relatively monotonous, dominated by maize (Table S1c). In addition, we sampled families distributed across the USA, including the greater metropolitan areas of St. Louis, Philadelphia and Boulder. The sampled populations included parents and siblings, and, in the USA and Malawi, monozygotic (MZ) and dizygotic (DZ) twin pairs. A total of 531 individuals (151 families) were studied: 115 individuals (34 families) from Malawi; 100 individuals (19 families) from Venezuela; and 316 individuals (98 families) from the USA (see Table S2 for subject characteristics; note that all except 35 adults and one child from the USA were explicitly recruited for this study).
DNA was extracted from a single fecal sample donated by each person. Variable region 4 (V4) of bacterial 16S rRNA genes present in each fecal community was amplified by PCR and the resulting amplicons sequenced on an Illumina HiSeq 2000 instrument (n=1,803,250±562,877 reads/fecal sample; 1,093,740,274 total reads, Table S2a) to define the phylogenetic types (phylotypes) present. Species-level bacterial phylotypes were defined as organisms sharing ≥97% nucleotide sequence identity (%ID) in the V4 regions of their 16S rRNA genes10. In addition, we characterized functions encoded in community DNA by performing multiplex shotgun 454 pyrosequencing of fecal DNA from a subset of 110 fecal samples, encompassing 43 families with members matched as closely as possible for age (155,890±87,083 reads/sample; total size of dataset, 5.9 Gb; Table S2b). The resulting shotgun reads were annotated with KEGG Orthology group (KO) assignments and with enzyme classification (EC) numbers (KEGG version 58).
Changes in the taxonomic/phylogenetic composition of fecal communities as a function of age and population
Many reports have examined the bacterial species content of the gastrointestinal tracts of infants and children within one population using culture-based methods. Far fewer studies have attempted to compare the gut communities of humans living in markedly different socio-economic, geographic, and cultural settings11,12. Culture-independent techniques have been used to define the gut microbiota at various points in postnatal development6,13, but have been limited either by the analytic methods used, by the low number of subjects examined, or by the scope of the populations surveyed. These studies have nonetheless provided important insights. Using 16S rRNA gene-based microarrays, Palmer et al14 observed considerable intra- and interpersonal variation in fecal bacterial community structures during the first year of life in 12 unrelated children and 1 twin pair. Interpersonal variation was less within the twin pair, and intrapersonal variation decreased as a function of age. Penders and coworkers15 performed a qPCR study of five bacterial taxa in the fecal microbiota of 1,032 Dutch infants at 1 month of age, and documented differences based on birth mode (Caesarian versus vaginal; also see ref 8).
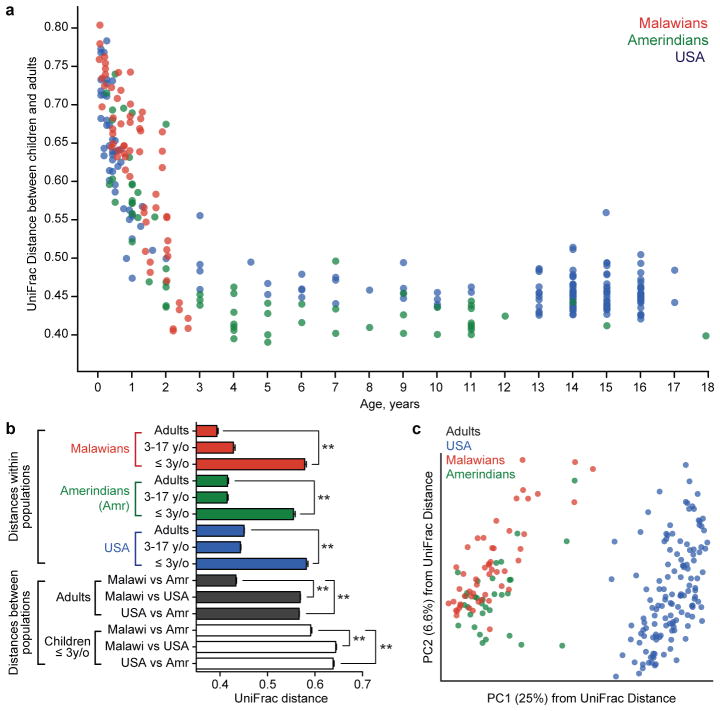
We collected bacterial V4-16S rRNA data from 326 individuals aged 0–17 years (83 Malawian, 65 Amerindian and 178 residents of the USA) plus 202 adults aged 18–70 years (31 Malawians, 35 Amerindians, and 136 residents of the USA). 16S rRNA datasets were first analyzed using UniFrac, an algorithm that measures similarity between microbial communities based on the degree to which their component taxa share branch length on a bacterial tree of life16. There were several notable findings. First, the phylogenetic composition of the bacterial community evolved towards an adult-like configuration within the 3-year period following birth in all three populations (Fig. 1a, Fig. S1). Second, interpersonal variation is significantly greater between children than between adults; this finding is robust to geography (Fig. 1b; also see ref. 4). Third, there were significant differences in the phylogenetic composition of fecal microbiota between individuals living in the different countries, with especially pronounced separation occurring between USA compared to Malawian and Amerindian gut communities; this was true for individuals aged 0–3 years, 3–17 years, and for adults (Fig. 1b, Table S3). Unsupervised clustering using Principal Coordinates Analysis (PCoA) of UniFrac distance matrices indicated that age and geography/cultural traditions primarily explain the variation in our dataset, where USA microbiota clustered separately from non-USA along principal coordinate 1 (Fig. 1c, S2). However, within the non-USA populations, separation between Malawians and Amerindians was also observed (along principal coordinate 3 in the case of adults, Fig. S2f). We did not find any significant clustering by a village for Malawians and Amerindians or by region within the USA. Fourth, bacterial diversity increased with age in all three populations (Fig. 2a,b). The fecal microbiota of USA adults was the least diverse compared to the two other populations (Fig. 2c, p<0.005, ANOVA with Bonferroni post-hoc test): these differences were evident in children older than 3 years of age (p<0.005, ANOVA with Bonferroni post-hoc test), but not in younger subjects.
Fig. 1. Differences in the fecal microbial communities of Malawians, Amerindians and residents of the USA at different ages.
(a) UniFrac distances between children and adults decrease with increasing age of children in each population. Each point shows an average distance between a child and all adults unrelated to that child but from the same country. Results are derived from bacterial V4-16S rRNA datasets. (b) Large interpersonal variations are observed in the phylogenetic configurations of fecal microbial communities at early ages. Malawian and Amerindian children and adults are more similar to one another than to USA children and adults. UniFrac distances were defined from bacterial V4-16S rRNA data generated from the microbiota of 181 unrelated adults (≥18 years old) and 204 unrelated children (n=31 Malawians 0.03–3 years old, 21 3–17 years old; 30 Amerindians 0.08–3 years old, 29 3–17 years old; 31 residents of the USA 0.08–3 years old, 62 sampled at 3–17 years of age). Abbreviations: * p<0.05; **p<0.005 (Student’s t-test with 1000 Monte Carlo simulations). See Table S3 for a complete description of the statistical significance of all comparisons shown in the Figure. (c) PCoA of unweighted UniFrac distances for the fecal microbiota of adults.
Fig. 2. Bacterial diversity increases with age in each population.
Number of observed 97%ID OTUs plotted against age for (a) all subjects, (b) during the first 3 years of life, and (c) adults (in the latter panel, average values ± SEM are plotted). *p<0.05, **p<0.005 (ANOVA with Bonferroni post-hoc test).
We next used the nonparametric Spearman rank correlation to determine which bacterial taxa change monotonically with increasing age within and between the 3 sampled populations. We only considered children who were breastfed and used datasets obtained from the V4 region of the 16S rRNA gene as well as datasets of shotgun pyrosequencing reads from the fecal microbiomes of the 110 sampled individuals [24 babies (0.6–5 months old), 60 children and adolescents (6 months to 17 years old) and 26 adults]. Shotgun reads were mapped to 126 sequenced human gut-derived microbial species (Table S4). The advantage of using these 126 gut microbes as a reference database is that spurious hits of shotgun microbiome reads to taxa that are not present in the gut are minimized. Nonetheless, when we repeated the entire analysis, blasting 1,280 genomes in KEGG, the results were similar (Fig. S3). Phylotypes belonging to Bifidobacterium longum exhibited significant decline in proportional representation with increasing age in all 3 populations (Fig. S3a). The majority (75±20%) of all shotgun and 16S rRNA-V4 sequences in all babies mapped to members of the genus Bifidobacterium. Bifidobacteria continued to dominate fecal communities throughout the first year of life although their proportional representation diminished during this period, in agreement with the results of several studies of small numbers of children4,6,7 (Fig. S3). Table S5 lists the species-level bacterial taxa whose representation changes significantly with age in all three populations, as well as species that change in a population-specific manner as defined from analysis of the shotgun sequencing data that were available from 110 of the 531 individuals.
We also used Random Forests, a supervised machine learning technique17, and the V4-16S rRNA datasets obtained from all 528 individuals to identify bacterial species-level operational taxonomic units (OTUs) that differentiate fecal community composition in children and adults within and between the three populations. The purpose of a classifier such as Random Forests is to learn a function that maps a set of input values or predictors (here, relative OTU abundances in a community) to a discrete output value (here, USA versus non-USA microbiota). Random Forests is a particularly powerful classifier that can exploit non-linear relationships and complex dependencies between OTUs. The measure of the method’s success is its ability to correctly classify unseen samples, estimated by training it on a subset of samples, and using it to classify the remaining samples (cross-validation). The cross-validation error is compared to the baseline error that would be achieved by always guessing the most common category. As an added benefit, Random Forests assigns an importance score to each OTU by estimating the increase in error caused by removing that OTU from the set of predictors. In our analysis, we considered an OTU to be highly predictive if its importance score was at least 0.001; all error estimates and OTU importance scores were averaged over 100 rarefactions at the same sample size for each community (305,631 sequences) in order to control for sequencing effort.
Random Forests analysis confirmed the dominance of Bifidobacterium in the baby microbiota (Table S6a). For adults, Random Forests revealed distinct community signatures for Western (USA) and non-Western individuals (baseline error=0.289, cross-validation error=0.011 ± 0.000). Of the 92 highly predictive species-level OTUs shown in Table S6b, 73 were over-represented in non-USA adults, and 23 of the 73 were assigned to the genus Prevotella. Malawians and Amerindians could also be distinguished from each other, although the difference was less extreme than the USA versus non-USA comparison (baseline error=0.407, cross-validation error=0.018 ± 0.009, 56 highly predictive OTUs, Table S6c). Only 28 OTUs distinguished USA and non-USA infants (Table S6d). Intriguingly, three OTUs assigned to genus Prevotella were overrepresented in the USA infant microbiota, unlike the result observed in adults (Table S6d). Twenty-one OTUs discriminated Malawian and Amerindian baby microbiomes, 18 of which were overrepresented in the former: the majority belonged to family Enterococcaceae (Table S6d). Thus, a Western (USA) lifestyle appears to affect the bacterial component of the gut microbiota systematically, although this influence is subtle compared to the high degree of variability observed in infants and children within each population (perhaps analogous to human genetic variability, in which variation among populations is small compared to variation within populations).
Confirming the importance of Prevotella as a discriminatory taxon, a recent study also showed that this genus was present in higher abundance in the fecal microbiota of children living in West Africa (Burkina Faso) compared to children living in Europe (Italy)11. Additionally, a member of this genus is one of three bacterial species that, in European adults, distinguishes strongly among three clusters, or enterotypes, of gut microbiota configurations that are claimed to be reproducible across Western adult populations18. Therefore, we asked whether the fecal microbiota of infants and adults in each of our three geographically distinct populations fell into natural discrete clusters19. We did not find strong evidence for discrete clustering (see Methods), but rather for variation driven in adults by a trade-off between Prevotella and Bacteroides (Fig. S4a). Including infants introduces a new, strongly supported gradient driven by Bifidobacteria, generally orthogonal to the Bacteroides/Prevotella trade-off (Fig. S4b). Clustering of sub-populations of increasing minimum age indicates that adult cluster membership is generally consistent, but that children between 0.6 years and 1 year of age may be clustered with adults or with younger children, depending on whether the younger children are included in the analysis (Fig. S4c).
In summary, this clustering analysis suggests that some features of normal variation in the bacterial composition of the gut microbiota, such as the Prevotella/Bacteroides trade-off, are highly reproducible even in human population subsets of reduced variability. However, a complete description of variation in the human gut microbiota will require a substantially broader cross-cultural and cross-age sampling. Importantly, the observed age- and geographic patterns were also detected with lower sequencing coverage (see Supplementary Results for a discussion of the influence of sequencing depth on performance of Random Forests and PCoA analyses (Fig. S5, S6), and our analysis of non-bacterial taxa that vary with age and population, Fig. S7).
Shared functional changes in the microbiome as children mature
Few studies have described changes in the gene content of the gut microbiome as a function of age: the largest study reported to date was carried out in 13 healthy Japanese individuals (5 children, the youngest 3 months old, plus 8 adults)4. Our shotgun sequencing dataset from 110 individuals allowed us to characterize the representation of functional gene groups [KEGG Orthology (KO) annotations and Enzyme Commission numbers (ECs)] in microbiomes representing broader age groups (youngest 3 weeks), and several distinct geographic locations/cultural traditions. We used Hellinger distance measurements to show that just as children are significantly more different from one another than are adults in terms of their fecal bacterial community phylogenetic structure, they are also more different in terms of their repertoires of microbiome-encoded functions, as defined by the proportional representation of EC and KO assignments. Moreover, as with UniFrac distances, Hellinger distances were greater between the USA and the other two populations at all ages sampled (Fig S8). Of interest is the concordance of patterns of covariance between the two data types: Procrustes analysis disclosed that the goodness of fit was significant (P<0.001 with 1,000 iterations) whether UniFrac (the most appropriate metric for 16S rRNA data) or Hellinger distances (for consistency with the method used on the KEGG EC and KO data) were used to reduce the OTU table (Fig. S9a,b plus data not shown). Annotation of shotgun reads from the microbiomes using the COG database produced similar concordance with 16S rRNA datasets (Fig. S9c).
When examining KEGG EC profiles across 110 fecal microbiomes, we obtained the remarkable result that there were no ECs identified as being unique to adults (n=26) or babies (less than 6 months old, n=24). Moreover, the total number of ECs found in adults was not significantly different compared to the total number of ECs scored in babies (sampling normalized to coverage in Fig. S10a). This finding was robust to culture/geography. The fraction of sequences with assignable KEGG EC annotations declined with increasing age in all 3 populations (Fig. S10b). This may be due to the increased complexity of the adult microbiome, with fewer representative species characterized by genome sequencing, genetic manipulation, or biochemically (also see Supplementary Results and Figs. S11, S12 for a comparison of our dataset to a published dataset of fecal microbiomes sampled from 124 adults living in Denmark and Spain2).
We used ShotgunFunctionalizerR20, a software tool designed for metagenomic analysis and based on a Poisson model, to identify 1,008 ECs whose proportional representation in fecal microbiomes differed significantly between all sampled breast-fed babies and all adults irrespective of their geographic location; 530 were significantly higher in adults (p<0.0001, Table S7). A prominent example of these shared age-related changes involves the metabolism of vitamins B12 (cobalamin) and folate. In contrast to folate, which is synthesized by both microbes and plants, cobalamin is produced primarily by microbes21. The gut microbiomes of babies are enriched in genes involved in the de novo biosynthesis of folate, while those of adults have a significantly higher representation of genes that metabolize dietary folate and its reduced form tetrahydrofolate (THF, Fig. S13, Table S7). Unlike de novo folate biosynthetic pathway components, which decrease with age, the proportional representation of genes encoding the majority of enzymes involved in cobalamin biosynthesis increase with age (Fig. S14, S15, Table S7). The folate and cobalamin pathways are linked functionally: methionine synthase (EC 2.1.1.13); this E.C., whose representation also increases with age, catalyzes formation of THF from 5-Methyl-THF and L-homocysteine and requires cobalamin as a cofactor (Fig. S13).
The low relative abundance of ECs involved in cobalamin biosynthesis in the fecal microbiomes of babies correlates with the lower representation of members of Bacteroidetes, Firmicutes, and Archaea in their microbiota (see Fig. S16 for Spearman correlation coefficients). While the biosynthetic pathway for cobalamin is well represented in the genomes of these organisms (Fig. S16), Bifidobacterium, Streptococcus, Lactococcus, Lactobacillus which dominate the baby gut microbiota (Table S5, Fig. S3), are deficient in these genes (Fig. S16). In contrast, a number of these early gut colonizers contain ECs involved in folate biosynthesis/metabolism (Fig. S16). The traditional view of the developing infant gut is that the principal change is in the representation of Bifidobacteria. Although differences in representation of Bifidobacteria contribute to this effect, differences in vitamin metabolism among the rest of the bacteria remain even when all Bifidobacteria reads were excluded (data not shown). These changes in vitamin biosynthetic pathway representation in the microbiome correlate with published reports indicating that blood levels of folate decrease and cobalamin increase with age22.
Besides cobalamin and folate, the relative abundance of ECs involved in the biosynthesis of vitamins B7 (biotin) (biotin synthase, EC2.8.16) and B1 (thiamine) (thiamine-phosphate diphosphorylase, EC2.5.1.3) are significantly higher in adult microbiomes compared to the microbiomes of babies (Fig. S17, Table S7). Together, these findings suggest that the microbiota should be considered when assessing the nutritional needs of humans at various stages of development.
Random Forests analysis asks a somewhat different statistical question from ShotgunFunctionalizeR: i.e., which genes or species are most discriminatory among different class labels, rather than which are most over/underrepresented, and tends to identify fewer features than does ShotgunFunctionalizeR when applied to the same data. Random Forests analysis yielded 107 ECs that best discriminate the adult and baby microbiomes (Table S7). These predictive ECs were among the most significantly different ECs determined by ShotgunFunctionalizeR and included ECs involved in the metabolism of cobalamin and folate (Table S7). In addition, Random Forests revealed that ECs involved in fermentation, methanogenesis and metabolism of arginine, glutamate, aspartate and lysine were higher in the adult microbiomes, while ECs involved in the metabolism of cysteine and fermentation pathways found in lactic acid bacteria [acetolactate decarboxylase (EC4.1.1.5) and 6-phosphogluconate dehydrogenase (EC1.1.1.44)] were represented primarily in baby microbiomes (Fig. S17).
Comparison of the representation of KOs, between baby and adult microbiomes, yielded essentially the same results as those reported with ECs. The only novel finding was the overrepresentation of KOs assigned to a wide variety of ABC transporters in baby microbiomes (Table S7b).
Population- and age-specific differences in the representation of microbiome functions
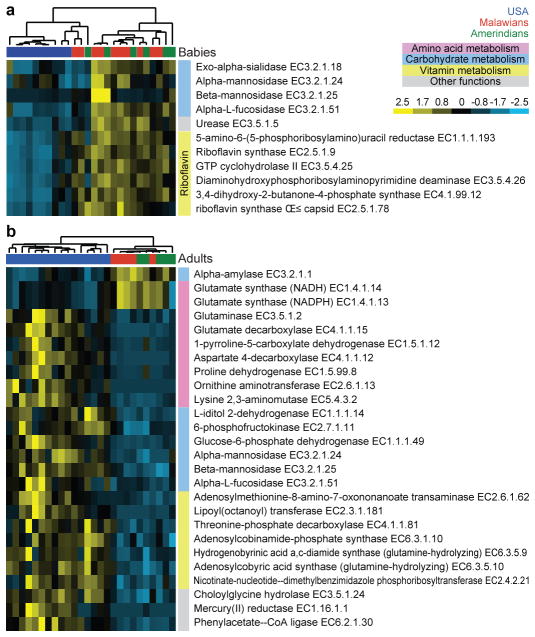
ShotgunFunctionalizeR, Random Forests and Spearman rank correlation analyses were all used to compare EC representation in fecal microbiomes as a function of predefined categories of geographic location and age. 476 ECs were identified as being significantly different in the USA versus Malawian and Amerindian breast-fed babies (p<0.0001, ShotgunFunctionalizeR; Table S8). The most prominent differences involved pathways related to vitamin biosynthesis and carbohydrate metabolism. Malawian and Amerindian babies had higher representation of ECs that were components of the vitamin B2 (riboflavin) biosynthetic pathway (Figs. 3a, S18). These differences were not evident in adults (Table S7). Riboflavin is found in human milk and in meat and dairy products. We did not measure the levels of these vitamins in mothers and in their breast milk in the sampled populations, although it is tempting to speculate the observed differences in baby microbiomes may represent an adaptive response to vitamin availability.
Fig. 3. Differences in the functional profiles of fecal microbiomes in the three study populations.
Examples of KEGG ECs that exhibited the largest differences, as determined by Random Forests and ShotgunFuntionalizeR analyses, in proportional representation between USA and Malawian/Amerindian populations. Shown are the relative abundances of genes encoding the indicated ECs (normalized by Z-score across all datasets). (a) UPGMA clustering of 10 USA, 10 Malawian and 6 Amerindian baby fecal microbiomes. (b) UPGMA clustering of 16 USA, 5 Malawian and 5 Amerindian adult fecal microbiomes.
Studies in gnotobiotic mouse models indicate that the ability of members of the microbiota to access host-derived glycans plays a key role in establishing a gut microbial community23,24. As expected4,5, compared to adults, baby microbiomes were enriched in ECs involved in foraging of glycans represented in mother’s milk and the intestinal mucosa (mannans, sialylated glycans, galactose, fucosyloligosaccharides; Table S7). A number of genes involved in utilizing these host glycans are significantly overrepresented in Amerindian and Malawian compared to USA baby microbiomes, most notably exo-alpha-sialidase and alpha-L-fucosidase (Fig. 3a, Table S8). These population-specific biomarkers may reflect differences in the glycan content of breast milk. In fact, the representation of these glycoside hydrolases decreases as Malawian and Amerindian babies mature and transition to a diet dominated by maize-, cassava-and other plant-derived polysaccharides. In contrast, alpha-fucosidase gene representation increases as USA infants age and are exposed to diets rich in readily absorbed sugars (Fig. S19d, Table S9).
Another biomarker that distinguishes microbiomes based on age and geography is urease (EC3.5.1.5). Urease gene representation is significantly higher in Malawian and Amerindian baby microbiomes and decreases with age in these two populations, unlike in the USA where it remains low from infancy through adulthood (Fig. 3a, Fig. S19e). Urea comprises up to 15% of the nitrogen present in human breast milk25. Urease releases ammonia that can be used for microbial biosynthesis of essential and nonessential amino acids26,27. Furthermore, urease plays a major role in nitrogen recycling, particularly when diets are deficient in protein28,29. Under conditions where dietary nitrogen is limiting, the ability of the microbiome to utilize urea would presumably be advantageous to both the microbial community and host. Urease activity has been characterized previously in Streptococcus thermophilus30. While most attribute urease to Helicobacter and Proteus spp., the relative abundance of members of these two genera was low (<0.05%) and not significantly different between the three populations. Our analysis of shotgun reads that matched to the 126 gut genomes revealed that the representation of five species that possess EC3.5.1.5 (Bacteroides cellulosilyticus WH2, Coprococcus comes, Roseburia intestinalis, Streptococcus infantarius and S. thermophilus) was significantly higher in Malawian and Amerindian compared to USA baby microbiomes (Table S5).
Additional support of the role of diet in shaping the infant gut microbiome comes from the differences detected between the breast-fed and formula-fed babies who were part of the USA infant twin cohort (see Supplementary Results and Figs. S2c, S8c, and S20).
Differences in adult fecal microbiomes associated with geography/cultural traditions
Annotation of the shotgun sequencing datasets yielded a total of 1,349 ECs in the 26 adults surveyed: ShotgunFunctionalizeR revealed that the representation of genes encoding 893 of these ECs were significantly different in USA versus Malawian/Amerindian fecal microbiomes (p<0.005 after multiple comparison correction; 433 overrepresented in USA samples). By contrast, at this threshold only 445 ECs were identified as different between Malawian and Amerindian adults (see Table S10 for a complete list). The Random Forests classifier revealed 52 ECs that were best at discriminating USA versus non-USA adult fecal microbiomes. These ECs were also identified by ShotgunFunctionalizeR as the most significantly different (Table S10).
A typical USA diet is rich in protein, while diets in Malawi and Amerindian populations are dominated by corn and cassava (Table S1). The differences between USA and Malawian/Amerindian microbiomes can be related to these differences in their diets. ECs whose representation are most significantly enriched in USA fecal microbiomes parallel differences observed in carnivorous versus herbivorous mammals31: ECs encoding glutamate synthase have higher proportional representation in Malawian and Amerindian adult microbiomes and are also higher in herbivorous mammalian microbiomes31 (Fig. 3b), while degradation of glutamine was overrepresented in USA as well as carnivorous mammalian microbiomes. Several ECs involved in the degradation of other amino acids were overrepresented in adult USA fecal microbiomes: aspartate (EC4.1.1.12), proline (EC1.5.99.8), ornithine (EC2.6.1.13) and lysine (EC5.4.3.2) (Fig. 3b), as were ECs involved in catabolism of simple sugars (glucose-6-phosphate dehydrogenase, 6-phosphofructokinase), sugar substitutes (L-iditol 2-dehydrogenase, which degrades sorbitol), as well as host glycans (alpha-mannosidase, beta-mannosidase, alpha-fucosidase, Fig. 3b). In contrast, alpha-amylase (EC 3.2.1.1), which participates in the degradation of starch, was overrepresented in the Malawian and Amerindian microbiomes, reflecting their corn-rich diet.
USA microbiomes also had significant overrepresentation of ECs involved in the vitamin biosynthesis [cobalamin (Fig. 3b, Fig. S10), biotin and lipoic acid (Fig. 3b)], the metabolism of xenobiotics [phenylacetate CoA ligase (EC 6.2.1.30) which participates in the metabolism of aromatic compounds) and mercury reductase (EC1.16.1.1)], and bile salt metaboism [choloyglycine hydrolase (EC3.5.1.24), perhaps reflecting a diet richer in fats) (Fig. 3b)].
Effects of kinship on the microbiome across countries
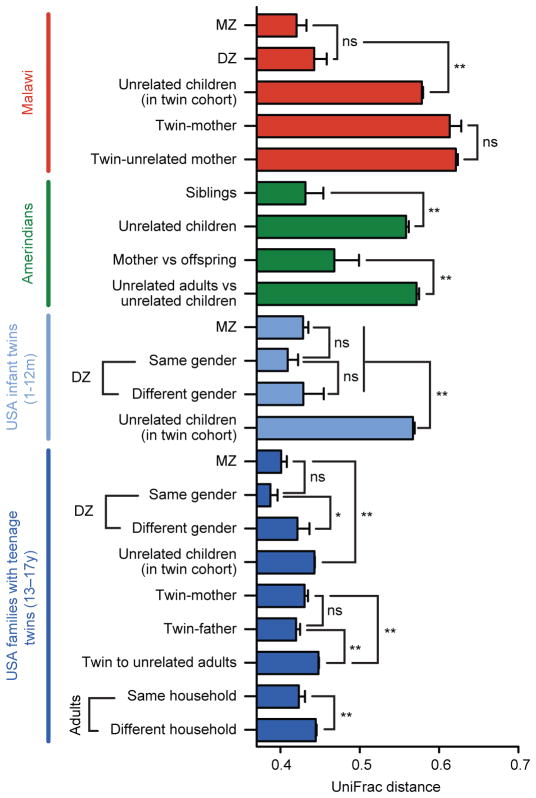
Differences in social structures may influence the extent of vertical transmission of the microbiota and the flow of microbes and microbial genes among members of a household. Differences in cultural tradition also affect food, exposure to pets and livestock, and many other factors that could influence how and from where a gut microbiota/microbiome is acquired. We previously observed that adult MZ twins are no more similar to one another in terms of their gut bacterial community structure than adult DZ twins32. This result suggests that the overall heritability of the microbiome is low. We confirmed that the phylogenetic architecture of the fecal microbiota of MZ Malawian co-twins ≤3 years of age is not more similar than the microbiota of similarly aged DZ co-twins (n=15 MZ and 6 DZ twin pairs). We found that this is also true for MZ and DZ twin pairs aged 1–12 months of age (n=16 twin pairs), as well as teenaged twins (13–17 years-old; n=50 pairs) living together in the USA (Fig. 4).
Fig. 4. Differences in the fecal microbiota between family members across the three populations studied.
UniFrac distances between the fecal bacterial communities of family members were calculated (n=19 Amerindian, 34 Malawian and 54 USA families with teenage twins). Mean ± SEM values are plotted. The UniFrac matrix was permutated 1000 times; p values represent the fraction of times permuted differences were greater than real differences: ns (not significant; p>0.05), * p<0.05, **p<0.005.
Although biological mothers are in a unique position to transmit an initial inoculum of microbes to their infants during and following birth, our analysis of mothers of teenage USA twins showed that their fecal microbiota were no more similar to their children than were those of biological fathers, and that genetically unrelated but co-habiting mothers and fathers were significantly more similar to one another microbially than were members of different families (Fig. 4; note that no fathers were sampled in Malawi and only 4 fathers in the Amerindian cohort). These latter observations emphasize the importance of a history of numerous common environmental exposures in shaping gut microbial ecology. Moreover, the similarity in overall pattern of the effects of kinship on microbial community structure suggests that despite the large influence of cultural factors on which microbes are present in both children and adults in each population, the bases for the degree of similarity among members of a family are consistent across the three populations studied.
Prospectus
Our results emphasize that it is essential to sample a broad population of healthy humans over time, both in terms of their age, geography and cultural traditions, in order to discover features of gut microbiomes that are unique to different locations and lifestyles. In addition, we need to understand how the pressures of westernization are changing the microbial parts of our genetic landscape - changes that potentially mediate the suite of pathophysiological states correlated with Westernization. Finally, given the need for global policies about sustainable agriculture and improved nutrition, it will be important to understand how we can match these policies not only to our varying cultural conditions but also to our varied gut microbiomes.
METHODS SUMMARY
Sample collection
Subjects were recruited for the present study using procedures approved by Human Studies Committees from Washington University in St. Louis, the University of Pennsylvania, the University of Colorado, Boulder, the University of Malawi, the University of Puerto Rico, and the Venezuelan Institute for Scientific Research (IVIC). Each fecal sample was frozen within 30 min of donation.
Multiplex DNA sequencing
Extracted genomic DNA was subjected to multiplex Illumina sequencing of V4 region of 16S rRNA genes, as well as multiplex 454 pyrosequencing of total community DNA. See Methods for further details about the analysis. DNA sequences can be found in MG-RAST server under accession numbers “qiime:850” for Illumina V4-16S rRNA, and “qiime:621” for microbiome shotgun sequences.
Supplementary Material
Acknowledgments
We thank Sabrina Wagoner, Jill Manchester for superb technical assistance, plus Brian Muegge, Andrew Grimm, Ansel Hsiao, Nicholas Griffin, and Philip Tarr for helpful suggestions, and Malick Ndao, Tara Tinnin and R. Mkakosya for patient recruitment and/or technical assistance. This work was supported in part by grants from the NIH (DK078669, T32-HD049338), St. Louis Children’s Discovery Institute (MD112009-201), the Howard Hughes Medical Institute, the Crohn’s and Colitis Foundation of America, and the Bill and Melinda Gates Foundation. Parts of this work utilized the Janus supercomputer, which is supported by National Science Foundation grant CNS-0821794, the University of Colorado, Boulder, the University of Colorado, Denver, and the National Center for Atmospheric Research.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions
T.Y., R.K. and J.I.G designed the experiments, M.M., I.T., M.G. D-B., M.C., M.M., G.H., A.C.H., A.P.A, R.K., R.N.B., C.A.L., C.L. and B.W. participated in patient recruitment, T.Y. generated the data, T.Y., F.E.R., J.C. M., J.R., J.K., J.G.C., D.K., R.K. and J.I.G. analyzed the results, T.Y., R.K. and J.I.G. wrote the paper.
References
- 1.Mueller S, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Research. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koenig JE, et al. Microbes and Health Sackler Colloquium: Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2010;108 (Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tannock GW. What immunologists should know about bacterial communities of the human bowel. Seminars in Immunol. 2007;19:94–105. doi: 10.1016/j.smim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peach S, Fernandez F, Johnson K, Drasar BS. The non-sporing anaerobic bacteria in human faeces. J Med Microbiol. 1974;7:213–221. doi: 10.1099/00222615-7-2-213. [DOI] [PubMed] [Google Scholar]
- 13.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 14.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penders J, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 16.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Micro Rev. 2011;35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 18.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristiansson E, Hugenholtz P, Dalevi D. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics. 2009;25:2737–2738. doi: 10.1093/bioinformatics/btp508. [DOI] [PubMed] [Google Scholar]
- 21.Krautler B. Vitamin B12: chemistry and biochemistry. Biochem Soc Trans. 2005;33:806–810. doi: 10.1042/BST0330806. [DOI] [PubMed] [Google Scholar]
- 22.Monsen AL, Refsum H, Markestad T, Ueland PM. Cobalamin status and its biochemical markers methylmalonic acid and homocysteine in different age groups from 4 days to 19 years. Clin Chem. 2003;49:2067–2075. doi: 10.1373/clinchem.2003.019869. [DOI] [PubMed] [Google Scholar]
- 23.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harzer G, Franzke V, Bindels JG. Human milk nonprotein nitrogen components: changing patterns of free amino acids and urea in the course of early lactation. Am J Clin Nutr. 1984;40:303–309. doi: 10.1093/ajcn/40.2.303. [DOI] [PubMed] [Google Scholar]
- 26.Metges CC, et al. Incorporation of urea and ammonia nitrogen into ileal and fecal microbial proteins and plasma free amino acids in normal men and ileostomates. Am J Clin Nutr. 1999;70:1046–1058. doi: 10.1093/ajcn/70.6.1046. [DOI] [PubMed] [Google Scholar]
- 27.Millward DJ, et al. The transfer of 15N from urea to lysine in the human infant. Br J Nutr. 2000;83:505–512. [PubMed] [Google Scholar]
- 28.Meakins TS, Jackson AA. Salvage of exogenous urea nitrogen enhances nitrogen balance in normal men consuming marginally inadequate protein diets. Clin Sci (Lond) 1996;90:215–225. doi: 10.1042/cs0900215. [DOI] [PubMed] [Google Scholar]
- 29.Langran M, Moran BJ, Murphy JL, Jackson AA. Adaptation to a diet low in protein: effect of complex carbohydrate upon urea kinetics in normal man. Clin Sci (Lond) 1992;82:191–198. doi: 10.1042/cs0820191. [DOI] [PubMed] [Google Scholar]
- 30.Mora D, et al. Characterization of urease genes cluster of Streptococcus thermophilus. J Appl Microbiol. 2004;96:209–219. doi: 10.1046/j.1365-2672.2003.02148.x. [DOI] [PubMed] [Google Scholar]
- 31.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufman L, Rousseeuw PJ. Finding groups in data : an introduction to cluster analysis. Wiley; 1990. [Google Scholar]
- 35.Rousseeuw PJ. Silhouettes - a Graphical Aid to the Interpretation and Validation of Cluster-Analysis. J Computational Appl Mathematics. 1987;20:53–65. [Google Scholar]
- 36.Tibshirani R, Walther G. Cluster validation by prediction strength. J Computational and Graphical Statistics. 2005;14:511–528. [Google Scholar]
- 37.Teal TK, Schmidt TM. Identifying and removing artificial replicates from 454 pyrosequencing data. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5409. [DOI] [PubMed] [Google Scholar]
- 38.Team, RDC. R Foundation for Statistical Computing. 2010. [Google Scholar]
- 39.Wiener ALAM. Classification and Regression by Random Forest. R News. 2002;2:18–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.