Abstract
Emiliania huxleyi, an environmentally important marine microalga, has a bloom- and-bust lifestyle in which massive algal blooms appear and fade. Phaeobacter gallaeciensis belongs to the roseobacter clade of α-Proteobacteria, whose populations wax and wane with that of E. huxleyi. Roseobacter are thought to promote algal growth by biosynthesizing and secreting antibiotics and growth stimulants (auxins). Here we show that P. gallaeciensis switches its secreted small molecule metabolism to the production of potent and selective algaecides, the roseobacticides, in response to p-coumaric acid, an algal lignin breakdown product that is symptomatic of aging algae. This switch converts P. gallaeciensis into an opportunistic pathogen of its algal host.
While bacteria use a vast repertoire of small molecules to sense and respond to the world around them, relatively few of these molecular signals and biosynthetic responses have been characterized in detail1–4. Most of the responsible molecules remain to be discovered, and these discoveries can lead to interesting chemical and biological insights5–7. Studying the intimate associations (symbioses) between bacteria and other organisms can be especially revealing, as the participants will have frequently evolved a distinctive set of molecules with specialized functions8. Some of the most abundant and least investigated of these symbioses occur in the oceans between members of the roseobacter clade and microscopic algae9.
Emiliania huxleyi, a microscopic alga covered with elaborate CaCO3 disks, drifts freely in the sunlit layers of the ocean where it and related marine phytoplankton photosynthesize nearly half of the planet’s O210. In addition to producing O2 and reduced organic molecules, E. huxleyi plays an important role in global carbon cycling by removing CO2 from the ocean and sequestering it as CaCO311,12. E. huxleyi can be found in most of the world’s oceans, and often forms massive blooms (>105 km2) with more than 106 cells/L, which because of their size and the reflectivity of CaCO3 in their coccoliths, can be easily detected by satellites13–15. During a bloom, E. huxleyi can account for 80–90% of the phytoplankton cells in the area, and bacteria belonging to the roseobacter clade, a diverse group within the α-Proteobacteria, account for up to 60% of the bacterial community9,16. Because of the correlation between the microalgal blooms and roseobacter predominance, associations between the two are thought likely.
Recent studies have shown that microalgae and roseobacter can be attracted to or repelled by each other, which suggests an intermittent symbiosis17–21. Since many microalgae cannot grow normally in marine settings without bacterial symbionts, bacterial colonization of microalgae clearly has a beneficial role22,23. The molecular basis of the bacterially conferred benefit is not well understood, but some studies suggest that roseobacter metabolites may be antibiotics and auxins that suppress the growth of potentially parasitic bacteria and promote algal growth, respectively20,24–27. The microalgae, in turn, could contribute nutrients and a suitable surface for roseobacter colonization and biofilm formation as roseobacters have host-associated lifestyles28. In one such example, the roseobacter species, Phaeobacter gallaeciensis, which associates with marine eukaryotes including algae20,29, produces the auxin phenylacetic acid (Fig. 1a, 1), and the potent broad spectrum antibiotics tropodithietic acid (TDA, Fig. 1a, 2) and its valence tautomer thiotropocin (Fig. 1a, 3)24,25,30. How this apparently mutualistic P. gallaeciensis-microalga symbiosis changes as algal growth inevitably wanes is unknown31,32. Here we report that p-coumaric acid (pCA, Fig. 1a, 4) a small molecule generated by E. huxleyi induces P. gallaeciensis BS107 to produce potent but selective algaecides, which we have named roseobacticides A and B. Roseobacticide biosynthesis likely involves an alternative use of the compounds employed in antibiotic and auxin production and transforms a bacterial mutualist into an opportunistic pathogen.
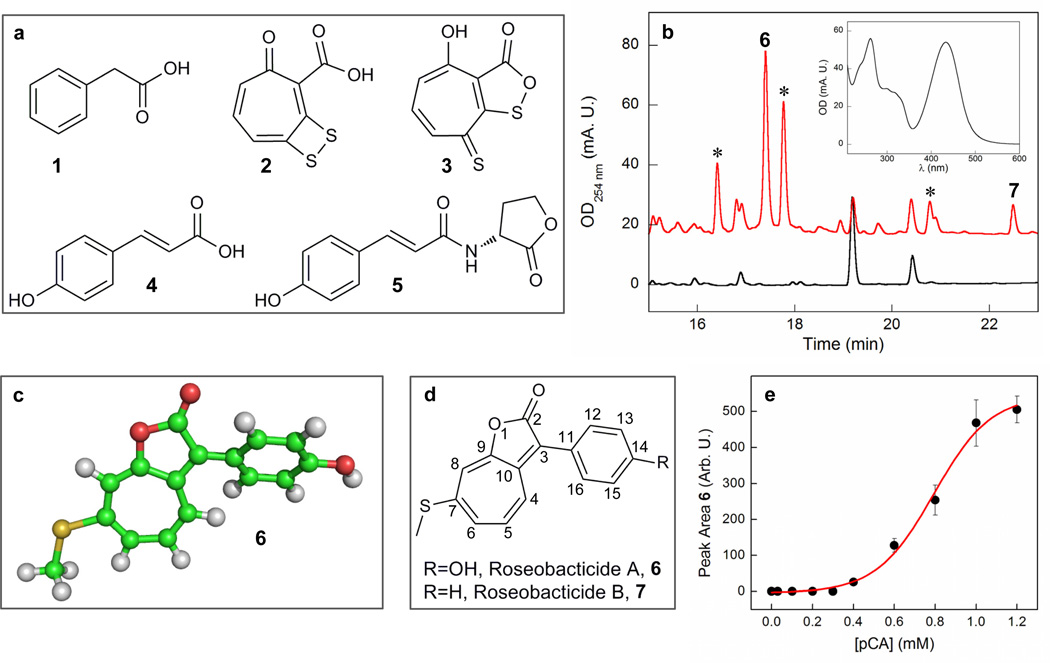
Figure 1.
Effect of pCA on secondary metabolites produced by P. gallaeciensis. (a) Structures of phenylacetic acid (1), TDA (2), its valence tautomer (3), pCA (4) and pCA-HSL (5). (b) HPLC-MS profile of the EtOAc extract of P. gallaeciensis BS107 cultures 72 h after inoculation in the absence (black trace) and presence (red trace) of 1 mM pCA. The starred peaks contain the 430 nm absorption feature typical for roseobacticides; their structures have yet to be determined. Inset, UV-visible absorbance spectrum of 6. (c) Crystal structure of 6 solved to 0.82 Å resolution. (d) Structures of 6 and 7 and their numbering schemes. (e) Dose response of 6 as a function of [pCA]. Each point is the average of two independent measurements. The red line is a dose-response fit and yields an EC50 of 0.79 ± 0.03 mM (see Supplementary Methods for details).
Results and discussion
We thought it likely that the algal-bacterial symbiosis would change as the microalgae aged. As algae senesce, their cell walls deteriorate and breakdown products are released into the surrounding medium. Since lignin and lignin components were recently identified in red, green and brown algal cells, lignin breakdown products such as p-coumaric acid (pCA, Fig. 1a, 4), which could indicate algal senescence, were tested as signals33–35. In addition to serving as a small molecule proxy of algal health, pCA has also been shown to be taken up by diverse bacteria – including the roseobacter Silicibacter pomeroyi – and used to biosynthesize p-coumaroyl-homoserine lactone (pCA-HSL, Fig. 1a, 5), a hybrid signaling molecule that can globally regulate gene expression36.
Bioinformatic and chemical analyses were employed to assess pCA biosynthesis in E. huxleyi. Bioinformatic analysis of the incomplete E. huxleyi genome (strain CCMP1516) showed orthologs for all five enzymes required to make H lignin from phenylalanine via pCA (Supplementary Table S1)37,38. The activity and function of these enzymes were confirmed by HPLC-MS analysis in two different E. huxleyi strains (CCMP1516 and CCMP372), which revealed pCA as a major secreted metabolite in these cultures (Supplementary Fig. S1). E. huxleyi is the first haptophyte that has been shown to produce lignin-like compounds.
We cultured three roseobacter strains – S. pomeroyi DSS-3, Ruegeria sp. R11, and P. gallaeciensis BS107 – in the presence of varying pCA concentrations (0.1–3 mM). At various time points, the cultures were extracted with ethyl acetate and analyzed by HPLC-MS (see Supplementary Methods). The HPLC-MS profiles from S. pomeroyi DSS-3 and Ruegeria sp. R11 cultures showed no significant pCA-dependent changes (Supplementary Fig. S2). However, in P. gallaeciensis BS107, pCA (1 mM) stimulated the production of a family of compounds characterized by a broad 430 nm peak in the UV-visible spectrum (Fig. 1b). In the absence of pCA, these compounds were not produced by P. gallaeciensis under any of the conditions we investigated (see Supplementary Methods). In addition, pCA regulated the production of only selected compounds; it did not, for example, affect TDA (2) production by P. gallaeciensis BS107 (Supplementary Fig. S3).
To examine the nature of the compounds induced in the presence of pCA further, two compounds were purified to homogeneity from 4 L cultures and named roseobacticide A (1.2 mg) and roseobacticide B (0.2 mg). High resolution MS analysis indicated molecular formulas of C16H12O3S and C16H12O2S for roseobacticide A and B, respectively. The low H/C ratio of 0.75 complicated structural elucidation by NMR methods. Consequently, roseobacticide A was crystallized by slow evaporation from CH2Cl2/hexanes and structurally characterized by single crystal X-ray diffraction analysis (Supplementary Table S2). This analysis provided the backbone connectivity, but some ambiguities in distinguishing atom types remained. The complete structure of roseobacticide A was finally solved by interpreting 1H, 13C, gCOSY, gHSQC and gHMBC spectra (Supplementary Fig. S4 & Table S3) in light of the template provided by the X-ray analysis (Fig. 2a, 6). With the structure of roseobacticide A in hand, the structure of roseobacticide B was easily characterized by analyzing 1-D and 2-D NMR data (Fig. 2b, 7; Supplementary Fig. S5 & Table S4). The structures of roseobacticides A and B have no close relatives among known natural products or synthetic molecules. They share a 1-oxaazulan-2-one bicyclic core with an 7-thiomethyl group and a 3-phenyl or 3-(p-hydroxyphenyl) substituent. The bicyclic part of roseobacticide A is planar, and its plane is rotated 33° around the C3–C11 bond relative to the plane of the p-hydroxyphenyl substituent (Fig. 2a).
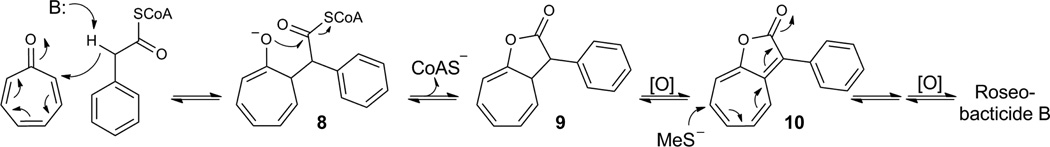
Figure 2.
A proposed biosynthesis for roseobacticides. Addition of the enolate of phenylacetyl CoA to the C2 of tropone gives 8, which after lactonization and release of CoA yields the [8+2] annulation product of the starting substrates (9). Oxidation, followed by 1,8-addition of MeSH to 10 and another oxidation gives 7. Phenylacetyl CoA is derived from 1, which along with tropone and MeSH have been shown to be produced by P. gallaeciensis BS10725. 6 may be obtained from a similar reaction scheme starting with the addition of p-hydroxyphenylacetyl CoA to tropone.
To assess pCA’s ability to elicit roseobacticide production, a dose-response analysis of the amount of 6 as a function of pCA concentration was carried out (Fig. 2c). Below 0.4 mM, no roseobacticides are produced. At and above 1.2 mM, production of 6 no longer increases indicating a switch-like regulation of roseobacticide production. The effective concentration of pCA within the biofilm between P. gallaeciensis and its host in an aging culture is difficult to estimate given the limited diffusion in this tiny region, but it certainly exceeds bulk solution measurements. In 10-day old E. huxleyi cultures (strain CCMP1516) grown in the laboratory, pCA concentration in bulk solution was ~25 µM (see Supplementary Fig. S1), but near the algal surface, it could be substantially higher.
How is the unusual roseobacticide core biosynthesized? Recent reports have noted that tropone, phenylacetic acid and methanethiol are all produced by P. gallaeciensis BS107, and these building blocks suggest a model for the biosynthesis of 7 (Fig. 2d) that could guide future studies25. In this model, tropone undergoes a favored 1,8-addition of the enolate of phenylacetyl-CoA, to give intermediate 8, followed by lactonization and release of CoA to yield intermediate 9, a formal [8+2] annulation product of the starting substrates39,40. A two-electron oxidation of 9 to 10, followed by 1,8-addition of methanethiol to C7 and another two-electron oxidation yields 7.
The molecules so far uncovered in this symbiosis are a tribute to phenylalanine’s versatility, as tropone and phenylacetic acid, likely precursors of the roseobacticides, are biosynthesized from phenylalanine, as is pCA, the induction signal, and virtually all the components of cell wall lignin. The conversion of Phe to tropone and to phenylacetic acid has been well-documented25,41. The known pathway from Phe to pCA involves Phe ammonia lyase (PAL) and cinnamic acid hydroxylase42,43. PAL has also been shown to reversibly transform pCA to Tyr42. An interesting feature of the biosynthetic pathway would be the use of exogenous pCA in the biosynthesis of 6 in analogy to the formation of pCA-containing acylhomoserine lactone signals by other bacteria (Fig. 1, 5).36 While the P. gallaeciensis genome does not encode a recognizable PAL homologue, there could be a functionally equivalent enzyme. Alternatively, pCA could be the signal, or part of a hybrid signal such as pCA-HSL (5), that leads to induction of roseobacticide biosynthetic genes36,44. Experiments to address these issues are currently underway.
To determine possible roles for roseobacticides in mediating the roseobacter-algal symbiosis, the effects of roseobacticides on bacteria and microalgae were tested. The half-maximal inhibitory concentrations (IC50 values) were determined for a panel of marine bacteria and B. subtilis 3610. No significant antibacterial activity was detected (IC50 > 0.16 mM, Supplementary Table S5). However, assays against a diverse selection of microalgae (the Prymnesiophytes E. huxleyi and Isochrysis sp., the green alga Tetraselmis suecica, the diatom Chaetoceros muelleri and the cryptomonad Rhodomonas salina) showed that roseobacticides had specific and potent algaecidal activity. Incubation of E. huxleyi with 6 or 7 resulted in cell lysis after 24 h, with cellular damage visible after 12 h (Fig. 3, a–c). The loss of chloroplasts suggests that roseobacticides may be acting directly on the chloroplast or that they are inducing apoptosis causing loss of cellular integrity. The IC50 values were determined by flow cytometry as it allowed thousands of cells to be rapidly counted for each concentration of roseobacticide tested, and 6 showed an IC50 of ~2.2 µM and 0.10 µM against E. huxleyi and R. salina, respectively (Supplementary Fig. S6 & Table S5). The other three algal strains were less susceptible with IC50 values > 35 µM, though C. muelleri showed major morphological changes upon incubation with 6 (Fig. 3, d–e). Because of the limited quantities of 7, it was only tested against E. huxleyi yielding an IC50 of 0.19 µM (Supplementary Fig. S6). Based on the recovered amounts of 6 and 7, their bulk solution concentration in the 4 L cultures would be ~1 µM and 0.2 µM, respectively. However, the concentration encountered by the algal host with a bacterial biofilm cannot be accurately determined.
Figure 3.
Activity of 6 against E. huxleyi and C. muelleri. (a–c) Light micrographs of E. huxleyi after exposure to methanol (solvent control) for 24 hr (a) or to 3.5 µM 6 for 12 hr (b) or 24 hr (c). (d–e) Light micrographs of C. muelleri after 24 h exposure to methanol (solvent control, d) or to 3.5 µM 6 (e). Panels a–e contain live samples viewed by light microscopy and are typical of these experiments; scale bar = 2 µm.
The production of pCA and dimethylsulfoniopropionate (DMSP) – a major algal metabolite that some roseobacters, including P. gallaeciensis, can use as a sole C- and S-source45 – by other micro- and macroalgae, and by terrestrial plants found in the marine environment46, suggests that P. gallaeciensis may interact with a variety of hosts in the ocean. Although there is no direct evidence for a naturally-occurring E. huxleyi-P. gallaeciensis symbiosis, several lines of support argue that they may interact. Their geographic ranges overlap, as both have been isolated from the same sites (the North Sea and the Tasman Sea)29,47,48. In addition, E. huxleyi is a prolific producer of DMSP49,50, which P. gallaeciensis can take up and transform45,51. Finally, the diverse primary and secondary metabolism of P. gallaeciensis and its quorum sensing and biofilm forming repertoire imply host interactions in agreement with the isolation of Phaeobacter spp. from a range of marine eukaryotes and algae.
The most parsimonious explanation for our findings is that there are two distinct phases in the P. gallaeciensis-algal interaction (Fig. 4). In the first phase, the young algal host is in a mutualistic association with P. gallaeciensis to which the bacteria contribute TDA antibiotics that protect the algal host from bacterial pathogens20, and phenylacetic acid that promotes algal growth25,26,52. In the alternative phase, increased concentrations of pCA, which could indicate elevated algal population density and/or algal senescence, signal P. gallaeciensis to produce roseobacticides from phenylacetic acid and the tropone precursor in TDA biosynthesis – a shift from growth- and health-promoting small molecules to toxins. This shift from mutualism to pathogenesis may occur to allow P. gallaeciensis rapid access to the plentiful food sources provided by the aging algal cells and allow the bacteria to rapidly dissociate from a dying host, disperse, and re-associate with healthy microalgae elsewhere. The roseobacter abundance observed at the conclusion of microalgal blooms supports both scenarios9,53.
Figure 4.
Proposed working model for the interaction between E. huxleyi and P. gallaeciensis. In the mutualistic phase (green), E. huxleyi is healthy and provides DMSP and a biofilm surface to P. gallaeciensis, which in turn produces algal growth promoters and antibiotics (1–3) to protect E. huxleyi. The interaction turns pathogenic (red) when E. huxleyi senesces and releases the algal breakdown product 4, which induces P. gallaeciensis to produce the potent algaecides 6 and 7.
Materials and Methods
General procedures
1H, 13C and 2-D NMR spectra for 6 and 7 were recorded in the inverse-detection probe of a Varian Inova spectrometer (600 MHz for 1H, 150 MHz for 13C). 13C NMR spectra were recorded on the same instrument with a broad-band probe. The 1-D/2-D NMR spectra for 6 and 7 were collected in a 3 mm Norell Select Series NMR tube (Sigma Aldrich) and a 1.7 mm NMR Capillary tube (Wilmad), respectively.
Cultivation of Roseobacter strains and HPLC-MS analysis
Roseobacter (Phaeobacter gallaeciensis BS107, Ruegeria sp. R11 and Silicibacter pomeroyi DSS3) and other marine bacterial strains (see Supplementary Table S5) were streaked out from frozen stocks and maintained on Marine Broth (Difco 2216) supplemented with 1.5% agar and incubated at 30°C for two days. A 50 mL test tube containing 5 mL of ½ strength yeast extract, tryptone and sea salt medium (YTSS, per L: 2 g yeast extract, 1.25 g tryptone, 20 g sea salt) was inoculated with each strain and grown overnight at 30°C on a horizontal rotating drum fermenter. A 500 mL Erlenmeyer flask containing 50 mL of YTSS medium and 0–3 mM pCA was inoculated with 0.5 mL of the overnight culture and incubated at 160 rpm and 30°C for 3 d. After 3 d, each culture was extracted once with an equal volume of ethyl acetate, dried in vacuo, resuspended in methanol and analyzed by HPLC-MS as described in Supplementary Methods.
Purification of roseobacticides A and B
The purification of 6 and 7 was carried out in the dark or under dim light. Four L of YTSS medium containing P. gallaeciensis BS107 and 1 mM pCA were cultured as described above. After 3 d, the culture was extracted once with an equal volume of ethyl acetate. The extract was dried over Na2SO4 and evaporated to dryness in vacuo. The residue was dissolved in CH2Cl2 and fractionated by flash silica gel chromatography (40 g, d=2.5 cm, l=20 cm). Roseobacticides were eluted with a gradient of 0–20% MeOH in CH2Cl2. The fractions containing roseobacticides, as monitored by the distinct broad 430 nm peak by UV-visible spectroscopy, were pooled, dried in vacuo, and purified further by reverse phase HPLC on a preparative Phenomenex C18 column (5 µm, 21.2 × 250 mm) operating at 12 mL/min using an isocratic elution of 35 % MeCN in H2O, followed by a gradient of 35–100 % MeCN in H2O over 40 min. 6 and 7 were then purified further using a semi-preparative Supelco Discovery HS C18 column (10 µm, 10 × 250 mm) operating at 3 mL/min with a gradient of 30–100 % MeOH in H2O over 35 min. They were further purified to homogeneity by reapplication onto the Supelco Discovery HS C18 column and elution with the same gradient. This procedure yielded 1.2 mg and 0.2 mg of 6 and 7, respectively.
Roseobacticide A
TLC (CH2Cl2:MeOH, 92:8 v/v): Rf = 0.55; UV/vis χmax 262 nm, 432 nm; HRMS (m/z): [M]+ calcd. for C16H12O3S, 285.0585; found 285.0578. See Supplementary Table S3 for NMR data. Roseobacticide B. TLC (CH2Cl2:MeOH, 92:8 v/v): Rf = 0.58; UV/vis χmax 262 nm, 316 nm, 430 nm; HRMS (m/z): [M]+ calcd. for C16H12O2S, 269.0637; found 269.0635. See Supplementary Table S4 for NMR data.
Assays of roseobacticide A and B against selected algal strains
The algaecidal effect of 6 and 7 was tested against five axenic microalgae (the Prymnesiophytes Emiliania huxleyi (CCMP372) and Isochrysis sp. (CCMP468), the green alga Tetraselmis suecica (CCMP908), the diatom Chaetoceros muelleri (CCMP1318) and the cryptomonad Rhodomonas salina (CCMP1319)). All strains were grown on L1 media except for CCMP1318, which was grown in L1+Si. All strains were maintained at 24 °C under 12 h:12 h light:dark illumination. To perform assays, algal cultures were diluted 1:1 with the medium they were grown in, then 1 mL aliquots were placed in a 48 well microtitre plate and treated with 10−2–10−9 mg/mL of 6 or 7, or alternatively with MeOH solvent, or medium (as controls) and incubated for 24 h at 24 °C under 12 h: 12 h light:dark illumination. Each treatment was monitored microscopically at 2, 12 and 24 h. Cell counts were determined by flow cytometry and compared to the solvent control.
Flow cytometry
For flow cytometry counts, duplicate 0.5 ml samples were fixed with 0.125% (vol/vol) glutaraldehyde in the dark for 10 min. Cells were then frozen in an ethanol dry ice bath and stored at −80 °C until analyzed. Cell concentrations were measured with an EPICS flow cytometer using red fluorescence and forward angle light scattering properties to identify the cells as previously described54.
Supplementary Material
Acknowledgements
We are thankful to Shao-Liang Zheng at the Center for Crystallographic Studies, Harvard University for solving the crystal structure of roseobacticide A, and to Yoko Saikawa and Brenda N. Goguen for helpful discussions. We also wish to thank the Chisholm lab for use of and help with their flow cytometer, and Joel B. Dacks for assistance with bioinformatic analyses. M. R. S. is a Novartis Fellow of the Life Sciences Research Foundation. R. J. C. is a Harvard Ziff Environmental Fellow. This work was supported by the Office of Naval Research grant N000141010447 and National Institutes of Health grants GM58213 and GM82137 to R. K. and grants CA24487 and GM086258 to J. C.
Footnotes
Author Contributions
M.R.S., R.J.C, R.K. and J.C. designed experiments and wrote the manuscript. M.R.S. performed roseobacticide isolation, structure elucidation and antibacterial assays, R.J.C. performed microscopy and flow cytometry for antialgal activity assays.
The authors declare no competing financial interest.
Supplementary information accompanies this paper at www.nature.com/naturechemistry.
Contributor Information
Roberto Kolter, Email: roberto_kolter@hms.harvard.edu.
Jon Clardy, Email: jon_clardy@hms.harvard.edu.
References
- 1.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt EW. Trading molecules and tracking targets in symbiotic interactions. Nat. Chem. Biol. 2008;4:466–473. doi: 10.1038/nchembio.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clardy J. Using genomics to deliver natural products from symbiotic bacteria. Genome Biol. 2005;6:232–235. doi: 10.1186/gb-2005-6-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott JJ, et al. Bacterial protection of beetle-fungus mutualism. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh DC, Poulsen M, Currie CR, Clardy J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh DC, Scott JJ, Currie CR, Clardy J. Mycangimycin, a polyene peroxide from a mutualist Streptomyces sp. Org. Lett. 2009;11:633–636. doi: 10.1021/ol802709x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Currie CR, et al. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science. 2003;299:386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez JM, et al. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel DA, Franz BA. Oceanography: Century of phytoplankton change. Nature. 2010;466:569–571. doi: 10.1038/466569a. [DOI] [PubMed] [Google Scholar]
- 11.Sabine CL, et al. The oceanic sink for anthropogenic CO2. Science. 2004;305:367–371. doi: 10.1126/science.1097403. [DOI] [PubMed] [Google Scholar]
- 12.Marsh ME. Regulation of CaCO(3) formation in coccolithophores. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003;136:743–754. doi: 10.1016/s1096-4959(03)00180-5. [DOI] [PubMed] [Google Scholar]
- 13.Holligan PM, Viollier M, Harbour DS, Camus P, Champagne-Phillippe M. Satellite and ship studies of coccolithophore production along a continental shelf edge. Nature. 1983;304:339–342. [Google Scholar]
- 14.Balch WM, Holligan PM, Ackleson SG, Voss KJ. Biological and optical properties of mesoscale coccolithophore blooms in the Gulf of Maine. Limnol. Oceanogr. 1991;36:629–643. [Google Scholar]
- 15.Holligan PM, et al. A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North Atlantic. Global Biogeochem. Cycles. 1993;7:879–900. [Google Scholar]
- 16.Everitt DA, Wright SW, Volkman JK, Thomas DP, Lindstrom EJ. Phytophlankton community compositions in the western equatorial Pacific determined from chlorophyll and carotenoid pigment distributions. Deep Sea Res. A. 1990;37:975–997. [Google Scholar]
- 17.Miller TR, Belas R. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 2006;8:1648–1659. doi: 10.1111/j.1462-2920.2006.01071.x. [DOI] [PubMed] [Google Scholar]
- 18.Kjelleberg S, et al. Do marine natural products interfere with prokaryotic AHL regulatory systems? Aquat. Mirob. Ecol. 1997;13:85–93. [Google Scholar]
- 19.Joint I, et al. Cell-to-Cell communication across the prokaryote-eukaryote boundary. Science. 2002;298:1207. doi: 10.1126/science.1077075. [DOI] [PubMed] [Google Scholar]
- 20.Rao D, et al. Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl. Environ. Microbiol. 2007;73:7844–7852. doi: 10.1128/AEM.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seymour JR, Simo R, Ahmed T, Stocker R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science. 2010;329:342–345. doi: 10.1126/science.1188418. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo Y, Imagawa H, Nishizawa M, Shizuri Y. Isolation of an algal morphogenesis inducer from a marine bacterium. Science. 2005;307:1598. doi: 10.1126/science.1105486. [DOI] [PubMed] [Google Scholar]
- 23.Keshtacher-Liebso E, Hadar Y, Chen Y. Oligotrophic bacteria enhance algal growth under iron-deficient conditions. Appl. Environ. Microbiol. 1995;61:2439–2441. doi: 10.1128/aem.61.6.2439-2441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geng H, Bruhn JB, Nielsen KF, Gram L, Belas R. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol. 2008;74:1535–1545. doi: 10.1128/AEM.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiel V, et al. Identification and biosynthesis of tropone derivatives and sulfur volatiles produced by bacteria of the marine Roseobacter clade. Org. Biomol. Chem. 2010;8:234–246. doi: 10.1039/b909133e. [DOI] [PubMed] [Google Scholar]
- 26.Ashen JB, Cohen JD, Goff LJ. GC-SIM-MS detection and quantification of free indole-3-acetic acid bacterial galls on the marine alga Prionitis lanceolata (Rhodophyta) J. Phycol. 1999;35:493–500. [Google Scholar]
- 27.Hold GL, Smith EA, Birbeck TH, Gallacher S. Comparison of paralytic shellfish toxin (PST) production by the dinoflagellates Alexandrium lusitanicum NEPCC253 and Alexandrium tamarense NEPCC407 in the presence and absence of bacteria. FEMS Microbiol Ecol. 2001;36:223–234. doi: 10.1111/j.1574-6941.2001.tb00843.x. [DOI] [PubMed] [Google Scholar]
- 28.Buchan A, Gonzalez JM, Moran MA. Overview of the marine roseobacter lineage. Appl. Environ. Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao D, Webb JS, Kjelleberg S. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl Environ Microbiol. 2005;71:1729–1736. doi: 10.1128/AEM.71.4.1729-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greer EM, Aebisher D, Greer A, Bentley R. Computational studies of the tropone natural products, thiotropocin, tropodithietic acid, and troposulfenin. Significance of thiocarbonyl-enol tautomerism. J. Org. Chem. 73:280–283. doi: 10.1021/jo7018416. (200) [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 32.Lutgendorff F, Akkermans LM, Soderholm JD. The role of microbiota and probiotics in stress-induced gastro-intestinal damage. Curr. Mol. Med. 2008;8:282–298. doi: 10.2174/156652408784533779. [DOI] [PubMed] [Google Scholar]
- 33.Delwiche CF, Graham LE, Thomson N. Lignin-like compounds and sporopollenin coleochaete, an algal model for land and plant ancestry. Science. 1989;245:399–401. doi: 10.1126/science.245.4916.399. [DOI] [PubMed] [Google Scholar]
- 34.Martone PT, et al. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 2009;19:169–175. doi: 10.1016/j.cub.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 35.Espineira JM, et al. Distribution of lignin monomers and the evolution of lignification among lower plants. Plant Biol. 2010 doi: 10.1111/j.1438-8677.2010.00345.x. in press. [DOI] [PubMed] [Google Scholar]
- 36.Schaefer AL, et al. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454:595–599. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- 37.Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 39.Rigby JH, Zbur Wilson J. Total synthesis of guaianolides: (+/−)-Dehydrocostus lactone and (+/−)-estafiatin. J. Am. Chem. Soc. 1984;106:8217–8224. [Google Scholar]
- 40.Nair V, Poonoth M, Vellalath S, Suresh E, Thirumalai R. An N-heterocyclic carbene-catalyzed [8+3] annulation of tropone and enals via homoenolate. J. Org. Chem. 2006;71:8964–8965. doi: 10.1021/jo0615706. [DOI] [PubMed] [Google Scholar]
- 41.Ismail W, et al. Functional genomics by NMR spectroscopy. Phenylacetate catabolism in Eschrichia coli. Eur. J. Biochem. 2003;270:3047–3054. doi: 10.1046/j.1432-1033.2003.03683.x. [DOI] [PubMed] [Google Scholar]
- 42.MacDonald MJ, D'Cunha GB. A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. 2007;85:273–282. doi: 10.1139/o07-018. [DOI] [PubMed] [Google Scholar]
- 43.Czichi U, Kindl H. Formation of p-coumaric acid and o-coumaric acid from L-phenylalanine by microsomal membrane fractions from potato: Evidence of membrane-bound enzyme complexes. Planta. 1975;125:115–125. doi: 10.1007/BF00388698. [DOI] [PubMed] [Google Scholar]
- 44.Lee YW, Jin S, Sim WS, Nester EW. The sensing of plant signal molecules by Agrobacterium: genetic evidence for direct recognition of phenolic inducers by the VirA protein. Gene. 1996;179:83–88. doi: 10.1016/s0378-1119(96)00328-9. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez JM, Kiene RP, Moran MA. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buchan A, Neidle EL, Moran MA. Diversity of the ring-cleaving dioxygenase gene pcaH in a salt marsh bacterial community. Appl. Environ. Microbiol. 2001;67:5801–5809. doi: 10.1128/AEM.67.12.5801-5809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. https://ccmp.bigelow.org/themes/ccmp/controls/search-map.php?genus=Emiliania&species=huxleyi&commonName=CommonNames&authenticStrain=NA&. [Google Scholar]
- 48.Brinkhoff T, et al. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 2004;70:2560–2565. doi: 10.1128/AEM.70.4.2560-2565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfe GV, Steinke M, Kirst GO. Grazing-activated chemical defence in a unicellular marine alga. Nature. 1997;387:894–897. [Google Scholar]
- 50.Wolfe GV, Steinke M. Grazing-activated production of dimethyl sulfide (DMS) by two clones of Emiliania huxleyi. Limnol. Oceanogr. 1996;41:1151–1160. [Google Scholar]
- 51.Newton RJ, et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4:784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- 52.Ashen JB, Goff LJ. Molecular identification of a bacterium associated with gall formation in the marine red alga Prionitis lanceolata. J. Phycol. 1996;32:286–297. [Google Scholar]
- 53.Lamy D, et al. Temporal changes of major bacterial groups and bacterial heterotrophic activity during a Phaeocystis globosa bloom in the eastern English Channel. Aquat. Mirob. Ecol. 2010;58:95–107. [Google Scholar]
- 54.Cavender-Bares KK, Frankel SL, Chisholm SW. A dual sheath flow cytometer for shipboard analyses of phytoplankton communities from the oligotrophic oceans. Limnol. Oceanogr. 1998;43:1383–1388. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.