Abstract
AIMS
To completely screen the SLCO1B3 gene in three distinct healthy Asian populations (Chinese, Malay and Indian, n = 168) and investigate the influence of haplotype-tag SNPs (htSNPs) on docetaxel disposition in 50 nasopharyngeal carcinoma patients.
METHODS
Genomic DNA of individuals was screened for SLCO1B3 polymorphisms by direct sequencing. htSNPs were derived based on the sequence clustering algorithm and profiled in the patients. Population based genetic association analysis was performed using Haplostats package implemented in R and PLINK.
RESULTS
A strong linkage disequilibrium pattern was detected across a total of 88 polymorphisms and 15-htSNPs were identified. The SLCO1B3 haplotypic region comprising seven htSNPs was found to be significantly associated with docetaxel clearance (P = 0.003). Conditional haplotype analyses revealed that the haplotypic constructs comprising the IVS4+76G>A, 699G>A(Met233Ile), IVS12-5676A>G, and *347_*348insA polymorphisms were critical determinants of variability in docetaxel disposition [clearance and area under the plasma concentration−time curve (AUC(0,∞)): r2 = 29% and 22%, respectively]. Patients harbouring the GAG*347insA haplotype were significantly associated with a 30% decrease in clearance and a 40% increase in AUC(0,∞) of docetaxel compared with patients harbouring the reference haplotype, GGA*347wt (P = 0.025 and 0.018, respectively). In contrast, a 50% higher clearance was observed in patients carrying the GAG*347wt haplotype compared with those with the reference haplotype (P = 0.002). The functional SLCO1B3 haplotypic constructs included the widely studied Met233Ile variant and *347_*348insA located in the putative miR-890 binding site in the 3′-untranslated region which may influence the transport characteristics of SLCO1B3.
CONCLUSIONS
This study highlights the importance of SLCO1B3 polymorphic variations in influencing docetaxel disposition in nasopharyngeal carcinoma patients.
Keywords: Asians, docetaxel, nasopharyngeal cancer, pharmacogenetics, SLCO1B3
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
SLCO1B3 is an influx transporter located at the hepatocyte basolateral membrane and it is involved in the uptake of a broad range of drug substrates including docetaxel.
The pharmacogenetics of SLCO1B3 is not well characterized and previous in vivo and in vitro studies reported conflicting results with regards to the functional effects of the limited number of SLCO1B3 polymorphisms that were studied.
Docetaxel displays a wide interindividual variability in its pharmacokinetics and pharmacodynamics and an understanding of SLCO1B3 pharmacogenetics might provide clinical benefits in guiding docetaxel dosing.
WHAT THIS STUDY ADDS
The SLCO1B3 gene was comprehensively screened in the local healthy Asian populations (n = 168). A strong linkage disequilibrium pattern was detected across a total of 88 polymorphisms and 15 haplotype-tag SNPs (htSNPs) were identified. These htSNPs were profiled in a cohort of Chinese nasopharyngeal cancer (NPC) patients (n = 50).
Genotypic-phenotypic analysis showed that a haplotypic construct comprising of four variants [IVS4+76G>A, 699G>A(Met233Ile), IVS12-5676A>G, and *347_*348insA] was the critical determinant of docetaxel disposition.
This study suggests that the comprehensive screening and haplotypic linkage analysis of SLCO1B3 can better elucidate its pharmacogenetic effects on interpatient variability of docetaxel and other putative drug substrates. Further studies are warranted in cancer patients belonging to other ethnic groups.
Introduction
OATP1B3, previously known as OATP-8 or liver-specific organic anion transporter-2 (LST-2) is a cellular membrane transport protein located at the basolateral membrane of hepatocytes. It is encoded by the solute carrier organic anion transporter 1B3 gene (SLCO1B3). The gene maps to chromosome 12p12 and spans across 106kbp [1]. OATP1B3 shares 80% homology with OATP1B1 (SLCO1B1) in amino acid sequence and they transport a range of common drug substrates such as methotrexate and pravastatin [1, 2]. However, OATP1B3 plays a distinctively pivotal role in transporting digoxin, docetaxel and paclitaxel [3–6]. SLCO1a/1b-/- mice were demonstrated to exhibit a pronounced increase in paclitaxel systemic exposure and decrease in liver uptake. This observation strongly highlights the role of OATP basolateral transporters in governing taxane exposure in vivo[7]. In addition, OATP1B3 is also expressed in the tumour cell and its role in drug sensitization had been documented [8, 9].
The SLCO1B3 gene is highly polymorphic and exhibits high variations across ethnic groups [3, 10, 11]. The allelic frequencies of the non-synonymous variants 334T>G (Ser112Ala; rs4149117) and 699G>A (Met233Ile; rs7311358) displayed a great degree of heterogeneity across diverse ethnic populations, ranging from 41% in African-Americans to approximately 71 to 90% in Caucasians and Chinese [10]. Previous in vitro investigations revealed that the functional effects of the OATP1B3 gene expressing the non-synonymous variants 334T>G (Ser112Ala; rs4149117) and 699G>A (Met233Ile; rs7311358) were substrate and cell-line specific [12, 13]. However, a recent study using HeLa cells demonstrated the 699G>A (Met233Ile; rs7311358) variant to display a significantly lower transport activity for the prototype substrates such as cholecystokinin and rosuvastatin [12]. Similar reduction in cholecystokinin transport activity was also observed when the variant haplotype consisting of 334T>G (Ser112Ala; rs4149117) and 699G>A (Met233Ile; rs7311358) was expressed [12]. In concordance with the observations above, transport assay studies revealed impaired uptake of mycophenolic acid glucuronide and digoxin into cells expressing the variant haplotype [14, 15]. Both variants were subsequently linked with a low hepatic uptake activity and clearance (CL) of mycophenolic acid glucuronide and digoxin in in vivo setting [14, 16]. In contrast to the above, a recent clinical study had demonstrated an association between 334T>G (Ser112Ala; rs4149117) and higher imatinib CL [17]. Importantly, the rare variants His520Pro, Gly522Cys (rs72559743) and Val560Ala (rs12299012) were previously documented to be associated with reduced protein expression and impaired transport activity, probably a consequence of their location in key regulatory regions of the transporter [12, 13]. These variants were, however, absent in our three distinct local Asian populations published previously [18].
Linkage patterns of SNPs vary widely between different populations and the observed phenotypic effects may be the consequence of several SNPs being in haplotypic linkage. Furthermore, most of the previous in vivo studies were restricted to selective SNPs [3, 4, 14, 17] which delineated the functional impact of the SNPs identified in in vitro model systems [12, 13, 19]. To date, no studies have comprehensively screened the SLCO1B3 gene and elucidated the linkage and haplotypic patterns of SLCO1B3 SNPs. In view of the importance of SLCO1B3 in drug disposition, we carried out a comprehensive screening of SLCO1B3 to (i) identify SNPs located at the 5′ UTR, exons, intronic-exonic boundaries and 3′ UTR in a cohort of three distinct healthy Asian subjects, and (ii) investigate the haplotypic impact of SLCO1B3 haplotype-tag SNPs (htSNPs) on the pharmacokinetics and pharmacodynamics of docetaxel in a cohort of Chinese nasopharyngeal cancer (NPC) patients. The pharmacogenomic influence of other candidate genes across the docetaxel biochemical pathway in Asian cancer patients has been recently published by our group [18]. The present study showed that SLCO1B3 haplotypes influenced docetaxel disposition and contributed to interpatient differences in docetaxel pharmacokinetics.
Methods
Subjects
A total of 168 healthy subjects (Chinese, Malay, Indian, n = 56 per ethnic group) and 50 Chinese NPC patients were recruited. Our previous study on elucidating the impact of candidate genes in the docetaxel biochemical pathway included 54 NPC patients (Chinese = 50, Malays = 4) [18]. Since the majority of the patients in the original cohort were Chinese and to ensure an ethnically homogenous patient population, we have excluded the four Malay patients from the present study. The ethnic group of the subjects was confirmed by verification against their National Registry Identification Cards. The patients were histologically confirmed for stage IVC or recurrent metastatic NPC who had failed one or two prior platinum-based combination chemotherapy regimens. The inclusion and exclusion criteria of this study were previously described elsewhere [20]. All participants provided written informed consent and the study was approved by the ethics review committee (EC reference number: 2004/409/B) of the National Cancer Centre, Singapore.
Docetaxel treatment and pharmacokinetics in NPC patients
Docetaxel (Taxotere; Sanofi-Aventis, Paris, France) was administered at a dose of 30 mg m−2 intravenously over 1 h on days 1, 8 and 15 every 28 days. Standard pre and post medications were uniformly administered to all patients and included i.v. dexamethasone 10 mg prior to the start of docetaxel infusion followed by oral dexamethasone 4 mg every 8 h for four doses started 8 h after the end of the docetaxel infusion. Blood samples for pharmacokinetic analyses were collected at the following time points: pre dose (0 h), 0.5 h and 1 h (end of infusion), then at 15 min, 30 min, 2 h, 4 h, 7 h and 23 h after the end of the docetaxel infusion in the first cycle. Blood samples were centrifuged at 2000 g for 10 min to separate the plasma and samples were immediately stored at −80°C until analysis. Docetaxel plasma concentrations were determined with slight modifications by use of a validated HPLC method [21]. In brief, 200 ng internal standard (paclitaxel; Bristol-Myers Squibb, NY, USA) and 1.6 ml of acetonitrile-n-butylchloride (1:4, v/v) were added to 200 µl of human plasma. The sample was mixed vigorously for 5 min, followed by centrifugation for 5 min at 4000 g at ambient temperature. The organic layer was collected and evaporated to dryness using a centrifugal concentrator at room temperature. The residue was reconstituted with 125 µl of methanol-water (1:1, v/v), and 100 µl was injected into the HPLC system (Waters, Milford, MA, USA). Separation was accomplished on a Spherisorb ODS2 column (5 µm, 250 × 4.6 mm; Waters, Milford, MA, USA) at a rate of 1.6 ml min−1. The elution was monitored by ultraviolet detection at a wavelength of 230 nm. Retention times for docetaxel and internal standard were 19.7 and 27.6 min, respectively. The standard curves were linear from 10–40000 ng ml−1 and the coefficients of variation for intraday and interday precision were lower than 7%. The pharmacokinetic parameters of docetaxel were determined using the non-compartmental modelling by WinNonLin version 2.1 (Pharsight, Mountain View, CA, USA).
Pharmacodynamic parameters
The percentage decrease in absolute neutrophil count, platelet count and haemoglobin count was defined as:
where the pre treatment value corresponded to the baseline value and nadir value denoted the lowest value measured in cycle 1. Response and toxicity evaluations were described in previous publication [20].
Pharmacogenetic analysis
Genomic deoxyribonucleic acid was isolated from the blood of healthy subjects using Puregene DNA isolation kit (Qiagen, Valencia, CA, USA). The exonic regions, exon-intron junctions and 2 kb of the 5′- and 3′- UTR of SLCO1B3 (UCSC accession number NM_019844) were screened for genetic polymorphisms by direct sequencing (primer sequences are available in Supplementary Table S1. The sequencing reactions were carried out using Applied Biosystems 3730 DNA Analyzer (Applied Biosystems Inc, Foster City, CA, USA).
Linkage disequilibrium and haplotype analysis
Linkage disequilibrium (LD) analyses, haplotype analyses and LD block construction were carried out by Haploview software v.3.32 (Daly lab, Broad Institute, MA, USA) [22]. Based on the LD pattern, the blocks were generated for tightly linked polymorphisms by using solid spine algorithm in Haploview [22]. Haplotypes for each ethnic group were inferred by maximum likelihood estimation based on an expectation-maximization algorithm. A sequence clustering algorithm was applied to construct genealogic trees in order to identify the stratifying htSNPs on the basis of nucleotide base difference between the major haplotypes (ClustalW; EBI, Cambridge, UK).
Statistical analysis
Fisher's exact test was applied to determine conformity with Hardy-Weinberg equilibrium. The inter-ethnic variability of the frequency of genotype distribution was evaluated using the Chi-square contingency test. Univariate analysis was adopted to assess the associations between patient covariates [age, gender, body surface area (BSA), ECOG, aspartate transaminase, alanine transaminase, albumin and total bilirubin] and pharmacokinetic parameters. General statistical analysis was carried out using SPSS Version 16; SPSS Inc., IL, USA. A P value of <0.05 was considered as statistically significant.
Population genetics analysis
Population based genetic association analysis was performed using Haplostats package (1.4.4v) implemented in R 2.12.1v and PLINK 1.02v [23, 24]. The asymptotic likelihood ratio test was applied to evaluate the individual association of SLCO1B3 genotypes with docetaxel disposition parameters. Haplotype association analysis was carried out using the haplowalk method implemented in R haplostats package [25]. Technically, this method adopts a moving window region comprising of ‘n’ SNPs (defined by adjacent pairs of ‘n’ loci across the entire region) and the phenotypic association with each window is evaluated using global haplotype score statistics (where n≠ 1). This score statistics allow the adjustment of the P value for the patient's covariates. In the present analysis, global haplotype score P values were calculated for each window region including 2–15 SLCO1B3 htSNPs to estimate the level of significant association with docetaxel disposition parameters. This P value was adjusted for the patients' covariates identified in univariate analysis. The global haplotype score test is asymptotically similar to the likelihood ratio test, but evaluates the haplotypic effects on phenotypic trait. The score statistics P value provides an overview of the variability contribution from the sub-haplotypic region [25]. Conditional haplotype analysis is a weighted maximum likelihood approach which accounts for potential ambiguity in the individuals [26]. This approach is applicable once the association signal has been detected and facilitates in determining the causal variants behind the association [26]. In our current analysis, the region of interest identified by the haplowalk procedure, was dissected iteratively for ‘n’ times and each association was estimated using an omnibus test P value (conditional haplotype association module in PLINK 1.02v) [23, 24]. The omnibus test P values and percentage contribution to variability (r2) were used for selecting the causal variants influencing the pharmacokinetics of docetaxel. The haplotype specific generalized linear model (haplo-GLM) implemented in the haplostats package was applied to estimate the regression coefficient and significant P value for individual haplotype effects. The haplo-GLM analyses perform a weighted approach to account for the regression coefficients and posterior probability of each haplotype pair using the expectation maximization algorithm. The population mean parameters specific to the haplotypes estimated from the GLM were used to calculate the percentage increase/decrease of docetaxel pharmacokinetic parameters between the haplotypes. A P value of <0.05 was considered as statistically significant, unless otherwise stated.
Results
Patient demographics
Table 1 summarizes the demographic information of the subjects in the present study. The majority of the patients were males (92%) compared with females (8%). The median age and BSA were 50 years (range 32–68 years) and 1.63 m2 (range 1.09–2.11 m2), respectively. A total of 208 cycles were administered and approximately half of the patients (46%) completed all six treatment cycles. Dexamethasone was uniformly prescribed to all patients as pre and post medications and there was no other concomitant therapy that could affect CYP3A4/5 activity was administered during cycle 1.
Table 1.
Demographic and clinical characteristics of Chinese NPC patients (n = 50)
| Characteristics (n = 50) | |
|---|---|
| Age (years) | 50 (32–68) |
| Gender | |
| Male | 46 (92%) |
| Female | 4 (8%) |
| BSA (m2) | 1.63 ± 0.19 |
| Number of cycles patients received | |
| 1 | 2 (4%) |
| 2 | 15 (30%) |
| 3 | 2 (4%) |
| 4 | 8 (16%) |
| 5 | 0 (0%) |
| 6 | 23 (46%) |
| Baseline clinical biochemistry | |
| AST (SGOT) (U/L) | 35.12 ± 31.62 |
| ALT (SGPT) (U/L) | 26.08 ± 20.27 |
| LDH* (U/L) | 479.11 ± 191.61 |
| Total bilirubin (µmol/L) | 10.90 ± 3.90 |
| Albumin (g/L) | 36.54 ± 4.23 |
n = 36.
SLCO1B3 pharmacogenetics in healthy Asian populations
SLCO1B3 genotype and allele frequencies in the healthy Chinese, Malay and Indian populations are appended in Supplementary Table S2. A total of 88 single nucleotide polymorphisms were identified from the complete screening of the SLCO1B3 genomic region. As depicted in Figure 1, two SNPs were located in 5′ UTR, nine in exonic regions, 59 in the intronic regions, 11 in the 5′ upstream region, one in 3′ UTR and six in the 3′ downstream region of the SLCO1B3 gene (Figure 1). There were fourteen novel SNPs with allelic frequencies ranging from 1% to 19% (Supplementary Table S2; correct as on 20/02/2011). Fisher's exact test showed that the SLCO1B3 genotype frequencies conformed to Hardy-Weinberg equilibrium. Significant interethnic differences in the genotypic distributions were detected in 74% (n = 65) of the polymorphisms (Supplementary Table S2, P < 0.05).
Figure 1.
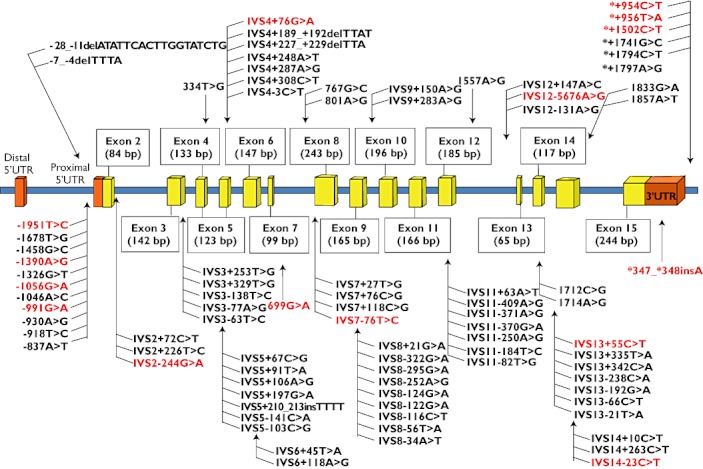
Graphical representation of identified polymorphisms in the SLCO1B3 gene. The locations of the identified polymorphisms (arrows) are indicated in relation to the promoter, exon and intron structures of SLCO1B3 (UCSC accession number NM_019844). The distal 5′UTR contains a non-coding exon 1. Exonic SNPs are labelled relative to the ATG translation initiation site in exon 2, with the A numbered as 1. The first nucleotide 5′ to the ATG site is −1, and the first nucleotide 3′ to the translation stop codon is *1. Intronic SNPs are labelled with the respective intronic number, followed by a + (downstream) or – (upstream) sign and the position in the intron, calculated from the nearest exon. Polymorphisms highlighted in red are the htSNPs selected and profiled in the patient cohort. The domain sizes are not drawn to scale
Linkage disequilibrium (LD) analysis in healthy subjects
The LD plots for Chinese, Malay and Indian ethnic groups are shown in Figure 2. Pairwise analyses for the identified SLCO1B3 SNPs showed high linkage among the SNPs located in the 5′ upstream region across all populations as well as in the region covering from intron 2 to 12 in the Chinese and Malay populations, and from intron 3 to exon 14 in the Indian population. In addition, SNPs located at the 3′ regulatory region were also found to be substantially linked in all populations. Based on the LD pattern, the SLCO1B3 gene was segregated into four blocks in Chinese and Malay subjects and three blocks in Indian subjects (Figure 2).
Figure 2.
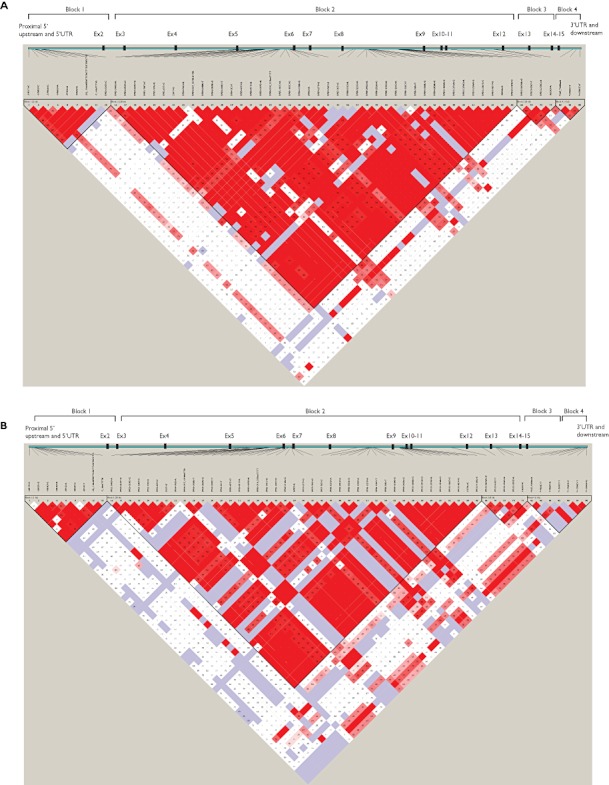
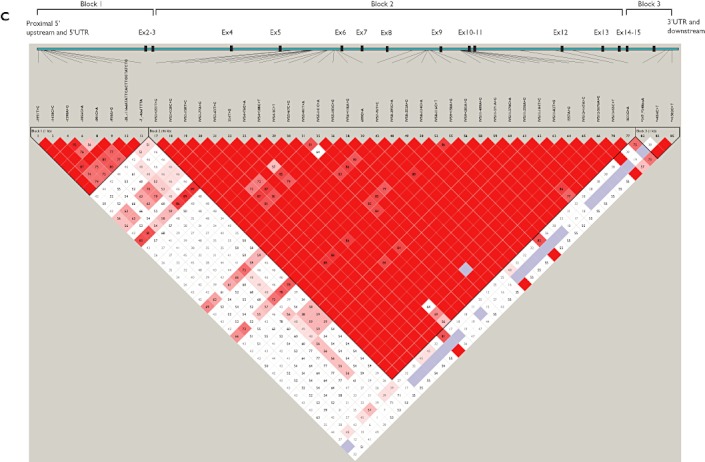
Linkage disequilibrium plot of SLCO1B3 polymorphisms in healthy (A) Chinese, (B) Malay and (C) Indian populations. LD values are denoted as (|D'|× 100). LD analyses, haplotype analyses and LD block construction were carried out by Haploview software v.3.32 (Daly lab, Broad Institute, MA, USA). The standard colour scheme in Haploview was chosen for LD display. Block 1 represented 5′ upstream and proximal UTR SNPs in all populations. Blocks 2 and 3 in the Chinese and Malay populations consisted of SNPs located between intron 2 and exon 14. The region represented by haplotype block 2 was the largest and comprised of 42, 41 and 33 exonic/intronic polymorphisms in the Chinese, Malay and Indian populations, respectively. Block 4 in the Chinese and Malay populations and block 3 in the Indian population consisted of SNPs from the 3′ UTR and downstream regions. The inter-block linkage was strong between blocks 2 and 3 (|D'| = 0.82–0.97), but moderate between blocks 1 and block 2 (|D'| = 0.55–0.78)
SLCO1B3 haplotypes and htSNPs in healthy Asian populations
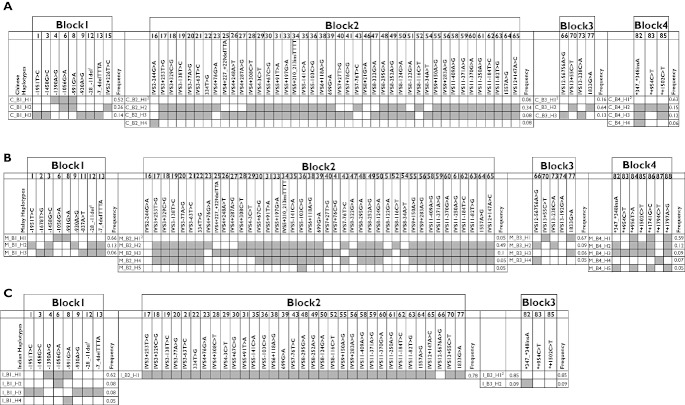
The inferred high frequency SLCO1B3 haplotypes (>5%) and the observed frequencies in each haplotype block across the three ethnic groups are shown in Figure 3. Cumulative frequencies of the high frequency haplotypes in the three ethnic groups ranged from 56% to 96%. A strong LD pattern and a high prevalence of variant allele haplotypes were observed as common phenomena across the blocks among the local Asian ethnic groups. Block 2 comprised the longest haplotype stretch with a high prevalence of variant alleles in all three populations. Interestingly, block 2 in Indian subjects comprised of only one high frequency haplotype (78%) which constituted of variant alleles at all locations, except for IVS12+5676A>C.
Figure 3.
SLCO1B3 haplotypes in healthy (A) Chinese, (B) Malay and (C) Indian populations. Haplotype analyses and LD block construction were carried out by Haploview software v.3.32 (Daly lab, Broad Institute, MA, USA).The shaded box represents the presence of variant. C, Chinese; M, Malays; I, Indians; B, Block; H, Haplotype. The haplotype name is designated with block and haplotype number as suffix, e.g. C_B1 represents haplotype block 1 identified in the Chinese subjects. 1ATATTCACTTGGTATCTG deletion at location −28 to −11. 2Denotes refererence haplotype
The linkage pattern was further examined by identifying the htSNPs which distinguished the major haplotypes in each block. Supplementary Figure S1 shows the genealogic tree of derived haplotypes and the corresponding htSNPs for each haplotype block among the three ethnic groups. A total of 10, 13 and four htSNPs were identified in Chinese, Malay and Indian populations, respectively. Collectively, a set of 15 htSNPs (−1951T>C, −1390A>G, −1056G>A, −991G>A, IVS2-244G>A, IVS4+76G>A, 699G>A, IVS7-76T>C, IVS12-5676A>G, IVS13+55C>T, IVS14-23C>T, *347_*348insA, *+954C>T, *+956T>A, *+1502C>T) that resolved the common haplotypes in the Asian cohort and highly specific to the Chinese population were profiled in the Chinese NPC patients.
Effects of SLCO1B3 haplotypes on docetaxel pharmacokinetics and pharmacodynamics in Chinese NPC patients
The values (mean ± SD) of the pharmacokinetics parameters of docetaxel in Chinese NPC patients (n = 50) are as follows: Cmax (µg ml−1) 1.12 ± 0.51, AUC(0,∞) (µg ml−1 h) 1.15 ± 0.45 and CL (l h−1) 48.20 ± 19.07. A wide degree of interpatient variability was observed in the docetaxel pharmacokinetics with approximately 6-, 4- and 5-fold variations in Cmax (range: 0.51–3.08 µg ml−1), AUC(0,∞) (range 0.56–2.14 µg ml−1 h) and CL (range 19.28–102.17 l h−1), respectively. The allelic frequencies of the htSNPs in healthy Chinese subjects and NPC patients (n = 50) were similar (Supplementary Table S2). Univariate analysis suggested docetaxel CL to be significantly affected only by age, while docetaxel AUC(0,∞) was found to be significantly influenced by albumin and ECOG status (P < 0.05 in each case, respectively).
Individual genotype association analysis using the asymptotic likelihood ratio test did not reveal any significant correlations between docetaxel pharmacokinetics and SLCO1B3 htSNPs. In order to elucidate the haplotypic effect of htSNPs on docetaxel pharmacokinetics, global haplotype score statistics were adopted to estimate the overall association of haplotypes and phenotypic traits using the haplowalk method [25]. The haplowalk method was previously applied to select the contributory genomic regions in relation to renal cell cancer susceptibility [27, 28]. This analysis revealed a sub-haplotypic region comprising of seven htSNPs [IVS4+76G>A(rs4149118), 699G>A(rs7311358), IVS7-76T>C(rs71583718), IVS12-5676A>G(rs11045585), IVS13+55C>T(rs4149153), IVS14-23C>T(rs78862986) and *347_*348insA(rs3834935)] to be significantly correlated with docetaxel CL (adjusted global haplotype score P = 0.003, Supplementary Figure S2). As the score test P value alone is an insufficient measure to estimate the effect of haplotype [25], we further performed conditional iterative haplotype analysis to identify the causal htSNPs from the sub-haplotypic region of seven htSNPs. This analysis identified four causal htSNPs [IVS4+76G>A (rs4149118), 699G>A (rs7311358), IVS12-5676A>G (rs11045585) and *347_*348insA (rs3834935)] that accounted for approximately 29% variability in docetaxel CL in Chinese NPC patients (P = 0.00071). Six major haplotypes (frequency > 5%) were inferred from the four focal htSNPs and accounted for a cumulative frequency of 93%. The haplotype AAA*347wt was the most frequently occurring haplotype in the patient population (44%).
A haplotype specific generalized linear model was employed to estimate the regression coefficient associated with docetaxel pharmacokinetics (Table 2). The haplotype GAG*347insA (frequency: 14%) was significantly associated with a 30% decrease in CL and a 40% increase in AUC(0,∞) of docetaxel compared with the reference haplotype GGA*347wt (frequency 13%) [GAG*347insA vs. GGA*347wt; mean ± SE CL (l h−1) 44.37 ± 6.26 vs. 58.92 ± 9.19, P = 0.025 and mean ± SE AUC(0,∞) (µg ml−1 h) 1.31 ± 0.16 vs. 0.91 ± 0.24, P = 0.018; Figure 4]. In contrast, a 50% higher CL was observed in patients harbouring the GAG*347wt haplotype (frequency: 5%) compared with carriers of the reference haplotype (GAG*347wt vs. GGA*347wt; mean CL (l h−1) 88.28 ± 8.77 vs. 58.92 ± 9.19, P = 0.002; Figure 4). No significant associations were observed between SLCO1B3 haplotypes and pharmacodynamic parameters.
Table 2.
Influence of four SNP SLCO1B3 haplotype constructs [IVS4+76G>A (rs4149118), 699G>A (rs7311358), IVS12−5676A>G (rs11045585) and *347_*348insA (rs3834935)] on docetaxel CL and AUC(0,∞)
| Haplotypes | Coefficient | Standard error | Mean value | P value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IVS4 +76G>A | 699G>A | IVS12 −5676A>G | *347_*348 insA | Frequency | CL | AUC(0,∞) | CL | AUC(0,∞) | CL (l h−1) | AUC(0,∞) (ug ml−1 h) | CL | AUC(0,∞) |
| G | G | A | wt | 0.13 | 58.92 | 0.91 | 9.19 | 0.24 | 58.92 | 0.91 | 0 | 0 |
| A | A | A | wt | 0.44 | −9.1 | 0.21 | 5.48 | 0.14 | 49.82 | 1.12 | 0.104 | 0.153 |
| G | A | G | ins | 0.14 | −14.55 | 0.40 | 6.26 | 0.16 | 44.37 | 1.31 | 0.025* | 0.018* |
| G | A | A | wt | 0.08 | 4.19 | −0.16 | 7.38 | 0.20 | 63.11 | 0.75 | 0.573 | 0.427 |
| G | G | A | ins | 0.08 | 3.02 | −0.16 | 7.30 | 0.18 | 61.94 | 0.75 | 0.681 | 0.38 |
| G | A | G | wt | 0.05 | 29.36 | −0.22 | 8.77 | 0.22 | 88.28 | 0.69 | 0.002* | 0.314 |
| Minor haplotypes (<5%) | 0.07 | −12.92 | 0.13 | 7.71 | 0.21 | 46 | 1.04 | 0.101 | 0.541 | |||
P values were obtained from the haplotype specific generalized linear model implemented in the R haplostats package. AUC(0,∞), area under plasma concentration–time curve from time zero to infinity; CL, plasma clearance.
indicates a significant P value (<0.05).
Figure 4.
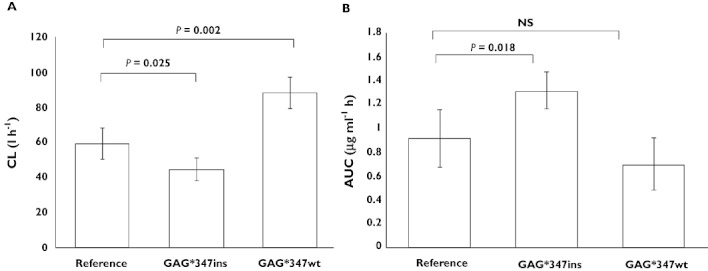
Effects of GAG*347insA and GAG*347wt in comparison with the reference haplotype (GGA*347wt) on docetaxel (A) CL and (B) AUC(0,∞). Patients carrying the GAG*347insA haplotype had significantly lower CL and higher AUC(0,∞) compared with those with the reference haplotype. In contrast, patients harbouring the GAG*347wt haplotype had significantly higher CL compared with those with the reference haplotype. Each bar represents the mean ± SE of the docetaxel pharmacokinetic parameters. P values were obtained from the haplotype specific generalized linear model implemented in the R haplostats package. AUC(0, ∞), area under plasma concentration−time curve from time zero to infinity; CL, plasma clearance; NS, non-significant
Discussion
The present study revealed the SLCO1B3 gene to be highly polymorphic and to display significant interethnic variations among the healthy Chinese, Malay and Indian populations. Similar to Japanese and Caucasian populations, complete linkage was found between 334T>G (Ser112Ala; rs4149117) and 699G>A (Met233Ile; rs7311358) in the local Asian populations [10, 29]. The inferred haplotypes in each Asian ethnic group varied in terms of haplotype structure, haplotype frequency and htSNPs (Figures 2 and 3). The major haplotype block of the Indian subjects (block 2) displayed a low degree of haplotypic diversity in comparison with Chinese and Malay subjects, probably due to the relatively lower recombination events and therefore stronger LD among the SNPs in the Indian populations. Consistently, strong LD pattern and high interethnic differences in the genetic profile of SLCO1B3 suggest that a pharmacogenetic-based approach could provide valuable insights into the interethnic and interindividual variations in disposition of SLCO1B3 substrates.
To date, this is the first study to evaluate comprehensively the effect of SLCO1B3 pharmacogenetics on docetaxel disposition. Previous in vitro findings were inconsistent with regards to the effects of non-synonymous polymorphisms [334T>G (Ser112Ala; rs4149117 and 699G>A (Met233Ile; rs7311358)] on the transport characteristics of prototypical substrates, probably due to cell-line and substrate specific effects [3, 12, 13, 15]. The results of the in vivo studies delineating the impact of these SNPs on taxane disposition were also inconsistent [3, 4]. In this study, we evaluated the associations of SLCO1B3 genetic variants with docetaxel disposition in Chinese NPC patients. Our patients received a once weekly docetaxel regimen and a wide degree of interindividual variability was inherent in the pharmacokinetic parameters. The pharmacokinetic values in this study were in concordance with the previous report in various populations employing a similar dosing regimen to ours [30]. In line with our observations, a variation of up to 10-fold in docetaxel clearance was previously reported in the Caucasian population [31].Dexamethasone is a known PXR inducer that may potentially affect the expression level of CYP3A4/5 and hence impact on the metabolizing capacity of the CYP3A4/5 enzymes [32]. However, it is unlikely that dexamethasone would have influenced the variation in docetaxel disposition as it was uniformly administered to all participating patients.
Our present investigations based on the likelihood ratio test did not reveal any significant association of individual genotypes for the aforementioned polymorphisms and docetaxel disposition parameters. In our earlier study investigating the influence of candidate polymorphisms across the biochemical pathway of docetaxel, we identified a significant pair-wise association between the htSNP IVS12-5676A>G (rs11045585) and docetaxel pharmacokinetics [18]. To make this self-contained, our previous findings exhibited that the patients with the homozygous variant genotype of IVS12-5676A>G (rs11045585) were associated with higher AUC(0,∞) and lower CL of docetaxel compared with those harbouring reference or heterozygous genotype. No pharmacodynamic effect of this SNP was identified in our previous findings, although this SNP was shown to correlate with a higher incidence of neutropenia in the Japanese patients [33]. Since the dosing regimen and the pharmacokinetic data of the patients were not available in the Japanese study, it is unclear if the discrepancy could have been due to the differences in the dosing regimen between two studies. As postulated in our previous manuscript, the phenotypic effect of IVS12-5676A>G (rs11045585) may have been due to its being in haplotypic association with other unknown functional polymorphisms in the SLCO1B3 gene [18]. Notably, haplotype analyses have been recognized to be more robust than single marker analyses for identifying a genomic region that is enriched for phenotype-relevant causal variant(s) [26, 34]. Thus to elucidate further the pharmacogenetic impact of SLCOB3 on docetaxel disposition, a complete screening of the gene was carried out in the present study. A strong linkage disequilibrium pattern was detected across the SLCO1B3 SNPs in Asian populations and therefore, a haplotype-based approach was employed to delineate the genotypic-phenotypic associations.
Our study demonstrated that patients carrying the haplotype GAG*347insA had significantly lower CL and higher AUC(0,∞) of docetaxel compared with those harbouring the reference haplotype GGA*347wt, suggesting a reduced hepatic uptake of docetaxel by SLCO1B3. This haplotype is represented by the presence of the wild-type allele of intronic SNP IVS4+76G>A (rs4149118), as well as the variant alleles of the non-synonymous SNP 699G>A (rs7311358), intronic SNP IVS12+5676A>G (rs11045585) and *347_*348insA (rs3834935). The in silico model predicted that the 699G>A coding amino acid is located at the third extracellular loop of the transporter which has the potential to affect substrate specificity [35]. In addition, the variant allele in IVS12+5676A>G (rs11045585) was previously demonstrated to be linked with docetaxel-induced neutropenia [33] and altered docetaxel disposition [18]. The 3′UTR variant *347_*348insA is computationally predicted to fall in the microRNA miR-890 target binding site [36] and thus, it can be hypothesized that an insertion A might affect the miRNA binding leading to altered mRNA stability [37]. Interestingly, the GAG*347wt haplotype was associated with a significantly higher docetaxel CL compared with the effect of reference haplotype. This GAG*347wt haplotype displayed a distinct opposite effect (approximately two-fold difference) to that of the aforementioned GAG*347insA haplotype. As *347insA is the distinguishing SNP between these two haplotypes, this observation probably highlights the plausible role of the 3′UTR SNP to alter phenotypic effect. Nevertheless, the local haplotypic background (GAG-) probably imparts a major role for this altered phenomenon, as another haplotypic construct GGA*347insA (frequency 8%) consisting of *347insA did not exert a significant effect on docetaxel pharmacokinetics. It is worth investigating the haplotypic-phenotypic associations in in vitro models to ascertain the functional impact of the identified SLCO1B3 haplotypic constructs in the present study.
Our results suggest that the genotypic effects of SLCO1B3 on docetaxel pharmacokinetics are probably attributable to the local haplotypic background of these four key variants. A majority of the previous studies had focused on the variants 334T>G (Ser112Ala; rs4149117) and 699G>A (Met233Ile; rs7311358) which had resulted in inconsistent genotypic-phenotypic relationships [3, 4]. The inconsistency in SLCO1B3 mediated effects could have been due to an incomplete analysis of the highly polymorphic SLCO1B3 gene in patients of diverse ethnic origin. Remarkably, a strong LD pattern was detected across the variants in the SLCO1B3 gene in the present study. Hence, the linkage effect of the region may have been overlooked in the previous studies. As the sub-haplotypic construct consists of the coding, intronic and 3′UTR variants, the phenotypic effects of the haplotypes may likely be mediated through interactions involving post-transcriptional modifications and protein coding. The limitations of the present study are that the evaluation of the gene-dose relationship and the population-specific effect of the haplotypes were not possible. Our results indicate that the haplotype association analysis may better explain the variability in docetaxel disposition and warrant further investigations in other ethnic populations as well as patients receiving a different docetaxel regimen.
In conclusion, this study comprehensively screened for the genetic variations in SLCO1B3 and established its linkage and haplotype patterns in three distinct Asian ethnic groups in Singapore. The significant relationships of SLCO1B3 haplotypes with docetaxel pharmacokinetics highlight the involvement of SLCO1B3 pharmacogenetics in influencing the interindividual variability in docetaxel disposition. Future studies should be carried out to investigate the effects of the identified haplotypic constructs on the pharmacokinetics of putative substrates in distinct ethnic groups. Moreover, further studies are required to elucidate the functional characterization of the haplotype construct in relation to SLCO1B3 mediated docetaxel transport which would provide further insights into the mechanistic basis underlying alterations in docetaxel disposition.
Acknowledgments
The authors thank Ho Pey Ying, Ismail Osman, Soh Xin Jie, Lyn Goh Mei Ming and Nur Liyanah Bte Md Zaffre for their help in the sequencing work. This study was supported by grants from the National Medical Research Council Singapore NMRCB1011 and NMCCG10122.
Conflict of Interests
The authors declare no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1
SLCO1B3 htSNPs in the healthy Asian populations: (A) Chinese, (B) Malays and (C) Indians. A sequence clustering algorithm was applied to construct genealogic trees in order to identify the stratifying htSNPs on the basis of nucleotide base differences between the major haplotypes (ClustalW; EBI, Cambridge, UK). Haplotype name is designated with block and haplotype number as suffix, e.g. C_B1_H1 represents haplotype 1 identified in block 1 of the Chinese subjects.
Figure S2
Haplowalk analysis of the 15 SLCO1B3 htSNPs withdocetaxel CL. A region of seven SLCO1B3 htSNPs(IVS4+76G>A, 699G>A, IVS7-76T>C, IVS12-5676A>G,IVS13+55C>T, IVS14-23C>T and *347_*348insA; as highlighted inred) was identified to be significantly associated with docetaxelCL at P = 0.003. This test was performed with the haplowalkmethod implemented in the R haplostats package. The score testP value was adjusted for age, BSA, ECOG and albumin.
Table S1
Primer sequences and annealing temperature.
Table S2
Genotype and allele frequencies of SLCO1B3 polymorphismsin healthy Asian populations (n = 56 each for Chinese,Malays and Indians) and Chinese NPC patients (n = 50).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
REFERENCES
- 1.König J, Cui Y, Nies AT, Keppler D. Localization and genomic organization of a new hepatocellular organic anion transporting polypeptide. J Biol Chem. 2000;275:23161–8. doi: 10.1074/jbc.M001448200. [DOI] [PubMed] [Google Scholar]
- 2.Hagenbuch B, Meier PJ. The superfamily of organic anion transporting polypeptides. Biochim Biophys Acta. 2003;1609:1–8. doi: 10.1016/s0005-2736(02)00633-8. [DOI] [PubMed] [Google Scholar]
- 3.Smith NF, Marsh S, Scott-Horton TJ, Hamada A, Mielke S, Mross K, Figg WD, Verweij J, McLeod HL, Sparreboom A. Variants in the SLCO1B3 gene: interethnic distribution and association with paclitaxel pharmacokinetics. Clin Pharmacol Ther. 2007;81:76–82. doi: 10.1038/sj.clpt.6100011. [DOI] [PubMed] [Google Scholar]
- 4.Baker SD, Verweij J, Cusatis GA, van Schaik RH, Marsh S, Orwick SJ, Franke RM, Hu S, Schuetz EG, Lamba V, Messersmith WA, Wolff AC, Carducci MA, Sparreboom A. Pharmacogenetic pathway analysis of docetaxel elimination. Clin Pharmacol Ther. 2009;85:155–63. doi: 10.1038/clpt.2008.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith NF, Acharya MR, Desai N, Figg WD, Sparreboom A. Identification of OATP1B3 as a high-affinity hepatocellular transporter of paclitaxel. Cancer Biol Ther. 2005;4:815–8. doi: 10.4161/cbt.4.8.1867. [DOI] [PubMed] [Google Scholar]
- 6.Kullak-Ublick GA, Ismair MG, Stieger B, Landmann L, Huber R, Pizzagalli F, Fattinger K, Meier PJ, Hagenbuch B. Organic anion-transporting polypeptide B (OATP-B) and its functional comparison with three other OATPs of human liver. Gastroenterology. 2001;120:525–33. doi: 10.1053/gast.2001.21176. [DOI] [PubMed] [Google Scholar]
- 7.Van de Steeg E, van Esch A, Wagenaar E, van der Kruijssen CMM, van Tellingen O, Kenworthy KE, Schinkel AH. High impact of OATP1A/1B transporters on in vivo disposition of the hydrophobic anticancer drug paclitaxel. Clin Cancer Res. 2011;17:294–301. doi: 10.1158/1078-0432.CCR-10-1980. [DOI] [PubMed] [Google Scholar]
- 8.Abe T, Unno M, Onogawa T, Tokui T, Kondo TN, Nakagomi R, Adachi H, Fujiwara K, Okabe M, Suzuki T, Nunoki K, Sato E, Kakyo M, Nishio T, Sugita J, Asano N, Tanemoto M, Seki M, Date F, Ono K, Kondo Y, Shiiba K, Suzuki M, Ohtani H, Shimosegawa T, Iinuma K, Nagura H, Ito S, Matsuno S. LST-2, a human liver-specific organic anion transporter, determines methotrexate sensitivity in gastrointestinal cancers. Gastroenterology. 2001;120:1689–99. doi: 10.1053/gast.2001.24804. [DOI] [PubMed] [Google Scholar]
- 9.Muto M, Onogawa T, Suzuki T, Ishida T, Rikiyama T, Katayose Y, Ohuchi N, Sasano H, Abe T, Unno M. Human liver-specific organic anion transporter-2 is a potent prognostic factor for human breast carcinoma. Cancer Sci. 2007;98:1570–6. doi: 10.1111/j.1349-7006.2007.00570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laitinen A, Niemi M. Frequencies of single-nucleotide polymorphisms of SLCO1A2, SLCO1B3 and SLCO2B1 genes in a Finnish population. Basic Clin Pharmacol Toxicol. 2011;108:9–13. doi: 10.1111/j.1742-7843.2010.00605.x. [DOI] [PubMed] [Google Scholar]
- 11.Boivin AA, Cardinal H, Barama A, Pichette V, Hébert MJ, Roger M. Organic Anion Transporting Polypeptide 1B1 (OATP1B1) and OATP1B3: Genetic variability and haplotype analysis in White Canadians. Drug Metab Pharmacokinet. 2010;25:508–15. doi: 10.2133/dmpk.dmpk-10-sh-046. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz UI, Meyer zu Schwabedissen HE, Tirona RG, Suzuki A, Leake BF, Mokrab Y, Mizuguchi K, Ho RH, Kim RB. Identification of novel functional organic anion-transporting polypeptide 1B3 polymorphisms and assessment of substrate specificity. Pharmacogenet Genomics. 2011;21:103–14. doi: 10.1097/FPC.0b013e328342f5b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letschert K, Keppler D, König J. Mutations in the SLCO1B3 gene affecting the substrate specificity of the hepatocellular uptake transporter OATP1B3 (OATP8) Pharmacogenetics. 2004;14:441–52. doi: 10.1097/01.fpc.0000114744.08559.92. [DOI] [PubMed] [Google Scholar]
- 14.Picard N, Yee SW, Woillard JB, Lebranchu Y, Le Meur Y, Giacomini KM, Marquet P. The role of organic anion-transporting polypeptides and their common genetic variants in mycophenolic acid pharmacokinetics. Clin Pharmacol Ther. 2009;87:100–8. doi: 10.1038/clpt.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada A, Sissung T, Price DK, Danesi R, Chau CH, Sharifi N, Venzon D, Maeda K, Nagao K, Sparreboom A, Mitsuya H, Dahut WL, Figg WD. Effect of SLCO1B3 haplotype on testosterone transport and clinical outcome in Caucasian patients with androgen-independent prostatic cancer. Clin Cancer Res. 2008;14:3312–8. doi: 10.1158/1078-0432.CCR-07-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsujimoto M, Dan Y, Hirata S, Ohtani H, Sawada Y. Influence of SLCO1B3 gene polymorphism on the pharmacokinetics of digoxin in terminal renal failure. Drug Metab Pharmacokinet. 2008;23:406–11. doi: 10.2133/dmpk.23.406. [DOI] [PubMed] [Google Scholar]
- 17.Yamakawa Y, Hamada A, Nakashima R, Yuki M, Hirayama C, Kawaguchi T, Saito H. Association of genetic polymorphisms in the influx transporter SLCO1B3 and the efflux transporter ABCB1 with imatinib pharmacokinetics in patients with chronic myeloid leukemia. Ther Drug Monit. 2011;33:244–50. doi: 10.1097/FTD.0b013e31820beb02. [DOI] [PubMed] [Google Scholar]
- 18.Chew SC, Singh O, Chen X, Ramasamy RD, Kulkarni T, Lee EJD, Tan EH, Lim WT, Chowbay B. The effects of CYP3A4, CYP3A5, ABCB1, ABCC2, ABCG2 and SLCO1B3 single nucleotide polymorphisms on the pharmacokinetics and pharmacodynamics of docetaxel in nasopharyngeal carcinoma patients. Cancer Chemother Pharmacol. 2011;67:1471–8. doi: 10.1007/s00280-011-1625-9. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz U, Dixit S, Leake B, Kim R. Identification and functional characterization of OATP1B3 allelic variants using transfected hela cells. Drug Metab Rev. 2006;38:237. [Google Scholar]
- 20.Ngeow J, Lim WT, Leong SS, Ang MK, Toh CK, Gao F, Chowbay B, Tan EH. Docetaxel is effective in heavily pretreated patients with disseminated nasopharyngeal carcinoma. Ann Oncol. 2011;22:718–22. doi: 10.1093/annonc/mdq425. [DOI] [PubMed] [Google Scholar]
- 21.Loos WJ, Verweij J, Nooter K, Stoter G, Sparreboom A. Sensitive determination of docetaxel in human plasma by liquid-liquid extraction and reversed-phase high-performance liquid chromatography. J Chromatogr B Biomed Sci Appl. 1997;693:437–41. doi: 10.1016/s0378-4347(97)00089-3. [DOI] [PubMed] [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S. PLINK (1.02) [Internet]. Available at http://pngu.mgh.harvard.edu/purcell/plink/ (last accessed 30 June 2011)
- 25.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–6. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 27.Karami S, Brennan P, Rosenberg PS, Navratilova M, Mates D, Zaridze D, Janout V, Kollarova H, Bencko V, Matveev V, Szeszenia-Dabrowska N, Holcatova I, Yeager M, Chanock S, Menashe I, Rothman N, Chow WH, Boffetta P, Moore LE. Analysis of SNPs and haplotypes in vitamin D pathway genes and renal cancer risk. PLoS ONE. 2009;4:e7013. doi: 10.1371/journal.pone.0007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore LE, Brennan P, Karami S, Menashe I, Berndt SI, Dong LM, Meisner A, Yeager M, Chanock S, Colt J, Schwartz K, Davis F, Zaridze D, Mattveev V, Janout V, Kollarova H, Bencko V, Navratilova M, Szeszenia-Dabrowska N, Mates D, Holcatova I, Boffetta P, Chow WH, Rosenberg PS, Rothman N. Apolipoprotein E/C1 locus variants modify renal cell carcinoma risk. Cancer Res. 2009;69:8001–8. doi: 10.1158/0008-5472.CAN-09-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsujimoto M, Hirata S, Dan Y, Ohtani H, Sawada Y. Polymorphisms and linkage disequilibrium of the OATP8 (OATP1B3) gene in Japanese subjects. Drug Metab Pharmacokinet. 2006;21:165–9. doi: 10.2133/dmpk.21.165. [DOI] [PubMed] [Google Scholar]
- 30.Cox MC, Low J, Lee J, Walshe J, Denduluri N, Berman A, Permenter MG, Petros WP, Price DK, Figg WD, Sparreboom A, Swain SM. Influence of garlic (Allium sativum) on the pharmacokinetics of docetaxel. Clin Cancer Res. 2006;12:4636–40. doi: 10.1158/1078-0432.CCR-06-0388. [DOI] [PubMed] [Google Scholar]
- 31.Baker SD, Sparreboom A, Verweij J. Clinical pharmacokinetics of docetaxel : recent developments. Clin Pharmacokinet. 2006;45:235–52. doi: 10.2165/00003088-200645030-00002. [DOI] [PubMed] [Google Scholar]
- 32.Pascussi J, Drocourt L, Gerbal-Chaloin S, Fabre J, Maurel P, Vilarem M. Dual effect of dexamethasone on CYP3A4 gene expression in human hepatocytes. Eur J Biochem. 2001;268:6346–57. doi: 10.1046/j.0014-2956.2001.02540.x. [DOI] [PubMed] [Google Scholar]
- 33.Kiyotani K, Mushiroda T, Kubo M, Zembutsu H, Sugiyama Y, Nakamura Y. Association of genetic polymorphisms in SLCO1B3 and ABCC2 with docetaxel-induced leukopenia. Cancer Sci. 2008;99:967–72. doi: 10.1111/j.1349-7006.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scherag A, Jarick I, Grothe J, Biebermann H, Scherag S, Volckmar AL, Vogel CIG, Greene B, Hebebrand J, Hinney A, Sorensen TIA. Investigation of a genome wide association signal for obesity: synthetic association and haplotype analyses at the melanocortin 4 receptor gene locus. PLoS ONE. 2010;5:e13967. doi: 10.1371/journal.pone.0013967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier-Abt F, Mokrab Y, Mizuguchi K. Organic anion transporting polypeptides of the OATP/SLCO superfamily: identification of new members in nonmammalian species, comparative modeling and a potential transport mode. J Membr Biol. 2005;208:213–27. doi: 10.1007/s00232-005-7004-x. [DOI] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2007;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra PJ, Humeniuk R, Longo-Sorbello GSA, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. PNAS. 2007;104:13513–8. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.