Abstract
Alterations in genome sequence and structure contribute to somatic disease, affect the fitness of subsequent generations and drive evolutionary processes. The critical roles of highly accurate replication and efficient repair in maintaining overall genome integrity are well known, but the more localized stability costs associated with transcribing DNA into RNA molecules are less appreciated. Here we review the diverse ways that the essential process of transcription alters the underlying DNA template and thereby modifies the genetic landscape.
Introduction
Changes to genomic DNA can be in the form of mutations that alter the primary sequence or rearrangements that alter chromosome structure. Most mutations and rearrangements are assumed to arise either randomly during the process of genome duplication or in response to DNA damage, and multiple repair pathways have evolved to maintain such changes at an acceptably low level. The consequences of too much genetic instability are particularly evident in humans, where a loss of repair capacity is associated with cancer predisposition syndromes and with aging. Whereas replication involves making precisely one copy of each DNA strand throughout the genome, the transcription of DNA into RNA products is a comparatively non-uniform process; it affects only defined segments of the genome, it typically copies only one strand of the DNA, and it occurs at highly variable rates.
The first indication that transcription might do more than just passively copy the DNA template came from bacterial studies in the 1970s demonstrating that exogenous mutagens were more efficient at inducing mutations if the reporter were highly transcribed 1,2. It was not until 15 years later that transcription was demonstrated to also stimulate spontaneous mutagenesis in eukaryotes, specifically in budding yeast 3. This phenomenon, which locally and permanently alters the primary sequence of the DNA template, is referred to as transcription-associated mutagenesis or TAM. It should be noted that TAM is not to be confused with so-called “transcriptional mutagenesis,” which refers to the production of mutant mRNAs and proteins through transient alterations in the DNA template 4. The discovery of transcription-associated recombination (TAR) came through identification of HOT1 in yeast, a sequence that stimulates mitotic recombination by promoting high levels of transcription 5,6. Transcription thus has the potential to modify the genetic landscape by locally altering mutation rates, by stimulating loss of heterozygosity and by generating diverse types of rearrangements that include deletions, duplications, inversions and translocations.
Studies of TAM and TAR typically require a selective system to identify rare mutants and recombinants, respectively, as well as regulation of the system by a promoter whose activity can be tightly controlled. Depending on the system used, the difference between low- and high-transcription levels can be orders of magnitude. Microorganisms such as Escherichia coli and Saccharomyces cerevisiae have been the experimental organisms of choice for practical reasons: rapid growth, ease of genetic manipulation, well-defined replication origins and availability of reporter systems. In general, specific mechanisms of TAM and TAR have been deduced by studying the effects of depleting individual proteins involved in DNA and/or RNA metabolism. This is trivial to do through standard gene-targeting techniques in microorganisms and now analogous studies in metazoans have become more feasible through the development of RNA interference technologies.
Recent efforts have moved away from simple descriptive analyses of TAM and TAR, and have focused on understanding how transcription destabilizes the underlying DNA template. The level of transcription is clearly important; in microorganisms, the rate of TAM is directly proportional to transcription rate 7,8, and a similar proportionality has been reported for TAR in mammalian cells 9. An early conceptual link between TAM and TAR derived from the fact that mutation and recombination each reflect a normal mechanism for dealing with DNA damage. The initial assumption was that there might be a single source of the transcription-associated damage underlying TAM and/or TAR. Indeed, early studies indicated that TAM primarily reflects damage to the nontranscribed strand of the DNA template, whereas TAR is largely due to transcription-replication conflicts. Recent studies have made it apparent, however, that there are multiple mechanisms that contribute to TAM and TAR. Importantly, transcription can affect stability of the template by mechanisms that are separate from DNA replication, potentially making transcription-associated alterations a key contributor to genetic changes in nondividing cells.
In the following sections, we begin by considering the significance of replication-transcription conflicts to genetic instability and how persistent association of the RNA transcript with the template DNA strand exacerbates these conflicts. Next, we summarize recent studies that highlight how the primary sequence of actively transcribed DNA, in particular its propensity to form non-B secondary structures, contributes to instability. Finally, our current knowledge of factors that contribute to TAM will be summarized. An important point to be borne in mind throughout is that even though an experimental observation or mechanism may currently be limited to a single organism, the high evolutionary conservation of DNA structure and of basic DNA metabolic processes makes it likely that documented mechanisms of TAM and TAR will be broadly applicable.
Transcription and replication collisions
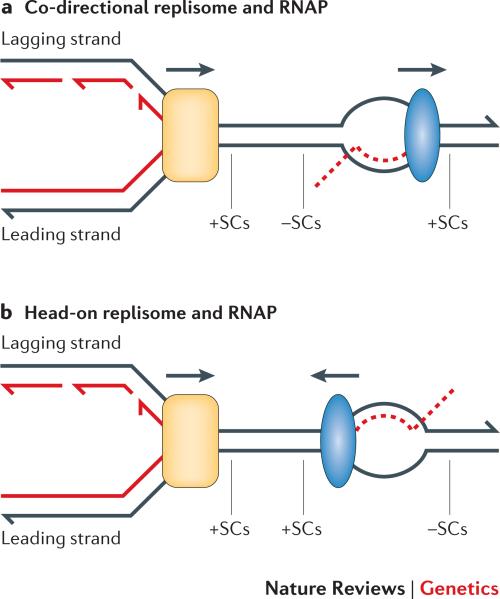
Transcription and replication occur on the same template, making conflicts between these two processes inevitable. Whereas replication copies both strands of duplex DNA, only one strand is typically transcribed by RNA polymerase (RNAP); the transcribed strand can thus correspond to either the leading- or lagging-strand template of replication (Figure 1). When the leading strand of replication is the transcribed strand, the replication and transcription forks move in the same direction; when the lagging strand is the template for transcription, the forks converge. We will refer to the resulting RNAP-replisome conflicts as co-directional and head-on collisions, respectively. In addition to direct conflicts between transcription and replication, positive supercoils accumulate ahead of both machineries, which poses an additional problem for head-on collisions. Such unresolved helical stress can trigger replication fork reversal 10, giving rise to a “chickenfoot” structure that can be enzymatically processed into a recombination-initiating double-strand break 11.
Figure 1. Co-directional and head-on orientations of RNA polymerase and the replisome.
a | In the co-directional orientation, the transcribed strand (TS) is the leading-strand template for replication. b | In the head-on orientation, the TS is the lagging-strand template. Positive supercoils (+SCs) accumulate ahead of replisome; +SCs and negative supercoils (–SCs) accumulate ahead of and behind RNAP, respectively. Nascent DNA and RNA are indicated as solid and dashed red lines, respectively; arrowheads are at the 3′ ends of DNA strands. RNA polymerase is in blue and the replisome in yellow.
The cost of head-on collisions
The relevance of RNAP-replisome collisions to genome instability has been extensively reviewed 11-13, and the general consensus is that head-on collisions are more destabilizing because they impede replication to a greater extent than co-directional collisions. An early indication that head-on collisions are more problematic came from the striking co-directional orientation of all seven of the highly expressed ribosomal RNA (rRNA) operons in the E. coli genome 14, a pattern that has been observed in over 80 prokaryotic species 15. Importantly, it was recently demonstrated that reversing the orientation of rRNA genes in Bacillus subtilis negatively affects fitness 16,17. In budding yeast, rRNA transcription units are insulated from head-on collisions by replication fork barriers, which physically block forks from entering the distal end of transcription units 18. Finally, within 50 kb of putative human replication origins, it has been estimated that genes transcribed co-directionally with replisome movement outnumber by 8 fold those with a head-on orientation 19. Although genome instability can result from transcription-associated disruptions in replication, it should be noted that the disruption of transcription caused by replication also can be potentially costly for fitness.
What happens when a replication fork approaches an actively transcribed gene? At least in in vitro studies with purified E. coli components, a replisome moving in either direction appears to dissociate RNAP from the template as it passes 12. As for the effect on replication, 2D gel analysis of forks has detected discrete replisome pausing with the head-on, but not with the co-directional, RNAP-replisome orientation in both E. coli 20 and yeast 21-23. More recently, ChIP-chip analysis of sequences preferentially associated with a yeast replicative DNA polymerase was used to infer positions of slow fork movement throughout the genome 24. These sites were positively correlated with transcription, although in this case, a replication-fork slowing was evident regardless of the relative orientation of transcription and replication. The 2D gel and ChIP-chip results, however, should not be considered contradictory, as the resolution of pausing is very different in the two types of assays.
TAR as a replication-dependent event
It is well known that replication roadblocks can be bypassed and replication at collapsed/broken forks re-established through homologous recombination 25. It is, therefore, not surprising that TAR has been linked to DNA replication. In yeast, for example, cell cycle-regulated promoters were used to show that transcription only in S-phase was able to stimulate recombination 22. A similar S-phase connection has been inferred in mammalian studies, in which TAR was detected only in cycling cells 26. In the case of yeast, the recombination-promoting potential of co-directional versus head-on collisions between the replisome and RNAP has been specifically examined. Consistent with a greater impairment of replication fork movement by head-on collisions, TAR was enhanced in the head-on relative to co-directional orientation 22,23,27. A similar orientation-specific effect has been reported in an analysis of transcription-associated gross chromosomal rearrangements in yeast 28, and with transcription-associated deletions in a plasmid-based E. coli assay 29. Despite multiple examples of greater instability with head-on than with co-directional replisome-RNAP collisions, it should be noted that the relative instability reverses in E. coli when DNA polymerase encounters a stalled rather than a processive RNAP elongation complex 30.
An additional point to consider with respect to TAR concerns the recent demonstration that highly expressed yeast genes are tethered to the nuclear pore, a phenomenon referred to as “gene gating” 31. This association facilitates mRNA export but creates an orientation-independent topological barrier that hinders replication fork progression 32. One function of the replication checkpoint is to sever this connection, preventing topologically driven fork reversal and allowing replication to continue. Whether such gene gating is relevant to TAR has not been specifically examined.
Is replication fork direction relevant to TAM?
In addition to stimulating recombination, an interesting possibility is that the transcription-associated slowing/pausing of replication forks might also lead to recruitment of translesion synthesis DNA polymerases. These polymerases are specialized to bypass lesions that stall replicative DNA polymerases and can have extraordinarily low fidelity on undamaged DNA templates 33. Their inappropriate recruitment would thus be expected to be mutagenic, and could be a potential source of TAM. Although there is as yet no compelling connection between replication-fork direction and TAM in eukaryotes, a higher rate of mutagenesis with head-on than with co-directional RNAP-replisome movement has been reported in B. subtilis 17. It is also intriguing to note that reversing the direction of replication through a highly expressed mutation reporter subtly affects the spectrum of TAM in yeast 7.
Transcription-associated R-loops
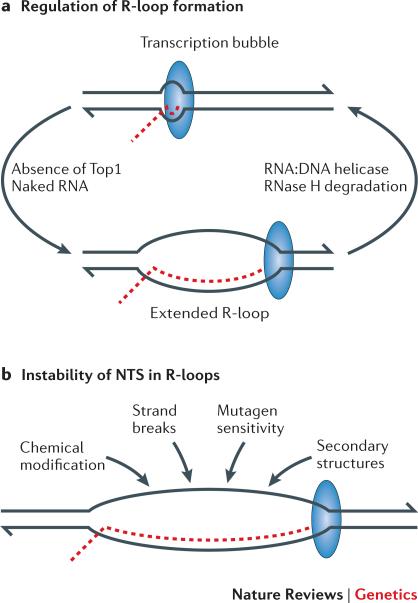
RNA exits through a channel in RNAP as it is synthesized, thereby disrupting its base pairing with the complementary DNA template strand. The nascent RNA has the potential to anneal back to the transcribed strand (TS), however, creating a stable RNA:DNA hybrid and leaving the non-transcribed strand (NTS) exposed as an extended, single strand of DNA 34-36. This structure, called an R-loop, can be over 1 kb in length in highly transcribed genes (Figure 2).
Figure 2. Factors that promote and remove R-loops during transcription.
a | R-loop formation is favored by the negative supercoils that accumulate in the absence of Top1 and by naked RNA that fails to be engaged immediately after transcription. In bacteria, the coupling between transcription and translation prevents the accumulation of naked RNA. In eukaryotes, RNA is co-transcriptionally assembled into ribonucleoprotein particles for splicing and/or nuclear transport. R-loops can be actively unwound by an RNA:DNA helicase or the RNA component degraded by RNase H. b | Factors expected to affect the exposed nontranscribed strand within R-loops. DNA strands are in black, with 3′ ends indicated by the half-arrowheads; the RNA transcript is in red; and the large blue oval corresponds to RNA polymerase.
Preventing the accumulation of R-loops
In bacteria, mRNA is immediately engaged in translation by ribosomes as it exits RNAP. When transcription and translation become uncoupled, transcription rapidly terminates, thereby blocking the expression of all downstream genes in polycistronic operons. This phenomenon, known as “polarity,” not only prevents the energy costs associated with the continued production of untranslated mRNA, it also blocks the accumulation of naked RNA that can potentially anneal back to the template DNA strand to form an R-loop 36. In eukaryotes, where transcription and translation occur in separate cellular compartments, nascent RNAs are co-transcriptionally assembled into ribonucleoprotein particles (RNPs) that promote splicing and/or nuclear export. While these cotranscriptional processes undoubtedly increase the efficiency of mRNA processing/transport, they also, as in prokaryotes, prevent the accumulation of naked RNA. Even in those cases where the RNA is the final gene product (e.g., rRNA and tRNA), either extensive formation of RNA secondary structure or rapid association with proteins discourages reannealing of the transcript to the DNA template.
In addition to co-transcriptional RNA engagement, R-loops are also held in check by the activity of topoisomerase I (Top1; Box 1), an enzyme that relaxes superhelical stress in duplex DNA. During transcription, positive and negative supercoils accumulate ahead of and behind RNAP complexes, respectively, forming “twin domains” of helical stress (Figure 1) 37. Positive supercoils can impede further DNA unwinding, whereas excessive negative supercoiling imparts single-stranded character to duplex DNA and thereby promotes R-loop formation. In murine cells, Top1 also limits R-loop formation through the regulation of RNA splicing and RNP assembly factors 38.
If the mechanisms that normally prevent R-loop formation fail, there are back-up mechanisms in place to remove these structures. This is a major function of the RNAse H class of enzymes, which specifically degrades the RNA component of RNA:DNA hybrids 39. The growth defect associated with a Top1 deficiency, for example, can be rescued by overexpression of RNase H in bacterial or human cells 38,40. Finally, the Sen1 helicase of budding yeast and its human homolog senataxin, as well as the RecG helicase of E. coli, have been implicated in unwinding R-loop structures 41-43.
R-loops and genetic instability
The connection between R-loops and TAR initially emerged through study of the hpr1 hyper-recombination mutant of yeast. Transcript elongation was shown to be impaired in hpr1 mutants 44 and this defect, together with the hyper-recombination phenotype, was suppressed by RNase H overproduction 45. Hpr1 is a subunit of THO complex, which interacts with the TREX complex to facilitate mRNA export 46. A general model that emerged from these studies is that, in the absence of THO/TREX components, the nascent RNA fails to assemble properly into RNPs. This in turn promotes R-loop formation, which impedes transcript elongation, causes conflicts with replication and promotes recombination 35. On a more global scale, recent ChIP-chip analysis has demonstrated that THO components preferentially associate with active ORFs genome-wide 47. In the absence of the THO complex, replisome movement is slower through these regions, and RNase H overexpression suppresses this effect.
A recent genome-wide screen in yeast has further broadened the involvement of RNA biogenesis in R-loop formation beyond THO/TREX, and further suggests that the associated DNA damage may sometimes occur outside the context of DNA replication 48. Finally, in the actively transcribed rDNA locus of yeast, both R-loops and extended regions of single-stranded DNA accumulate in the absence of Top1 and impede transcription 49,50. Importantly, loss of Top1 is associated with the accumulation of extrachromosomal rDNA circles formed via homologous recombination 51, providing an additional connection between R-loops and TAR.
In higher eukaryotes, the relevance of R-loop formation to genome instability was first documented in chicken DT40 cells, where depletion of the splicing factor ASF/SF2 resulted in the accumulation of R-loops and elevated genome rearrangements 52. A genome-wide screen for functions relevant to genome maintenance in human cells has uncovered roles for diverse RNA-processing factors, with perturbations in RNA biogenesis again being associated with R-loop formation 53. Although the THO/TREX complex was not picked up in this analysis, directed depletion of individual components in human cells has demonstrated evolutionary conservation of its role in RNP biogenesis and genome stability 54.
A potential way to minimize the genome-destabilizing effects of R-loops in eukaryotes is to temporally separate transcription and replication during the cell cycle. This is not an option, however, for very large genes whose complete transcription requires more than a single cell cycle. Indeed, a subset of common fragile sites of human chromosome breakage map to very long genes, and fragility has been associated with transcription and R-loop formation during S phase 55. In the context of maintaining a stable genome, R-loops are clearly pathological structures. It should be noted, however, that there are at least two instances where these structures are physiologically relevant. In bacteria, the RNA component of an R-loop is typically used as a primer to initiate DNA synthesis, and it could have a similar function in restarting replication forks in eukaryotes. In addition, R-loops are important in the vertebrate immune system, where they are proposed to play a role in facilitating class-switch recombination (Box 2) 56.
Transcription and non-B DNA structures
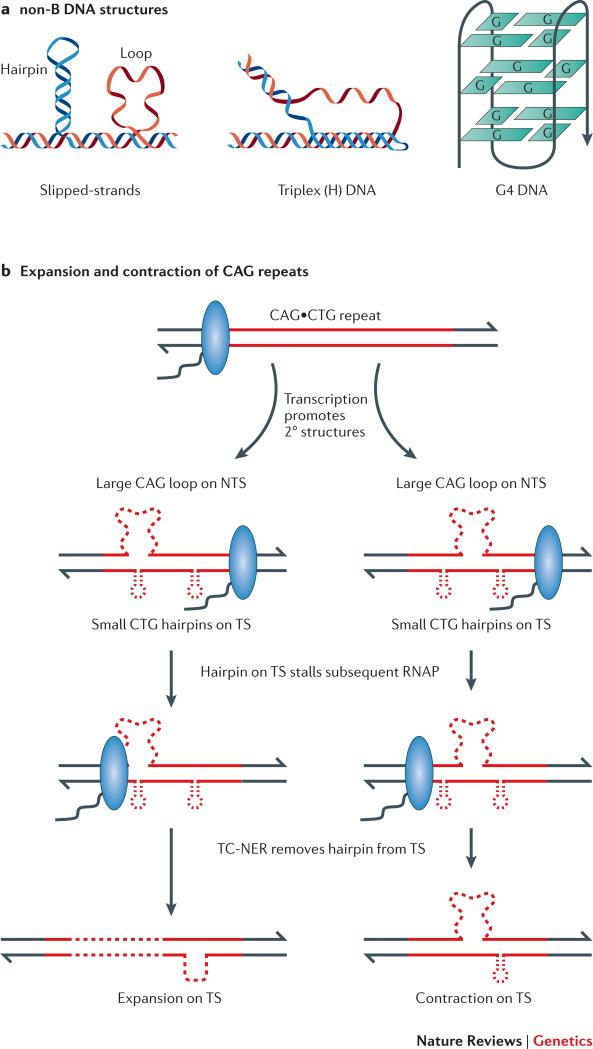
All DNA transactions (replication, repair, recombination and transcription) require the transient separation of complementary strands, providing the opportunity for single-stranded DNA to assume non-canonical, non-B DNA structures (Figure 3a). During transcription, the single-stranded character of the negatively supercoiled region that accumulates behind RNAP also can facilitate the formation of non-B structures. R-loops likewise facilitate the accumulation of non-canonical DNA structures on the NTS, and non-B DNA on the NTS promotes and stabilizes R-loops. In this section, we consider the destabilizing potential of two types of sequences known to form non-B DNA structures: guanine-rich sequences and trinucleotide repeats.
Figure 3. non-B DNA structures and genome instability.
a | Representative non-B DNA structures are illustrated. b | Transcription through CAG•CTG repeats promotes the formation of slipped-strand structures, which subsequently stall RNA polymerase (RNAP) and lead to recruitment of the nucleotide excision repair (NER) machinery. Transcription-coupled NER removes the portion of the transcribed strand containing the RNAP-blocking hairpin; the resulting gap is filled in using the nontranscribed strand (NTS) as a template. Depending on the location of loops on the NTS relative to the removed hairpin, the repair event will either expand or contract the trinucleotide repeat.
Co-transcriptional G4 DNA and TAR
G-rich sequences can form stable, non-B structures known as G-quadruplex or G4 DNA, which is comprised of a stacked array of G quartets (Figure 3a); a G quartet is a planar structure in which four guanines are paired through Hoogsteen hydrogen bonds 57. An involvement of G4 DNA in genome instability is suggested by class-switch recombination in the vertebrate immune system, a process that requires transcription and occurs between GC-rich “switch” regions adjacent to the constant region gene segments of the immunoglobulin heavy-chain locus (Box 2). A distinguishing feature of the switch regions is the asymmetric distribution of guanines and cytosines between the two DNA strands; in the physiological orientation, the G-rich strand is the NTS. When switch regions are highly expressed in vitro or in bacterial cells, bubbles containing G4 DNA opposite an R-loop (a “G-loop”) have been observed by electron microscopy 58. Co-transcriptional G-loops have been shown to block transcription in vitro 59, and presumably would be a potent block to replication as well. Consistent with possible relevance for R-loops and/or G-loops in class-switch recombination, inversion of the switch region relative to the promoter converts the G-rich strand to the transcription template and eliminates most class-switch recombination 60.
We recently introduced a murine switch-region fragment into the yeast genome, and examined the effects of transcription and of fragment orientation on stability, using recombination as a read out 61. Recapitulating observations in the immune system, recombination was enhanced by transcription and was further elevated when the G-rich strand was the NTS. Consistent with a causal role for co-transcriptional R-loop/G-loop formation, loss of either Top1 or RNase H activity exacerbated the instability of switch-region sequences. Similarly, studies in E. coli have demonstrated an orientation-dependent effect of a switch-region sequence on TAR, which was accompanied by replication fork slowing and was suppressed by RNase H overexpression 62.
Is G4 DNA relevant to transcription-associated genome instability beyond the specialized case of class-switch recombination? It seems likely, although only the destabilizing effect of G4 DNA in the context of DNA replication has been examined to date. For example, the GC-rich, human CEB1 minisatellite, which efficiently forms G4 DNA in vitro, is highly unstable when introduced into the yeast genome. CEB1 becomes more unstable upon loss of the Pif1 helicase, which is capable of disrupting the G4 DNA formed by CEB1 in vitro 63. In addition, ChIP-chip analysis has uncovered sequences with G4-forming potential as preferential sites of yeast Pif1 association and as sites of replication slowing 64.
Instability of trinucleotide repeats
The expansion of trinucleotide repeats (TNRs) is responsible for at least 20 neurodegenerative and neuromuscular diseases, including Fragile X syndrome (CGG•CCG), Huntington's disease (CAG•CTG), and Friedreich's ataxia (GAA•TTC) 65. Significantly, only those TNRs capable of forming non-B DNA structures have been implicated in disease, leading to belief that their formation is a critical, early step that initiates instability 66. Large germline expansions of TNRs have been most extensively studied and are thought to arise during replication; such expansions account for the initial disease appearance and the increase in severity in subsequent generations, a phenomenon referred to as “anticipation”. TNR expansions that occur in somatic cells have attracted less attention, but may be relevant to disease severity and progression, especially in non-dividing tissue 67. Expansions in non-dividing cells are, by definition, replication independent and recent work exploring transcription-associated TNR instability will be briefly summarized below.
Transcriptional effects on TNR instability have been characterized in human cell lines using CAG•CTG repeats, which can form stable slipped-hairpin structures (Figure 3a) 68. Such structures on either the TS or the NTS are sufficient to stall RNAP in vitro 69. Key to the analysis of CAG•CTG instability in human cells has been the development of a transcriptionally regulated reporter that allows the direct selection of contraction events 70. Induction of transcription elevates contractions approximately 15-fold in this system, and this property has been used to identify proteins that enhance or suppress instability. When considered together, the data are consistent with the model shown in Figure 3b, in which transcription promotes the formation of hairpins, which can stall the passage of subsequent RNAPs to trigger “gratuitous” transcription-coupled nucleotide excision repair (TC-NER). Depending on the location of slip-outs relative to the stalled RNAP, the repair process can lead to either contractions or expansions of CAG•CTG repeats 69,71.
Only contractions can be selected in the mammalian experimental system noted above, but a Drosophila-based system has been used to identify transcription-associated expansions of CAG•CTG repeats, and these likewise depend on TC-NER 72. It seems likely that similar, transcription-based instability mechanisms will also apply to other classes of TNRs. GAA•TTC repeats, for example, have the propensity to form three-stranded H-DNA or triplex DNA (Figure 3a) 73; they promote R-loop formation in bacterial cells 74 and are destabilized by transcription in mammalian cells 75,76. To date, there have been no reports of transcriptional effects on CGG•CCG stability.
Analyses of expressed sequences in mammalian cells indicate that both strands of DNA are often transcribed 77. Therefore, recent work has examined the effect of simultaneously transcribing through CAG•CTG repeats in both directions 78. Significantly, convergent transcription led to greater instability than would be predicted by summing the individual effects of forward and reverse transcription 79,80, and was an efficient trigger of apoptosis 79.
Transcription-associated DNA damage and TAM
The disabling of DNA damage repair or bypass pathways in yeast elevates spontaneous TAM in defined reversion assays, implicating transcription-associated damage as a causative factor 3,81. Striking synergistic effects of transcription and exogenous mutagens on TAR have also been reported, again consistent with an enhanced accumulation of damage in highly transcribed DNA 82. Unexpectedly, recent work has also found that Top1 activity, which reduces R-loop associated TAR, can be an important source of TAM 83,84.
Chemical modification of DNA biases TAM to the NTS
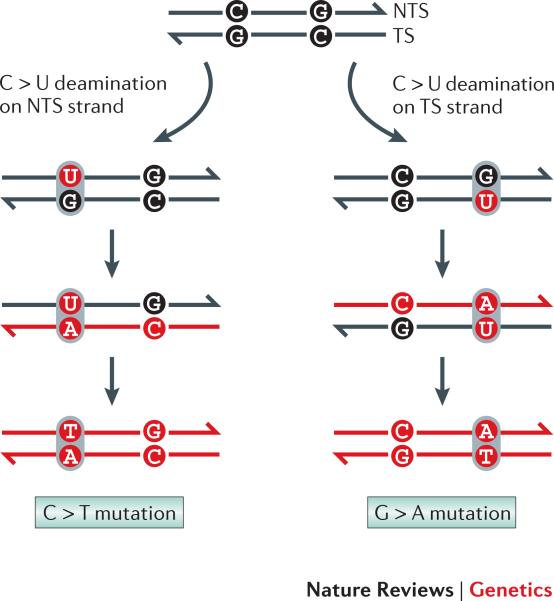
In reversion assays, all types of base substitutions appear to be stimulated by transcription in yeast 85 and bacterial cells 85-87; a large variety of transcription-associated insertions/deletions additionally have been detected in yeast 7,81. The strand-of-origin of a given mutation is impossible to deduce in wild-type cells, but it can be assigned with confidence in some repair-defective backgrounds. In the absence of uracil DNA glycosylase, for example, CG > TA transitions can be assumed to result from cytosine deamination to uracil rather than from damage to guanine on the complementary strand (Figure 4). In E. coli, such mutations are strongly biased to cytosines located on the NTS when transcription is highly activated 88,89, as are G > T mutations associated with oxidation or loss of guanine 90. Because chemical modifications occur much more often in single-stranded than in duplex DNA 91, it has been argued that the enhanced single-stranded character of the NTS is an important contributor to TAM.
Figure 4. Deducing the strand on which mutations arise.
In the absence of uracil removal, deamination of C on the nontranscribed strand (NTS) leads to C > T mutations (note: by convention, DNA sequences are read from the NTS, which has the same sequence as the mRNA). In contrast, deamination of C on the transcribed strand results in G > T mutations.
An NTS-related bias for spontaneous TAM has not been examined in yeast, but it has been demonstrated that enzymatic deamination of cytosine to uracil primarily targets cytosines located on the NTS of active genes in THO mutants 92. Such deamination is required for the somatic hypermutation of immunoglobulin genes, a specialized process in which the initiating event accesses primarily the NTS through a transcription-dependent process (Box 2). A much more general, strand-specific effect of transcription has been inferred through whole-genome sequencing. A comparative analysis of nine mammalian genomes, for example, reported a bias for A > G relative to T > C mutations on the NTS 93, with the degree of asymmetry correlating with the gene expression level in the germline 94. More recently, an examination of somatic “passenger” mutations in rapidly evolving tumor cells has suggested that a similar asymmetry is generated during somatic divisions 95.
The strand asymmetries associated with TAM could reflect the transient exposure of small regions of the NTS within the transcription bubble that moves with RNAP and/or the formation of more extensive regions behind the transcription machinery. Although the negative supercoils formed in the wake of RNAP would be expected to impart single-stranded character to both strands, the associated formation of R-loops would confine the exposed DNA to the NTS. A more refined model has been proposed in which secondary structures formed on the NTS leave only specific bases exposed in an unpaired and vulnerable state 96. An algorithm has been developed to predict the most likely sites of potential damage, and its predictive value demonstrated using a bacterial reporter 97. Intriguingly, this analysis has been extended to the human p53 gene, in which the predicted frequency of base exposure positively correlates with the frequency of mutation at the 12 most mutable p53 sites in human tumors 98.
An alternative, but not necessarily mutually exclusive, model for the biased accumulation of mutations on the NTS is that transcription-coupled nucleotide excision repair (TC-NER) preferentially removes lesions from the TS 99. A TC-NER dependent modulation in mutation spectra, for example, has been reported for E. coli treated with alkylating agents 100 or exposed to UV irradiation 101. In yeast, we recently demonstrated that TC-NER likewise alters spontaneous mutation patterns in a highly transcribed gene 85. Finally, TC-NER has been invoked to explain a mutagen-induced NTS bias for mutations in the p53 tumor suppressor gene in human cells 102. Although it is generally accepted that transcription biases mutations to the NTS, a reversal in this pattern has been reported following the treatment of mouse ES cells with UV 103,104.
Top1 activity as a source of TAM
In repair-competent yeast cells, the class of forward mutations increased most by transcription is comprised of short deletions (2-5 bp). These signature deletions are uniquely dependent on the activity of Top1 and presumably reflect its recruitment to relieve the supercoiling associated with transcriptionally active DNA 83,84. Recent analyses suggest that Top1-dependent mutations have two distinct causes (see Box 1). The first reflects an irreversible trapping of Top1 on DNA during its normal cleavage-ligation cycle, while the second reflects Top1-mediated cleavage at a ribonucleotide monophosphate (rNMP) misincorporated into DNA 105. Whether both types of Top1 cleavage product generate the observed deletions through a common intermediate or via distinct mechanisms is not known.
Transcription-associated changes in DNA composition
We recently reported that transcription in yeast alters the nucleotide composition of the underlying template in a very unexpected way: specifically increasing the direct incorporation of dUMP in place of dTMP 106. This was discovered through genetic analyses of a distinctive TAM signature associated with reduced efficiency of the base excision repair pathway, which is specialized to repair abasic sites. Significantly, the TAM signature required an ability to excise uracil from the DNA backbone, indicating that uracil levels increase in highly transcribed DNA and that its removal is responsible for most abasic sites. Although the excess uracil could be derived from cytosine deamination, TAM decreased dramatically upon overexpression of the yeast dUTPase, an enzyme that specifically hydrolyzes dUTP. Because this lowers the dUTP:dTTP ratio in the nucleotide pool and thereby disfavors dUTP incorporation into DNA, the results imply that the elevated uracil is derived from direct dUTP incorporation associated with transcription.
In higher eukaryotes, the removal of uracils from the DNA backbone is a critical and early step in the molecular events associated with somatic hypermutation and class-switch recombination within immunoglobulin genes (Box 2). Although directed cytosine deamination by the enzyme AID is the major source of uracil during these processes, direct incorporation of dUTP by DNA polymerase has not yet been ruled out as a minor contributor 107. It was recently reported, for example, that uracil can be detected at positions of thymines in the variable region of mouse immunoglobulin genes 108. Another study concluded, however, that uracil in hypermutating immunoglobulin genes was only found at positions of cytosines 109. Given the results in yeast and the generally universal effects of transcription on genome instability, it is expected that direct incorporation of dUTP into highly transcribed DNA will extend to other systems.
Precisely how transcription affects the nucleotide pool remains to be answered. One possibility is that a locally higher, transcription-associated dUTP concentration forces more frequent uracil incorporation by DNA polymerases. Spatially, a subnuclear localization of highly transcribed genes might be involved. Temporally, unscheduled DNA synthesis occurring outside of S phase, such as that involved in repair of transcription-associated damage, might be responsible for the elevated dUTP incorporation. It is known, for example, that expression of yeast dUTPase gene is up-regulated during S phase 110, thereby lowering dUTP levels within the dNTP pool and reducing its incorporation into DNA during genome duplication.
Conclusions and future directions
In this review, we have summarized current knowledge of the diverse ways that transcription locally stimulates recombination and/or mutagenesis: via collisions with the replication machinery, formation of co-transcriptional R-loops, facilitation of non-B DNA structure formation, engagement of Top1 activity, promotion of DNA damage and alteration in DNA base composition. In future studies, it will be important to determine how conserved a given mechanism is and to determine its contribution relative to other transcription-related mechanisms that affect genome stability. Even if a given mechanism is a relatively rare contributor to TAM and/or TAR in “normal” cells, its importance may shift under some conditions. This may be particularly relevant during the evolution of tumors, where there may already be underlying deficiencies in DNA repair/checkpoint processes. In addition, both the levels of endogenous mutagens and global transcription profiles are expected to fluctuate in response to environmental conditions. Environmental influences on transcription have the potential to alter the evolutionary landscape by targeting changes to genes where they are most likely to be beneficial in terms of promoting growth. In bacterial cells, transcription thus provides a mechanism for adaptive/stress-induced mutagenesis 8,111; in higher eukaryotes, a similar process could contribute to tumor evolution. An important take-home lesson from the results obtained to date is that transcription likely alters the genetic landscape of all organisms on an evolutionary time scale.
In closing, we would like to mention other connections between transcription and genome stability that were not specifically dealt with in this review. In addition to coupling of nucleotide excision repair pathway to transcription (TC-NER), proteins involved in both single- and double-strand break repair have been isolated as part of a complex with human RNAP II 112. Are these proteins possibly there to deal specifically with damage caused by transcription, or is transcription (in the form of damage-blocked RNAP complexes) used as a sensor to target repair where it is most urgently needed for proper gene expression? Alternatively, does transcription provide an efficient way to detect and deal with damage that can potentially impede DNA polymerases before the damage actually causes a replication-associated problem?
A potential twist to the TAR story comes from studies in budding yeast demonstrating that the DNA synthesis associated with homologous recombination is much more error-prone than that associated with genome duplication 113-115. An intriguing possibility is that some TAM might simply be a byproduct of DNA synthesis associated with TAR. Although most of the stimulatory effects of transcription on homologous recombination likely come through the generation of recombination-initiating lesions 116, this is not the entire story. Transcription additionally seems to enhance access to the repair template in both budding yeast and human cells 117,118. In addition to the process of transcription, the primary transcript itself may be relevant to recombination processes. Studies in yeast have demonstrated, for example, that spliced transcripts can be reverse transcribed by endogenous enzymes into cDNAs, which can then be used to correct chromosomal mutations 119. Such a mechanism has been invoked to explain why most yeast genes are devoid of introns 120; in metazoans, random rather than homology-based insertion of cDNAs has long been assumed to be the source of intron-less and promoter-less pseudogenes. Finally, studies have shown that small RNAs transformed into yeast cells can be directly used to template the repair of chromosomal DSBs 121.
With respect to TAM, only the tip of the iceberg has been touched in terms of what types of damage accumulate in highly transcribed DNA. Importantly, TAM can provide a replication-independent source of mutations that may be particularly relevant in post-mitotic cells or under defined starvation conditions. We expect and look forward to the discovery of additional mechanisms of transcription-associated mutagenesis and recombination as more systems are developed to study these fascinating processes.
Box 1 | Roles of Top1 in TAR and TAM.
Top1 relaxes the superhelical stress generated when DNA strands are unwound during transcription. Top1 nicks one strand of the DNA, forming a covalent linkage with one end. The intact strand passes through the nick, and then Top1 reseals the nick.
Top1 activity constrains TAR: In the absence of Top1, negative supercoils accumulate behind RNAP, giving the DNA single-stranded character and promoting the formation of R-loops. Top1 also regulates the activity of RNA splicing factors and RNP assembly, thereby preventing the accumulation of the naked RNA required for R-loop formation. R-loops are a primary cause of TAM, presumably through their interference with replication fork progression.
Top1 activity promotes TAM: Top1 can become trapped during its cleavage-ligation cycle, giving rise to a nick with the enzyme covalently attached to one end. This irreversible complex is likely processed into a small gap. If the gap is within a short tandem repeat, misalignment between the complementary strands can bring the ends together to facilitate ligation and create a deletion intermediate 83. If an rNMP is present at the Top1 cleavage site, the 2’-OH of ribose attacks the covalent Top1-DNA linkage to completely release Top1 122. Although rNMP-dependent deletions might occur by a mechanism similar to that proposed for a trapped Top1 cleavage complex, an alternative possibility is that strand rejoining occurs via a second Top1 cleavage-ligation reaction 123.
Box 2 | Programmed instability in the immune system124.
Somatic hypermutation (SHM): Antigen-induced stimulation of B cells activates programmed mutagenesis of the heavy and light chain variable segments of immunoglobulin genes, which results in the production of high-affinity antibodies during an immune response. Transcription is crucial during SHM because it allows the single-strand specific enzyme AID (activation induced deaminase) to access and deaminate cytosines on NTS. Uracils in the resulting U:G mispairs are excised by a uracil DNA glycosylase, leading to the accumulation of abasic (AP) sites. Mutations are introduced either during the gap-filling reaction that follows the excision of AP sites or when replication bypasses the non-informative AP sites. Highly mutagenic translesion synthesis DNA polymerases are likely used during gap filling as well as during AP-site bypass. AID-dependent mutagenesis can also occur at other highly expressed, non-immunoglobulin genes in hyper-mutating B cells.
Class-switch recombination (CSR): CSR occurs between GC-rich switch regions that precede the constant segments of heavy-chain genes, which specify different classes of immunoglobulins (IgA, IgG, etc.). CSR requires AID, transcription and components of the nonhomologous end-joining (NHEJ) pathway. Current models suggest that transcription-dependent, AID-initiated DSB form in two switch regions, with loss of the intervening DNA occurring when the broken ends are ligated by NHEJ. This irreversible process joins a new constant segment to the variable region of a heavy-chain gene. Interestingly, the breakpoints of NHEJ-derived translocations in B-cells preferentially localize to transcribed regions of the genome 125.
Acknowledgments
This work has been supported by grants from the National Institutes of Health.
Glossary Terms
- Transcribed and nontranscribed DNA strands (TS and NTS, respectively)
The TS is used as the template to make RNA. The complementary NTS has the same sequence as the RNA (except that it contains thymine instead of uracil); it is often referred to as the coding strand and is the strand whose sequence is standardly given.
- Replisome
The multi-protein complex (machine) that contains all of the proteins/enzymes required for DNA replication. This includes the DNA polymerases, factors that increase the processivity of DNA synthesis and a helicase to unwind duplex DNA.
- Two-dimensional (2D) gels
2D gels are used to visualize replication fork progression across a defined segment of DNA 126. DNA is separated by size in the first dimension and by shape in the second; the fragment of interest is visualized by Southern blot analysis. Linear fragments run on a diagonal; fragments that run off the diagonal correspond to replicating or branched molecules.
- ChIP-chip
DNA that interacts with a given protein is immunoprecipitated from cell extracts (“ChIP”). The precipitated DNA is labeled and hybridized to a microarray (“chip”), where signals above background reflect sequences preferentially immunoprecipitated/interacting with the protein of interest.
- Transcription-coupled nucleotide excision repair (TC-NER)
TC-NER is a specialized subpathway of the NER pathway that is initiated specifically in response to an RNA polymerase arrested by damage on the DNA template 99. The net effect is more efficient NER-directed repair of lesions on the transcribed than on the nontranscribed strand of active genes.
- Mutation assays
Forward mutation assays select for loss of a gene function and can detect any change in the DNA sequence that inactivates the encoded product, which is usually a protein. Reversion assays start with a mutant allele, typically containing a change in a single base pair or the insertion/deletion of a single base pair, and select for restoration of gene function. The change that restores gene function is usually limited to the position of the original mutation. It is typcially easier to select for restoration of gene function that to select for loss of gene function.
References
- 1.Savic DJ, Kanazir DT. The effect of a histidine operator-constitutive mutation on UV-induced mutability within the histidine operon of Salmonella typhimurium. Mol. Gen. Genet. 1972;118:45–50. doi: 10.1007/BF02428331. [DOI] [PubMed] [Google Scholar]
- 2.Herman RK, Dworkin NB. Effect of gene induction on the rate of mutagenesis by ICR-191 in Escherichia coli. J. Bacteriol. 1971;106:543–550. doi: 10.1128/jb.106.2.543-550.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [This work, using budding yeast as a model system, was the first demonstration of transcription-associated mutagenesis in eukaryotic cells.] [DOI] [PubMed] [Google Scholar]
- 4.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chem. Rev. 2006;106:474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 5.Keil RL, Roeder GS. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 6.Voekel-Meiman K, Keil RL, Roeder GS. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [This paper identifies the recombination stimulating sequence HOT1 as the rDNA promoter in budding yeast, thereby providing the first link between transcription and mitotic recombination.] [DOI] [PubMed] [Google Scholar]
- 7.Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst) 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reimers JM, Schmidt KH, Longacre A, Reschke DK, Wright BE. Increased transcription rates correlate with increased reversion rates in leuB and argH Escherichia coli auxotrophs. Microbiology. 2004;150:1457–1466. doi: 10.1099/mic.0.26954-0. [DOI] [PubMed] [Google Scholar]
- 9.Nickoloff JA. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol. 1992;12:5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postow L, et al. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 11.Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair (Amst) 2007;6:981–993. doi: 10.1016/j.dnarep.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Pomerantz RT, O'Donnell M. What happens when replication and transcription complexes collide? Cell Cycle. 2010;9:2537–2543. doi: 10.4161/cc.9.13.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- 14.Ellwood M, Nomura M. Chromosomal locations of the genes for rRNA in Escherichia coli K-12. J. Bacteriol. 1982;149:458–468. doi: 10.1128/jb.149.2.458-468.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guy L, Roten CA. Genometric analyses of the organization of circular chromosomes: a universal pressure determines the direction of ribosomal RNA genes transcription relative to chromosome replication. Gene. 2004;340:45–52. doi: 10.1016/j.gene.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 16.Wang JD, Berkmen MB, Grossman AD. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:5608–5613. doi: 10.1073/pnas.0608999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010;6:e1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brewer BJ, Fangman WL. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 19.Huvet M, et al. Human gene organization driven by the coordination of replication and transcription. Genome Res. 2007;17:1278–1285. doi: 10.1101/gr.6533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirkin EV, Mirkin SM. Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol. 2005;25:888–895. doi: 10.1128/MCB.25.3.888-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 22.Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. Embo J. 2005;24:1267–1276. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [In this paper, the ChIP-chip technique was used to identify sites of DNA Pol2 occupancy, which are presumed to reflect replication pausing, throughout the budding yeast genome. Highly transcribed ORFs were highly enriched among these sites, indicating that transcription can impede replication.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox MM, et al. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 26.Gottipati P, Cassel TN, Savolainen L, Helleday T. Transcription-associated recombination is dependent on replication in Mammalian cells. Mol Cell Biol. 2008;28:154–164. doi: 10.1128/MCB.00816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Loza MC, Wellinger RE, Aguilera A. Stimulation of direct-repeat recombination by RNA polymerase III transcription. DNA Repair (Amst) 2009;8:620–626. doi: 10.1016/j.dnarep.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Sikdar N, Banerjee S, Zhang H, Smith S, Myung K. Spt2p defines a new transcription-dependent gross chromosomal rearrangement pathway. PLoS Genet. 2008;4:e1000290. doi: 10.1371/journal.pgen.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilette D, Ehrlich SD, Michel B. Transcription-induced deletions in Escherichia coli plasmids. Mol. Microbiol. 1995;17:493–504. doi: 10.1111/j.1365-2958.1995.mmi_17030493.x. [DOI] [PubMed] [Google Scholar]
- 30.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blobel G. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bermejo R, et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146:233–246. doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguilera A. The connection between transcription and genomic instability. Embo J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 36.Gowrishankar J, Harinarayanan R. Why is transcription coupled to translation in bacteria? Mol. Microbiol. 2004;54:598–603. doi: 10.1111/j.1365-2958.2004.04289.x. [DOI] [PubMed] [Google Scholar]
- 37.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [This paper proposed the “twin domain” model of transcription-associated DNA helical stress.] [DOI] [PubMed] [Google Scholar]
- 38.Tuduri S, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baaklini I, Hraiky C, Rallu F, Tse-Dinh YC, Drolet M. RNase HI overproduction is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol Microbiol. 2004;54:198–211. doi: 10.1111/j.1365-2958.2004.04258.x. [DOI] [PubMed] [Google Scholar]
- 41.Mischo HE, et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudolph CJ, Upton AL, Briggs GS, Lloyd RG. Is RecG a general guardian of the bacterial genome? DNA Repair (Amst) 2010;9:210–223. doi: 10.1016/j.dnarep.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 43.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chavez S, Aguilera A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [This paper highlights the role of co-transcriptionally formed RNA:DNA hybrids (R-loops) in TAR by showing that degradation of the nascent mRNA by RNase H1 or a self-cleaving hammerhead ribozyme can suppress hyper-recombination in budding yeast hpr1 mutants.] [DOI] [PubMed] [Google Scholar]
- 46.Rondon AG, Jimeno S, Aguilera A. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim Biophys Acta. 2010;1799:533–538. doi: 10.1016/j.bbagrm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Gomez-Gonzalez B, et al. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011;30:3106–3119. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.French SL, et al. Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol Cell Biol. 2011;31:482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christman MF, Dietrich FS, Fink GR. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- 52.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [This paper demonstrated the role of mRNA splicing complex assembly in suppressing TAR in chicken DT40 and mammalian cells.] [DOI] [PubMed] [Google Scholar]
- 53.Paulsen RD, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 56.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 57.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 58.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [This work demonstrated that a highly transcribed immunoglobulin switch region sequence forms G-loop structures visible by electron microscope both in vitro and intracellularly in E. coli.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belotserkovskii BP, et al. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci U S A. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shinkura R, et al. The influence of transcriptional orientation on endogenous switch region function. Nat. Immunol. 2003;4:435–441. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 61.Kim N, Jinks-Robertson S. Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. DNA Repair (Amst) 2011 doi: 10.1016/j.dnarep.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gan W, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lopes J, et al. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 66.McMurray CT. DNA secondary structure: a common and causative factor for expansion in human disease. Proc Natl Acad Sci U S A. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swami M, et al. Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 2009;18:3039–3047. doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petruska J, Arnheim N, Goodman MF. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salinas-Rios V, Belotserkovskii BP, Hanawalt PC. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic Acids Res. 2011;39:7444–7454. doi: 10.1093/nar/gkr429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [This work showed that contraction of CAG triplet repeats in human cells requires transcription and components of defined DNA repair pathways.] [DOI] [PubMed] [Google Scholar]
- 71.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [Using a Drosophila model system, this work demonstrated that CAG repeat expansion requires transcription and is mediated by TC-NER.] [DOI] [PubMed] [Google Scholar]
- 73.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grabczyk E, Mancuso M, Sammarco MC. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ditch S, Sammarco MC, Banerjee A, Grabczyk E. Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 2009;5:e1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rindler PM, Bidichandani SI. Role of transcript and interplay between transcription and replication in triplet-repeat instability in mammalian cells. Nucleic Acids Res. 2011;39:526–535. doi: 10.1093/nar/gkq788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yelin R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 78.Lin Y, Wilson JH. Transcription-induced DNA toxicity at trinucleotide repeats: double bubble is trouble. Cell Cycle. 2011;10:611–618. doi: 10.4161/cc.10.4.14729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin Y, Leng M, Wan M, Wilson JH. Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol Cell Biol. 2010;30:4435–4451. doi: 10.1128/MCB.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakamori M, Pearson CE, Thornton CA. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)•(CAG) repeats. Hum. Mol. Genet. 2011;20:580–588. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morey NJ, Greene CN, Jinks-Robertson S. Genetic analysis of transcription-associated mutation in Saccharomyces cerevisiae. Genetics. 2000;154:109–120. doi: 10.1093/genetics/154.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garcia-Rubio M, Huertas P, Gonzalez-Barrera S, Aguilera A. Recombinogenic effects of DNA-damaging agents are synergistically increased by transcription in Saccharomyces cerevisiae. New insights into transcription-associated recombination. Genetics. 2003;165:457–466. doi: 10.1093/genetics/165.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lippert MJ, et al. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci U S A. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [This and the following reference describe transcription-dependent short deletions at tandem repeats and demonstrate that these events require Top1 activity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim N, Jinks-Robertson S. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2010;30:3206–3215. doi: 10.1128/MCB.00308-10. [This paper showed that uracil-derived AP sites and the ensuing mutagenesis are highly stimulated by transcription in budding yeast. The uracil reflects direct incorporation of dUTP in place of dTTP rather than cytosine deamination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hudson RE, Bergthorsson U, Ochman H. Transcription increases multiple spontaneous point mutations in Salmonella enterica. Nucleic Acids Res. 2003;31:4517–4522. doi: 10.1093/nar/gkg651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klapacz J, Bhagwat AS. Transcription-dependent increase in multiple classes of base substitution mutations in Escherichia coli. J Bacteriol. 2002;184:6866–6872. doi: 10.1128/JB.184.24.6866-6872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beletskii A, Bhagwat AS. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [This work demonstrated that in E. coli, C to T mutations resulting from spontaneous deamination of cytosines occur more frequently on the nontranscribed strand of an active gene.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beletskii A, Bhagwat AS. Correlation between transcription and C to T mutations in the non-transcribed DNA strand. Biol Chem. 1998;379:549–551. [PubMed] [Google Scholar]
- 90.Klapacz J, Bhagwat AS. Transcription promotes guanine to thymine mutations in the non-transcribed strand of an Escherichia coli gene. DNA Repair (Amst) 2005;4:806–813. doi: 10.1016/j.dnarep.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 91.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 92.Gomez-Gonzalez B, Aguilera A. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc. Natl. Acad. Sci. USA. 2007;104:8409–8414. doi: 10.1073/pnas.0702836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Green P, Ewing B, Miller W, Thomas PJ, Green ED. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet. 2003;33:514–517. doi: 10.1038/ng1103. [This and the following paper together demonstrated a strand-specific mutation bias in mammalian genes that correlates with expression level.] [DOI] [PubMed] [Google Scholar]
- 94.Majewski J. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am J Hum Genet. 2003;73:688–692. doi: 10.1086/378134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rubin AF, Green P. Mutation patterns in cancer genomes. Proc Natl Acad Sci U S A. 2009;106:21766–21770. doi: 10.1073/pnas.0912499106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wright BE, Reschke DK, Schmidt KH, Reimers JM, Knight W. Predicting mutation frequencies in stem-loop structures of derepressed genes: implications for evolution. Mol. Microbiol. 2003;48:429–441. doi: 10.1046/j.1365-2958.2003.t01-1-03436.x. [DOI] [PubMed] [Google Scholar]
- 97.Schmidt KH, Reimers JM, Wright BE. The effect of promoter strength, supercoiling and secondary structure on mutation rates in Escherichia coli. Mol. Microbiol. 2006;60:1251–1261. doi: 10.1111/j.1365-2958.2006.05166.x. [DOI] [PubMed] [Google Scholar]
- 98.Wright BE, et al. The roles of transcription and genotoxins underlying p53 mutagenesis in vivo. Carcinogenesis. 2011 doi: 10.1093/carcin/bgr177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 100.Iannone R, et al. Mutation spectra analysis suggests that N-(2-chloroethyl)-N'-cyclohexyl-N-nitrosourea-induced lesions are subject to transcription-coupled repair in Escherichia coli. Mol Carcinog. 1997;19:39–45. [PubMed] [Google Scholar]
- 101.Li BH, Ebbert A, Bockrath R. Transcription-modulated repair in Escherichia coli evident with UV-induced mutation spectra in supF. J Mol Biol. 1999;294:35–48. doi: 10.1006/jmbi.1999.3265. [DOI] [PubMed] [Google Scholar]
- 102.Moriya M, et al. TP53 Mutational signature for aristolochic acid: an environmental carcinogen. Int. J. Cancer. 2011;129:1532–1536. doi: 10.1002/ijc.26077. [DOI] [PubMed] [Google Scholar]
- 103.Hendriks G, et al. Gene transcription increases DNA damage-induced mutagenesis in mammalian stem cells. DNA Repair (Amst) 2008;7:1330–1339. doi: 10.1016/j.dnarep.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 104.Hendriks G, et al. Transcription-dependent cytosine deamination is a novel mechanism in ultraviolet light-induced mutagenesis. Curr Biol. 2010;20:170–175. doi: 10.1016/j.cub.2009.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim N, Jinks-Robertson S. dUTP incorporation into genomic DNA is linked to transcription in yeast. Nature. 2009;459:1150–1153. doi: 10.1038/nature08033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Neuberger MS, et al. Somatic hypermutation at A.T pairs: polymerase error versus dUTP incorporation. Nat Rev Immunol. 2005;5:171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- 108.Roche B, Claes A, Rougeon F. Deoxyuridine triphosphate incorporation during somatic hypermutation of mouse VkOx genes after immunization with phenyloxazolone. J Immunol. 2010;185:4777–4782. doi: 10.4049/jimmunol.1001459. [DOI] [PubMed] [Google Scholar]
- 109.Maul RW, et al. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol. 2011;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 111.Pybus C, et al. Transcription-associated mutation in Bacillus subtilis cells under stress. J. Bacteriol. 2010;192:3321–3328. doi: 10.1128/JB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maldonado E, et al. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 113.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Strathern JN, Shafer B, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gonzalez-Barrera S, Garcia-Rubio M, Aguilera A. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics. 2002;162:603–614. doi: 10.1093/genetics/162.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saxe D, Datta A, Jinks-Robertson S. Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol. Cell. Biol. 2000;20:5404–5414. doi: 10.1128/mcb.20.15.5404-5414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schildkraut E, Miller CA, Nickoloff JA. Transcription of a donor enhances its use during double-strand break-induced gene conversion in human cells. Mol. Cell. Biol. 2006;26:3098–3105. doi: 10.1128/MCB.26.8.3098-3105.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Derr LK, Strathern JN. A role for reverse transcripts in gene conversion. Nature. 1993;361:170–173. doi: 10.1038/361170a0. [DOI] [PubMed] [Google Scholar]
- 120.Fink GR. Pseudogenes in yeast? Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- 121.Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 123.Henningfeld KA, Hecht SM. A model for topoisomerase I-mediated insertions and deletions with duplex DNA substrates containing branches, nicks, and gaps. Biochemistry. 1995;34:6120–6129. doi: 10.1021/bi00018a015. [DOI] [PubMed] [Google Scholar]
- 124.Maizels N. Immunoglobulin gene diversity. Annu. Rev. Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 125.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]