Abstract
Previously, we have shown that human umbilical cord blood stem cell (hUCBSC) treatment downregulate cyclin D1 in glioma cells. To study the cell cycle progression and investigate the upstream molecules regulating cyclin D1 expression, we analyzed the involvement of extracellular signal-regulated kinase (ERK) and its functionality after treatment with hUCBSC. We observed downregulation of pERK after hUCBSC treatment at both transcriptional and translational levels. Increased translocation of ERK from cytoplasm to the nucleus was observed in glioma cells, whereas hUCBSC cocultures with glioma cells showed suppressed nuclear translocation. This finding suggests that hUCBSC regulates ERK by suppressing its phosphorylation at phospho-Thr202/Tyr204 retarding pERK nuclear translocation. ERK promoter analysis has shown c-Myc binding sites, indicative of possible transcriptional interactions that regulate cyclin D1 and ERK expression levels. Treatment of U251 and 5310 glioma cells with U0126, a MEK/ERK inhibitor receded pERK and c-Myc levels. In another experiment, U251 and 5310 cells treated with 10074-G5, c-Myc/Max inhibitor displayed reduction in pERK and c-Myc levels suggestive of a positive feedback loop between ERK/c-Myc/Max molecules. In the present study, we show that glioma cells exhibit abundant c-Myc expression and increased c-Myc/Max activity. In contrast, the glioma cells cocultured with hUCBSC demonstrated high Mad1 expression that competitively binds to Max to repress the c-Myc/Max mediated gene transcription. Our studies thus elucidate the potential role of hUCBSC in controlling glioma cell cycle progression and invasion by limiting Max binding to c-Myc, thus regulating the expression of glioma cell cycle and invasion associated molecules such as ERK, integrins via increased levels of Mad1 expression.
Introduction
Glioblastoma multiforme, an aggressive primary brain tumor, displays extensive infiltration into the surrounding brain tissue, thereby making it resistant to existing therapeutic strategies. There is currently no optimal treatment due to tumor invasiveness and an inability to deliver therapeutic agents directly to the tumor site. This lack of effective, targeted delivery mechanisms has especially slowed down the development of novel gene therapy strategies [1]. A better understanding of the molecular mechanisms that contribute to the biology of gliomas is crucial for developing effective treatment strategies. The extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway has been a popular target of cancer therapeutics because of its overexpression in many malignancies. ERK is involved in the control of fundamental cellular processes such as cell proliferation, survival, differentiation, apoptosis, motility, and metabolism [2]. ERK/MAP kinases drive cell cycle progression through modulation of cyclin D1 expression and associated cyclin-dependent kinase activity. The ERK/MAPK pathway mediates signal transfer from cell surface receptors to ERK/MAPK that is then distributed to different effectors that induce Ras activation. Activation of Ras further recruits Raf kinases to the cell membrane where they are sequentially activated, inducing a signal transduction cascade that includes the MEK/ERK kinase mitogen activated protein kinase (MEK), ERK, and ribosomal S6 kinase along with a set of transcription factors like CREB, AP-1, and c-Myc [3]. MEKs phosphorylate ERK1 on threonine202 and tyrosine204 and ERK2 on threonine 185 and tyrosine 187 to activate their kinase activities [4].
Downstream targets for ERK include nuclear transcription factors like c-Myc that is involved in regulating cell growth [5]. Fastidious phosphorylation events regulating the Myc protein half-life involves hierarchical phosphorylation by ERK. The c-Myc protein (bHLH-ZIP family of transcription factors) induces cell proliferation by targeting and modulating transcriptional expression of genes including cyclin D1, cyclin D2, cyclin A, cyclin E, Cdk1, and Cdk4, leading to Cdk 4/6 activation associated with cell cycle G0–G1 progression [6–8]. Paradoxically, c-Myc promotes cell cycle progression through heterodimerization with its biological partner Myc associated factor X (Max) [9]. Sufficient accumulation of c-Myc/Max leads to the activation of transcription of genes like cyclin D1 and Cdk 4, resulting in DNA replication and cell cycle G1–S transition. Myc-Max heterodimers recognize the promoter hexameric palindromic sequence CACGTG (E-box) and activate transcription of genes related to cell growth and cell cycle activation. Conversely, in the presence of Mad proteins, Max forms heterodimers with Mad and acts to repress gene transcription by associating with the mSin3 co-repressor complex via histone deacetylation [10]. The levels of Myc–Max–Mad in the cells thus determine the activation or repression of the target genes.
The obstacles encountered in controlling the dysregulated mechanism of these pathways in glioblastoma have prompted researchers to employ innovative strategies to understand these high-grade brain tumors. Several researchers have found that human mesenchymal stem cells could be the basis for a potential brain tumor therapy. Moreover, human umbilical cord-derived mesenchymal stem cells are often regarded as an alternative cell source for cell transplantation and cell therapy due to their hematopoietic and mesenchymal potential. Recent evidence suggests that human umbilical cord blood stem cell (hUCBSC) migrate toward gliomas and track microscopic tumor deposits, infiltrating tumor cells within the brain. Previously, we have shown that hUCBSC regulate glioblastoma progression at the G0–G1 level by downregulating cyclin D1 and its associated partner kinases Cdk 4 and Cdk 6 [11]. To study in detail the mechanistic aspects of how and why the downregulation of cyclin D1 causes cell cycle arrest by hUCBSC, we targeted the upstream regulatory molecules ERK and c-Myc. In this present report, we present transcriptional expression of Mad in hUCBSC and increased Mad-Max complex in coculture experiments. Our present study focuses on the involvement of ERK with cell surface receptors and their respective ligands in the invasive phenotype of glioma cells mediated through integrins.
Materials and Methods
Antibodies and reagents
We used the following antibodies: rabbit anti-ERK1, mouse anti-pERK, mouse anti-MEK1, mouse anti-c-Myc, mouse anti-cyclin D1, rabbit anti-α9 integrin, rabbit anti-β9 integrin, mouse anti-α9β1 integrin, rabbit anti-GRB2, mouse anti-vinculin, mouse anti-Mad1, rabbit anti-Max, mouse anti-FAK, rabbit anti-paxillin, mouse anti-β-actin, and mouse anti-GAPDH (1:200). A c-Myc/Max interaction inhibitor (10074-G5) was obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). MEK protein inhibitor UO126 was obtained from Promega (Madison, WI). Recombinant ERK (rERK) was obtained from Sigma Aldrich (St. Louis, MO). ERK/MAPK Cell-Based Translocation Assay Kit was purchased from Cayman Chemicals (Ann Arbor, MI).
Culture of hUCBSC
hUCBSCs were collected upon delivery from healthy donors with informed consent according to the protocol approved by the University of Illinois College of Medicine at Peoria Institutional Review Board. hUCBSC were isolated using Ficoll-Paque (GE Health Care, Piscataway, NJ) density gradient centrifugation. The isolated cells were plated in 100-mm plates in knockout Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 10% knockout serum (Hyclone, Logan, UT), and 1% penicillin/streptomycin. When the adherent cells reached 20%–30% confluence, they were supplemented with Mesencult medium (Stem Cell Technologies, Vancouver, Canada) containing human mesenchymal stem cell stimulatory supplements (Stem Cell Technologies) and 1% penicillin/streptomycin (Invitrogen). For coculture experiments, hUCBSC and glioma cells were cultured at a ratio of 1:1. Cocultures of hUCBSC and U251 were grown in DMEM; cocultures of hUCBSC and 5310 were grown in RPMI-1640.
Glioma cell culture
U251 cells obtained from the National Cancer Institute (NCI) (Frederick, MD) were grown in DMEM supplemented with 10% FBS (Hyclone) and 1% penicillin-streptomycin (Invitrogen). Xenograft cell line 5310 (a kind gift from Dr. David James at University of California, San Francisco) was grown in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin at 37°C in a humidified atmosphere containing 5% CO2.
Western blot analysis
U251 and 5310 cells alone or in coculture with hUCBSC for 72 h were collected and lysed in RIPA buffer [50 mmol/mL Tris-HCl (pH 8.0), 150 mmol/mL NaCl, 1% octylphenoxypolyethoxyethanol, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)] containing 1 mM sodium orthovanadate, 0.5 mM phenylmethanesulfonyl fluoride, 10 μg/mL aprotinin, and 10 μg/mL leupeptin and then resolved via SDS-polyacrylamide gel electrophoresis (PAGE). After transfer onto nitrocellulose membranes, blots were blocked with 5% nonfat dry milk in phosphate-buffered saline (PBS) and 0.1% Tween-20. Blots were incubated with respective primary antibodies, followed by incubation with a HRP-conjugated secondary antibody. Immunoreactive bands were visualized using chemiluminescence enhanced chemiluminescence western blotting detection reagents on Hyperfilm-MP autoradiography film (Amersham, Piscataway, NJ). GAPDH or β-actin antibody was used to verify equal loading of proteins in all lanes. A similar protocol was repeated for all of the different treatments.
Reverse transcription–PCR analysis
Primer sequences were designed from human GenBank sequences using Primer 3 software (v.0.4.0). For real-time polymerase chain reaction (RT-PCR) analysis, total cellular RNA was isolated from control and hUCBSC-treated cancer cells using the RNeasy kit (Qiagen, Valencia, CA) and quantified by measuring absorbance at 260 nm. Total RNA was reverse transcribed into first strand cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN). We used the following sequences for the forward and reverse primers: ERK: 5′TACACCAACCTCTCGTACATCG 3′ (sense) and 5′CATGTCTGAAGCGCAGTAAGATT3′ (antisense); MEK1: 5′ GTCCAAAATGCCCAAGAAGA 3′ (sense) 5′ CCCACGATGTACGGAGAGTT 3′ (antisense); c-Myc: 5′ CTTTGCACTGGAACTTACAA 3′ (sense) 5′ CAGCTCGAATTTCTTCCAGA 3′ (antisense); cyclin D1: 5′ GAGGAAGAGGAGGAGGAGGA 3′ (sense) and 5′ GAGATGGAAGGGGGAAAGAG 3′ (antisense); Mad1: 5′CAGAAAAGCCGTTCACCAAATC3′ (sense) and 5′CGTCAGAGTCGCTCACACTG3′ (antisense); Max: 5′GAGAGCGACGAAGAGCAACC3′ (sense) and 5′GCACTTGACCTCGCCTTCT3′ (antisense); β-actin: 5′GGCATCCTCACCCTGAAGTA3′ (sense) and 5′GGGGTGTTGAAGGTCTCAAA3′ (antisense). Reverse transcriptase–PCR was set up using the PCR cycle [95°C for 5 min, (95°C for 45 s, 55–60°C for 45 s, and 72°C for 45 s)×35 cycles, 72°C for 10 min]. PCR products were resolved on a 1.6% agarose gel, visualized, and photographed under UV light. Each sample was measured in triplicate and normalized to GAPDH or β-actin gene expression.
Matrigel invasion assay
Matrigel invasion assay was used to assess cell invasive potential as previously described [12]. Cell culture medium supplemented with 10% FBS was added to the lower chamber to act as a chemoattractant. U251 and 5310 control cells, alone or in coculture with hUCBSC, were seeded at a density of 2×105 cells/well onto the upper inserts and then incubated at 37°C. After 24 h, the noninvasive cells were removed from the upper surface of the separating membrane by gentle scrubbing with a cotton swab, and the invading cells were fixed in 100% methanol and stained with Hema-3. Similar experimental procedures were used when we investigated the inhibitory effect of ERK shRNA and rERK-mediated invasion.
Immunohistochemical staining
Detection of ERK and pERK was performed in 5 μm sections of formalin-fixed, paraffin-embedded tissues of U251 and 5310 controls or treated with hUCBSC sections. Sections were deparaffinized and then subjected to antigen retrieval for 10 min at 90°C in 0.1 M sodium citrate buffer (pH 6.0). Next, sections were washed in PBS and blocked in 10% goat serum for 30 min. A 1:100 dilution of rabbit anti-ERK and mouse anti pERK antibodies in 10% goat serum was added to sections that were incubated overnight at 4°C. After washing in PBS, sections were incubated with HRP-conjugated goat anti-rabbit IgG and goat anti-mouse IgG respectively. These sections were washed in PBS and developed with diaminobenzidine (DAB) substrate (Sigma-Aldrich) to produce color. The specificity of ERK and pERK staining was confirmed by a lack of staining without primary antibody or an isotype-matched irrelevant antibody. After counterstaining with hematoxylin, sections were mounted, cleared, cover slipped, and examined using a confocal microscope. For immunofluorescence, sections were treated with primary antibodies overnight at 4°C and then treated with appropriate Alexa Fluor secondary antibodies at room temperature for 1 h. Negative controls were maintained either without primary antibody or using IgG. Antigen retrieval was performed in the citrate buffer (pH 6.0). All immunostained sections, which were stained with fluorescent antibodies, were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The sections were blind reviewed by a neuropathologist.
Construction of shRNA-expressing plasmid and transfection of shERK
The plasmid vector, pSilencer™ 4.1-CMV (Ambion, Austin, TX), was used in the construction of the shRNA-expressing vector, and human ERK sequence was used as the shRNA target sequence. Inverted repeat sequences were synthesized for ERK. The inverted repeats were laterally symmetrical, making them self-complimentary with a 9-bp mismatch in the loop region. This 9-bp mismatch would aid in the loop formation of the shRNA. Oligonucleotides were heated in a boiling water bath in 6× saline sodium citrate for 5 min and self-annealed by slow cooling to room temperature. The resulting annealed oligonucleotides were ligated to pSilencer at the BamHI and HindIII sites. For transfection, the cancer cells were cultured as previously mentioned. Cells at 60%–70% confluence in 100-mm tissue culture plates were transfected with 10 μg of shRNA-expressing plasmid constructs (shERK) using Fugene HD following the manufacturer's instructions (Roche). Similar methodology was followed when shFAK and specific siRNA to integrin β1 (Santa Cruz Biotechnology, Inc.) were used to transfect U251 and 5310 cell lines. For treatments with rERK, U0126, and 10074-G5, both U251 and 5310 cells were treated with 10 μM U0126 kinase inhibitors (Promega), 25 μM 10074-G5 (Santa Cruz Biotechnology, Inc.), or 50 ng rERK (Sigma) for 12 h.
Cell cycle analysis using flow cytometry
Progression of glioblastoma through different cell cycle phases was monitored by flow cytometric analysis of U251 and 5310 controls, shERK-transfected samples, rERK-treated samples, and shERK-transfected samples treated with rERK. DNA content was analyzed by staining with propidium iodide and carried out with a fluorescence-activated cell sorter (FACS Caliber flow cytometer; Becton Dickinson, San Jose, CA). The percentage of cells within G1, S, G2, and M phases was determined using CellQuest software. Approximately 10,000 events were counted for each analysis, and 3 to 4 independent experiments performed in triplicate were conducted for each group (n=3).
ERK/MAPK(Phospho-Thr202/Tyr204) phosphorylation/translocation assay
Approximately 1×105 cells of U251 or 5310 alone or in coculture with hUCBSC were seeded in 96-well plates using the ERK/MAPK (Phospho-Thr202/Tyr204) phosphorylation/translocation kit (Cayman Chemicals). Cells were washed with tris-buffered saline (TBS) (pH 7.4) and fixed using Cell-Based Assay Fixative Solution for 10 min. The cells were then treated with 0.1% Triton X-100 3 times for 5 min each. The cells were blocked with Cell-Based Assay Blocking Solution for 30 min and treated with ERK/MAPK (Phospho-Thr202/Tyr204) primary antibody (diluted 1:200 in tris-buffered saline and tween-20 [TBST]) for 1 h. Cells were washed with TBST and incubated with DyLight™ 549-conjugated goat anti-rabbit secondary antibody (diluted 1:100 in tris-buffered saline and tween-20 [TBST]) for 1 h. Cells were examined under a fluorescent microscope. Cell-based tamoxifen (20 μM) was used as a positive control.
Immunocytochemistry
Cells were grown in 2-well chamber slides, washed with PBS, fixed, permeabilized with ice-cold methanol, and blocked with 10% goat serum for 1 h. Cells were incubated with primary antibodies for either 2 h at room temperature or overnight at 4°C, washed with PBS, and incubated with fluorescent-labeled, species-specific secondary antibodies (Alexa Fluor®) for 1 h at room temperature. Before mounting, the slides were washed with PBS, incubated for 3 min with DAPI for nuclear staining, and analyzed under a microscope (Olympus BX61 Fluoview, Minneapolis, MN).
Immunoprecipitation analysis
Protein lysates were prepared from 3–5 mm3 pieces of frozen intracranial tumors. Approximately 200–400 μg of protein cell lysates were incubated at 4°C with 50 μL of Protein G/A beads (Miltenyi Biotec, Auburn, CA) and followed by sequential additions of 10 μL (2 μg) of pERK and Mad antibodies with end-to-end rotation overnight. The immunoprecipitates were then loaded onto “μ” columns (Miltenyi Biotec) and rinsed with lysis buffer. The columns were washed twice with 200 μL of lysis buffer. Preheated (95°C) 1×SDS gel loading buffer was loaded onto the column matrix using a fresh pipette tip and was incubated at room temperature for 5 min. After discharging the supernatant, 50 μL of 1×SDS gel loading buffer was added to the immunoprecipitates, and the supernatants were then collected and loaded into 10%–12% SDS-PAGE followed by electrophoretic transfer to nitrocellulose membranes for further analysis.
Intracranial administrations of glioma cells and hUCBSC in nude mice
The Institutional Animal Care and Use Committee of the University of Illinois College of Medicine at Peoria approved all surgical interventions and postoperative animal care. Glioma cells were intracerebrally injected into the right side of the brains of nude mice with 10 μL aliquots of U251 and 5310 (1×106) cells under isofluorane anesthesia with the aid of a stereotactic frame. hUCBSC were injected near the left side of the brain after a week of tumor implantation. The ratio of the hUCBSC to cancer cells was maintained at 1:1. Three weeks after tumor inoculation, 6 mice from each group were sacrificed by intracardiac perfusion, first with PBS and then with 4% buffered formalin. The removed brains were stored in 4% paraformaldehyde, processed, embedded in paraffin, and sectioned (5 μm thick) using a microtome. Paraffin-embedded sections were stained with hematoxylin and eosin to visualize tumor cells.
Statistical analysis
Quantitative data from cell counts, FACS analysis, western blot analysis, and other assays were evaluated for statistical significance using 1-way analysis of variance (ANOVA). Data for each treatment group are represented as mean±standard error of the mean and compared with other groups for significance by 1-way ANOVA using Graph Pad Prism version 3.02, a statistical software package.
Results
hUCBSC treatment downregulates ERK in glioma cells
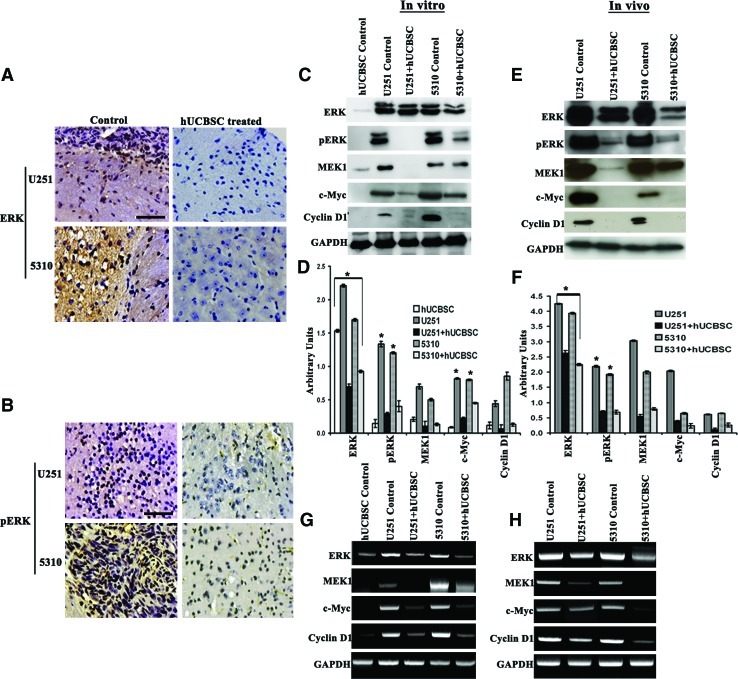
Positive expression of ERK and pERK was observed in U251 and 5310 tumors but this expression was reduced in the mice treated with hUCBSC. Immunohistochemistry analysis demonstrated that the ERK was localized in the cytoplasm whereas pERK mainly was localized in the nucleus of the glioma cells (Fig. 1A, B). Treatment with hUCBSC downregulated the expression of activated form, pERK. Negligible change was observed in the total ERK levels. Further our results obtained from the translocation/phosphorylation assay suggest that hUCBSC restricts ERK to the cytoplasm. The levels of pERK were significantly reduced in the hUCBSC cocultured samples when compared with U251 and 5310 control cells and the positive control tamoxifen (Supplementary Fig. S1C; Supplementary Data are available online at www.liebertonline.com/scd). To study the role of hUCBSC treatment on the cellular levels of ERK along with other critical cell cycle regulators including pERK, MEK1, c-Myc, and cyclin D1, we treated glioma cells with hUCBSC for 72 h. Cells were then subjected to western blot analyses. The results showed that hUCBSC treatment considerably reduced the expression levels of pERK and their downstream target molecules, MEK1, c-Myc, and cyclin D1 in both U251 and 5310 cells (Fig. 1C, D). Similar results were observed in the in vivo samples (Fig. 1E, F). Further RT-PCR analysis in vitro and in vivo samples reduced expression levels of ERK and its downstream molecules in the cocultured cells, when compared to the U251 and 5310 glioma controls (Fig. 1G, H). Taken together, these experiments aimed at studying the protein and mRNA levels clearly demonstrate that hUCBSC treatments regulate the expression levels of ERK and its regulatory molecules in U251 and 5310 glioma cells.
FIG. 1.
hUCBSC controls the expression levels of ERK and pERK in control and treated tumor brain sections. (A) IHC with ERK antibody in U251 and 5310 control tissue sections showing ERK cytoplasmic levels. Bar=200 μm. (B) IHC with pERK antibody detecting nuclear pERK levels in core tumor cells of controls. Bar=200 μm. (C, E) Immunoblot analysis to study the expression of ERK and its regulatory molecules in the cell lysates and tissue lysates of brains extracted from nude mice. Orthotopic intracranial tumors were established in nude mice by injecting glioma cells (U251 and 5310) and then treating with hUCBSC. Equal amounts of proteins (40 μg) from untreated (control) and hUCBSC-treated cell lysates and tissue lysates were loaded onto 8%–14% gels and transferred onto nitrocellulose membranes which were then probed with respective antibodies. GAPDH was used as a positive loading control. (D, F) The relative band intensities were measured by densitometry and normalized against the respective GAPDH signals. (G, H) Analysis of expression levels of ERK and other molecules in U251 and 5310 cell lines and mice tissues by semi-quantitative RT-PCR. GAPDH was used as an internal control for equal loading of the PCR products. Results presented in this study are the representative images of 3 independent experiments (n=3) and results are expressed as mean±SE. *P<0.05. hUCBSC, human umbilical cord blood stem cell; ERK, extracellular signal-regulated kinase; IHC, immunohistochemistry; RT-PCR, real-time polymerase chain reaction; SE, standard error. Color images available online at www.liebertonline.com/scd
hUCBSC reduce invasive potential of glioblastoma by downregulating ERK
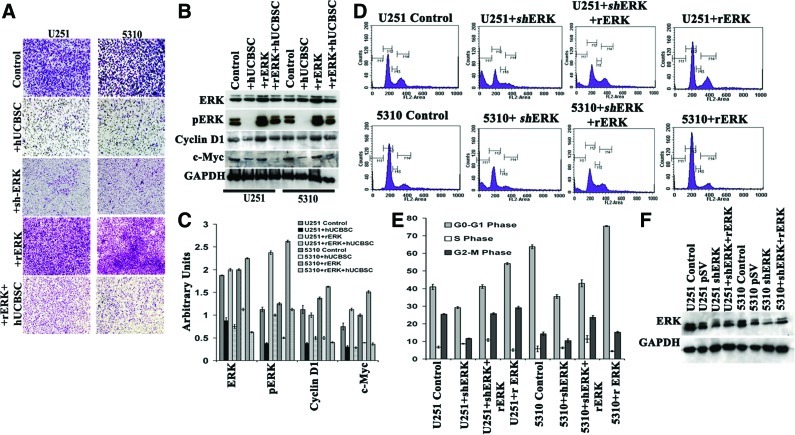
To test whether expression of ERK in glioblastoma cells drives cell invasion, we performed an in vitro cell Matrigel invasion assay. The invasion capacity of the U251 and 5310 glioma cells was reduced in the cells treated with hUCBSC treatment for 72 h (Fig. 2). Further, to show the role of ERK in glioma cell invasion we transfected U251 and 5310 cells with shRNA specific to ERK. When compared to the control U251 and 5310 cells, the shERK-transfected cells showed reduced invasion potential, suggesting that ERK inhibition is required to control invasion inferring that the ERK signaling pathway may be activated during glioblastoma cell invasion. We next treated the glioma cells with rERK (10 ng) for 12 h. The ability of the treated cells to potentially invade the Matrigel increased when compared with the control glioma cells, thereby indicating that ERK may be an important candidate molecule that regulates cell invasion. Likewise, we examined to what extent hUCBSC could inhibit the overexpressed ERK (rERK treatment). Interestingly, hUCBSC inhibited the invading capacity that significantly increased upon rERK treatment (Fig. 2A). Immunoblotting experiments with U251 and 5310 control cells showed increased expression of ERK, pERK, c-Myc, and cyclin D1; hUCBSC treatment reduced the expression levels of these proteins. rERK-treated glioma cells showed increased expression levels of ERK, pERK, c-Myc, and cyclin D1 when compared with the control cells. The rERK glioma cells treated with hUCBSC for 72 h showed decreased levels of ERK and its downstream regulatory molecules (Fig. 2B, C).
FIG. 2.
ERK shRNA retards invasion and causes accumulation of cells in the G0–G1 phase. (A) U251 and 5310 cells alone or in coculture with hUCBSC were transfected with ERK shRNA or recombinant ERK and were then allowed to invade Matrigel in the Boyden's chamber for 24 h and followed by Hema-3 staining. (B) Immunoblot analysis showing the expression levels of ERK, pERK, cyclin D1, and c-Myc in various treatment groups. (C) The relative band intensities were measured by densitometry and normalized against the respective GAPDH signals. (D) To determine changes in cell cycle in U251 and 5310 cells transfected with shRNA or treated with recombinant ERK, cells were trypsinized and stained with propidium iodide as per standard protocols. To determine cellular DNA content, the cells were sorted using a flow cytometer. Ten thousand cells were sorted per experiment with 3 replications. (E). Quantitative analysis for different phases of the cell cycle was done and graphically represented as percentage of controls. The values in all experiments are mean±standard deviation from 3 independent experiments (P<0.05). (F) One representative western blot showing the correlative expression levels as depicted in fluorescence-activated cell sorting analysis using different treatments. These data represent the average of 3 independent experiments. Color images available online at www.liebertonline.com/scd
The ERK protein is known to activate the transcription of several genes that are critical in maintaining proper cell cycle entry [13]. To study the presumptive role of ERK in regulating glioma cell cycle progression, U251 and 5310 glioma cells were subjected to FACS analysis. The results showed 41%, 64% of G0–G1 phase, 7%, 6% of S phase, and 26%, 15% of G2–M phase in the U251 and 5310 cells respectively, whereas shERK-transfected samples have shown 29%, 35% of G0–G1 phase, 8%, 6% of S phase, and 11%, 10% reduction in G2–M phase, respectively. Treatment with rERK after 12 h in U251 and 5310 control cells showed 55%, 75% of G0–G1 phase, 5%, 4.5% of S phase, and 30%, 15% of G2–M phase, whereas shERK-transfected samples treated with rERK showed 42%, 43% of G0–G1 phase, 11%, 11% of S phase, and 25.76%, 23.67% of G2–M phase, respectively (Fig. 2D, E). When compared with control cells, shERK-transfected samples showed decreased ERK expression as demonstrated by immunoblot analysis. Conversely, addition of rERK to shERK-transfected samples resulted in increased endogenous levels of ERK in U251 and 5310 cells (Fig. 2F).
ERK regulates integrin-mediated invasion
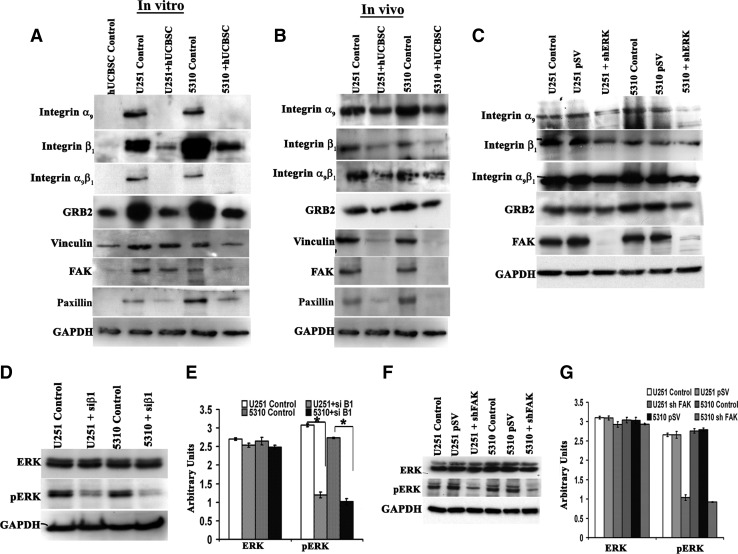
Previous studies have shown significant increase in the expression of integrins and integrin mediated human cancer cell invasion [14,15]. Based on these studies, we framed the present study to elucidate a possible mechanism involving ERK-mediated glioma invasion. Given the importance of invasion in glioblastoma progression, we investigated whether ERK expression could support this activity in U251 and 5310 cells. To test whether a functional correlation exists between the expression of ERK and upregulation of the integrins, we asked whether the hUCBSC treatment can regulate integrin-enhanced invasion. Western blot analysis of invasion associated molecules showed increased levels of α9 integrin, β1 integrin, α9β1 integrin, Grb2, vincullin, focal adhesion kinase (FAK), and paxillin in the glioma cells whereas cells cocultured with hUCBSC for 72 h exhibited significantly reduced expression levels of these molecules (Fig. 3A). Immunoblot analysis performed on control and hUCBSC-treated brain tumors confirm these findings, and the results obtained are in accord with the in vitro analysis (Fig. 3B). As a second test of the role of ERK in integrin-mediated invasion, we examined the effect of ERK gene knockdown. Expression levels of α9 integrin, β1 integrin, α9β1 integrin, Grb2, and FAK decreased in shERK-transfected glioma cells when compared with the control cells (Fig. 3C). Reduction of ERK probably abrogated integrin-mediated invasion of these cells.
FIG. 3.
Effect of hUCBSC treatment and shERK transfections on the expression of integrins in U251 and 5310 cells. (A) Western blotting of in vitro and in vivo samples. Briefly, cell lysates from in vitro samples (1×106 cells) were prepared from U251 and 5310 cells alone or in coculture with hUCBSC for 72 h. Cell lysates were prepared as previously described. Western blotting was carried out to determine the effect of hUCBSC on integrin α9, integrin β1, integrin α9β1, GRB2, vincullin, FAK, and paxillin. (B) In vivo expression was studied by loading equal amounts of protein (40 μg) from tissue lysates of untreated and treated mice brains onto 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The transferred proteins were then probed with respective antibodies. GAPDH served as the loading control. Each experiment was repeated 3 times. (C) Western blot analysis showing the effect of shERK on the protein expressions of various invasive molecules in U251 and 5310 cells (n=3). (D) Western blot analysis showing the effect of β1 siRNA constructs on the protein expression levels of ERK and pERK in U251 and 5310 cells. The expression levels of pERK were reduced with integrin β1 siRNA treatments (n=3). (E) Quantitative estimation of Fig. 3D. (F) Western blot analysis of ERK and pERK expression levels in control, pSV, and shFAK in U251 and 5310 cells using specific antibodies. (G) Quantitative analysis of ERK and pERK expression in U251 and 5310 cells as assessed by densitometry. Results from 3 independent experiments are shown as mean±SE (*P<0.05). GAPDH served as a loading control.
High expression of α9β1 integrin on malignant gliomas suggests its importance for pathological tumor progression. This integrin acts as a multifunctional receptor by interacting with various endogenous ligands possibly spreading tumor to nonaffected areas of the brain. Given the need for ERK activation and the importance of integrins as promoters of invasion, we next examined any functional relationship that may exist between ERK activation and the β1 integrin in the context of its knockdown by integrin β1 siRNA. Knockdown of the β1 integrin in both U251 and 5310 cells did not result in any change in total ERK levels but significantly downregulated pERK levels (Fig. 3D, E).
Studies on integrin clustering revealed the prominence of FAK and its subsequent activation of downstream effector molecules like ERK. To further confirm the role of FAK in ERK-mediated invasion, we treated U251 and 5310 cells with shERK. Western blot analysis showed FAK suppression with ERK knock down (Fig. 3C). We next treated U251 and 5310 cells with shRNA specific for FAK. We observed a transient decrease in the amounts of pERK whereas total ERK did not show any significant change (Fig. 3F, G). Immunocytochemical analysis of shERK-transfected U251 and 5310 cells probed with α9β1 integrin showed decreased expression levels that is indicative of the regulatory function of ERK (Supplementary Fig. S1B).
hUCBSC reduces the interaction of ERK with FAK and α9β1 integrin
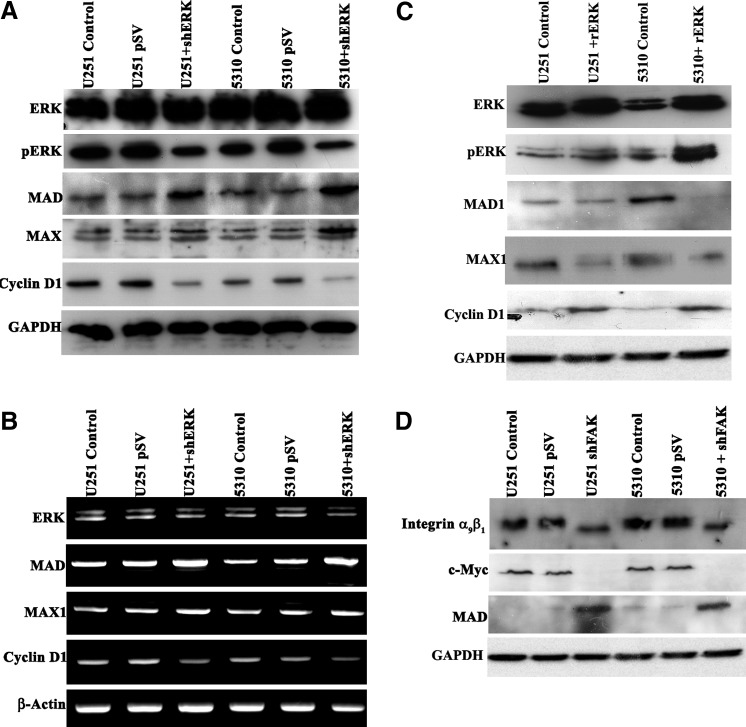
Our primary assumption was that ERK association with the α9β1 integrin mediated by FAK is integral to the mechanism by which ERK regulates glioblastoma invasion. Therefore, we sought to identify the possible association using immunoprecipitation experiments. As a first step, we tested the effect of shERK treatments on α9β1 integrin and FAK. Expression of both α9β1 integrin and FAK were observed to be reduced (Fig. 4A). Similar to shRNA treatments, coculture of U251 and 5310 cells with hUCBSC also showed reduction in the expression levels of both α9β1 integrin and FAK (Fig. 4B). Because we were specifically interested in the role of c-Myc on ERK, as our bioinformatic approach showed c-Myc putative binding sites in the ERK promoter (Acc No: NM_002745) (Supplementary Fig. S2), we also examined c-Myc in the present study. When glioma cells alone and in coculture with hUCBSC were immunoprecipitated with c-Myc and immunoblotted with pERK, MAX, and FAK antibodies, a reduction was observed in the expression levels of pERK, MAX, and FAK suggesting an interplay between these molecules (Fig. 4C). Previous studies have demonstrated that integrin signaling through FAK plays an important role in the regulation of cell cycle progression correlating with changes in the expression of cyclin D1 [16]. In another study, ERK acts downstream of Src-FAK signaling [17,18] and upstream to regulate FAK and Paxillin expression [19]. In the present study we found that c-Myc associates and interacts with FAK. This interaction may be viewed more as a general role in regulating cell cycle progression. We noticed that in shFAK-treated samples, c-Myc expression levels were reduced (Fig. 4D). These results suggest that ERK downregulation aided in reduced invasion and may involve critical players like FAK, c-Myc, and α9β1 integrin.
FIG. 4.
ERK binding to α9β1 integrin and FAK is downregulated by hUCBSC treatment. (A) Around 800 μg of total soluble protein were taken from U251 and 5310 control cells and shERK-transfected samples. The protein samples were incubated with 2 μg/μL of ERK antibody. The mixture containing total soluble protein and respective antibody was incubated with 50 μL of protein A/G agarose beads for 30 min in ice. The ERK immunoprecipitated samples were immunoblotted against FAK and integrin α9β1. (B) Eight hundred microgram of total soluble protein from U251 and 5310 control and hUCBSC-treated tissue lysates were incubated with 2 μg/μL of ERK antibody. The ERK-immunoprecipitated samples were immunoblotted against FAK and integrin α9β1. The same blots were stripped and re-probed with anti-ERK antibody. (C) Eight hundred microgram of total soluble protein from U251 and 5310 control and hUCBSC-treated tissue lysates were immunoprecipitated with c-Myc, and the samples were immunoblotted against pERK, MAX, and FAK. The same blots were stripped and re-probed with anti-c-Myc antibody. (D) U251 and 5310 control and hUCBSC-treated tissue lysates were immunoprecipitated with FAK and immunoblotted against c-Myc. The same blot was stripped and re-probed with anti-FAK antibody. These data are representative of at least 2 individual experiments.
Studies on c-Myc/Max and c-Myc/pERK using U0126 or 10074-G5
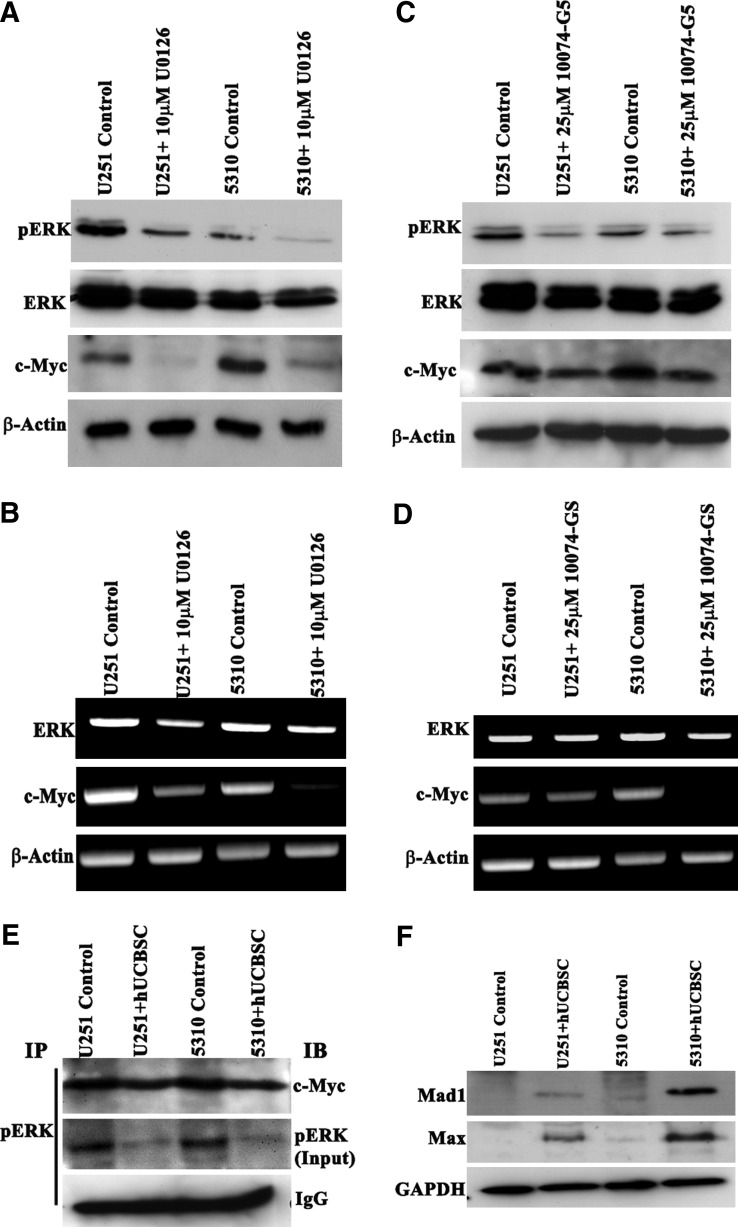
Since c-Myc expression was downregulated during the coculture treatment with hUCBSC (Fig. 1C, E), we sought to determine whether c-Myc functions as a target of the ERK pathway which might affect cyclin D1 activity. To evaluate this finding, we treated U251 and 5310 cells with 10 μM U0126 (MEK/ERK inhibitor) for 12 h followed by immunoblot analysis. U0126 induced c-Myc downregulation in the U0126-treated samples when compared with the untreated control cells. The level of pERK was markedly reduced, whereas the expression level of ERK was unaffected in the treated cells when compared with the controls (Fig. 5A). The RT-PCR analysis showed similar results compared with immunoblotting analysis (Fig. 5B). When we treated U251 and 5310 cells with 25 μM 10074-G5 (Myc-Max inhibitor) for 12 h, we observed reduced expression levels of pERK but no significant change in the expression levels of total ERK (Fig. 5C). RT-PCR analysis of 10074-G5-treated samples showed reduced expression of ERK and c-Myc (Fig. 5D). These results and promoter analysis demonstrate that c-Myc functionally regulates ERK (Supplementary Fig. S2). These results also show that U0126 and 10074-G5 regulate the expression levels of ERK and c-Myc at both the transcriptional and translational levels. Immunoprecipitation experiments aimed to study the interaction between pERK and c-Myc showed decreased expression levels of c-Myc, suggesting the existence of possible regulatory mechanism (Fig. 5E). Since we found that the 10074-G5 molecule inhibits c-Myc activity by blocking Max, we inferred its role in regulating ERK pathway. Interestingly, high levels of Max protein in the hUCBSC-treated brain tumors in our studies fueled the need to investigate other Max interacting molecules. Recent evidence suggests a functional correlation to Mad1 protein, which acts as an antagonist to c-Myc [20,21]. Western blot analyses performed to determine Mad activity showed increased expression levels in hUCBSC treated xenograft lysates when compared with controls (Fig. 5F). The result point out to a new regulatory mechanism of c-Myc and ERK activity, as illustrated by the increase in Mad and Max protein levels in the coculture experiments (Fig. 5F).
FIG. 5.
MEK/ERK inhibitor affects c-Myc expression, c-Myc-Max inhibitor; 10074-G5 inhibits pERK expression. (A) Cell lysates from U251 and 5310 cells either untreated or treated with 10 μM U0126 for 12 h were analyzed by immunoblotting with specific antibodies for indicated proteins. β-actin expression shows the equal loading of samples. (B) Semi-quantitative RT-PCR analysis was carried out to detect ERK and c-Myc in U0126 treatments. β-actin expression was used as a loading control. (C, D) U251 and 5310 cells were treated with 25 μM 10074-G5 for 24 h. Immunoblot of total lysates was performed using antibodies specific for the indicated proteins. Fold decrease over the control was obtained by RT-PCR analysis. (E) Eight hundred microgram of total soluble protein from U251 and 5310 control and hUCBSC-treated tissue lysates were immunoprecipitated with pERK, and the samples were immunoblotted against c-Myc. The same blots were stripped and re-probed with anti-pERK antibody. (F) Immunoblot showing the expression levels of Mad and Max in the coculture treatments and compared with control U251 and 5310 cells.
pERK co-localization with c-Myc and Mad in glioblastoma cells
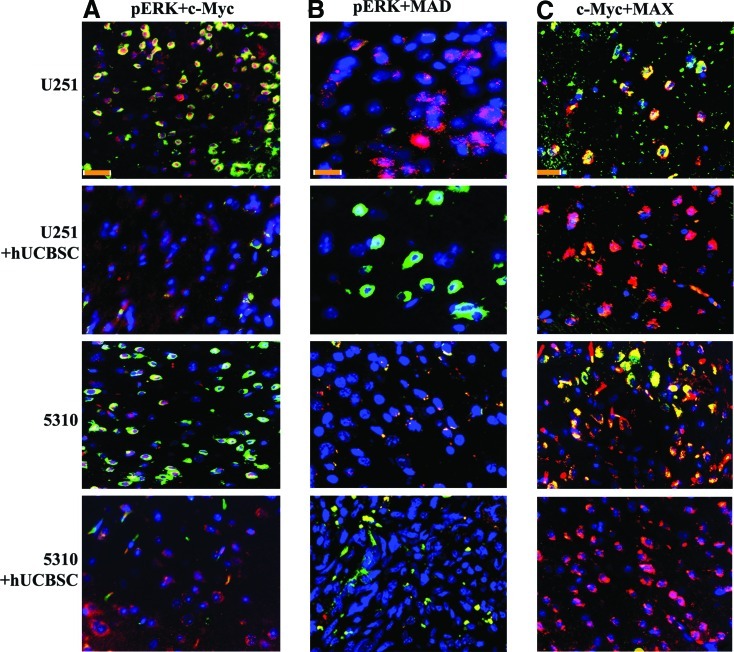
To examine mechanisms by which hUCBSC mediate suppression of cancer cell growth, we determined the expression levels of c-Myc and Mad in control and hUCBSC-treated tumor sections. For this study, we carried out co-localization experiments with pERK, c-Myc, pERK, and Mad antibodies. pERK and c-Myc were highly expressed in the control brain tumor sections as compared to hUCBSC-treated brain tumor sections (Fig. 6A). In another experiment, we checked the co-localization of pERK and Mad in both control and hUCBSC-treated mice brain tumor sections. pERK was highly expressed in both U251 and 5310 control brain tumors whereas Mad was expressed in hUCBSC-treated mice brains; co-localization of pERK and Mad was almost negligible (Fig. 6B). We then checked whether the expression of ERK in the control tumor was reduced with hUCBSC treatment. Immunohistochemical analysis was performed using CD81 (mesenchymal stem cell marker) and ERK antibody. ERK was highly expressed in the control tumors with negligible CD81 levels whereas hUCBSC-treated brain sections showed high expression levels of CD81 with reduced expression of ERK (Supplementary Fig. S1A). Based on the hypothesis that coordinated expression of c-Myc with Max protein may trigger transcription of c-Myc, and since deregulated expression of c-Myc is most commonly observed in glioma tumors, we tested the interaction of c-Myc with Max. Enhanced expression levels of both c-Myc and Max were observed in U251 and 5310 control cells, whereas their co-localized expression was observed to be reduced when treated with hUCBSC (Fig. 6C). Collectively, these results indicate that hUCBSC treatment regulates the expression and function of c-Myc, pERK, Mad, and Max in glioma cells.
FIG. 6.
Co-localization studies of pERK in control and hUCBSC-treated brains. Nude mice with pre-established intracranial human glioma tumors (U251 or 5310) were treated with hUCBSC by intracranial injection (2×105). The hUCBSC-treated and control brain sections were harvested, sectioned, and immunoprobed for the presence of pERK/c-Myc complex or pERK/Mad complex as well using appropriate secondary antibodies. Each experiment was performed in duplicate with each sample (n=6). (A) Four rows in the left panel show pERK (red) co-localization with c-Myc (green). (B) Four rows of middle panel show co-localization studies of pERK (red) with Mad (green). Bar=200 μm. (C) Four rows of right panel show co-localization studies of c-Myc (green) with Max (red). In all cases, 4′,6-diamidino-2-phenylindole was used to stain the nuclei. Bar=200 μm. Color images available online at www.liebertonline.com/scd
shERK increases levels of Mad and Max while rERK shows antagonistic expression
In the present study, we have shown that shERK treatment reduced the G0–G1 phase, which is in continuation with the results of our earlier report [11]. We hypothesize that ERK may regulate cyclin D1 activity through c-Myc. We next asked whether cell cycle regulatory proteins like c-Myc and cyclin D1 were modulated in shERK-treated cells by immunoblotting analysis. Figure 7A shows that shERK treatment in glioma cells induced decrease in pERK, c-Myc, and cyclin D1 levels when compared with the control cells. Moreover, Mad and Max protein levels were increased with shERK treatment. However, there was no significant change in the total ERK levels. RT-PCR analysis of shERK showed decreased transcript levels of ERK, c-Myc, and cyclin D1, and increased levels of Mad1 and Max (Fig. 7B). These results point to the existence of a pathway in which expression of Mad-Max proteins in the ERK shRNA treatments halts c-Myc and cyclin D1 activity, thereby mediating G0–G1 cell cycle arrest in glioblastoma control cells. Conversely, U251 and 5310 cells, when treated with 50 ng of rERK, showed increased levels of total ERK, pERK, cyclin D1, and c-Myc protein levels when compared with the control cells (Fig. 7C). As expected, Mad1 and Max levels were suppressed suggesting the antagonistic role of the Mad-Max heterodimer complex in deregulating c-Myc expression, thus delineating cyclin D1 activity. We next explored whether downregulation of FAK regulates the expression of α9β1 integrin and c-Myc. As shown in Fig. 7D, shFAK-transfected samples show significant reduction in the expression levels of both α9β1 integrin and c-Myc. In contrast, Mad1 protein expression was observed to be significantly higher in the shFAK samples. However, the mechanism, through which FAK downregulation increases Mad1 expression, is not clear.
FIG. 7.
Effects of shERK and rERK on the expression of c-Myc-related proteins in glioblastoma cell lines. Downregulation of ERK by shERK in U251 and 5310 glioblastoma cell lines. (A) Total lysates from the indicated tumor cell lines transfected with shERK for 72 h were analyzed by immunoblotting with anti-ERK, anti-phospho ERK, anti-c-Myc, anti-cyclin D1, anti-Mad, and anti-Max antibodies. (B) RT-PCR analysis was done to detect the expression levels of above mentioned proteins. (C) U251 or 5310 glioblastoma cell lines were treated with 50 ng of rERK for 12 h and were analyzed with specific antibodies for indicated proteins. GAPDH expression shows the equal loading of samples. Similar results were obtained in 3 different experiments. (D) Total lysates from the indicated tumor cell lines transfected with shFAK for 72 h were analyzed by immunoblotting with anti-c-Myc, anti-integrin α9β1, and anti-Mad antibodies.
Mad exhibits more affinity towards Max in hUCBSC cocultures
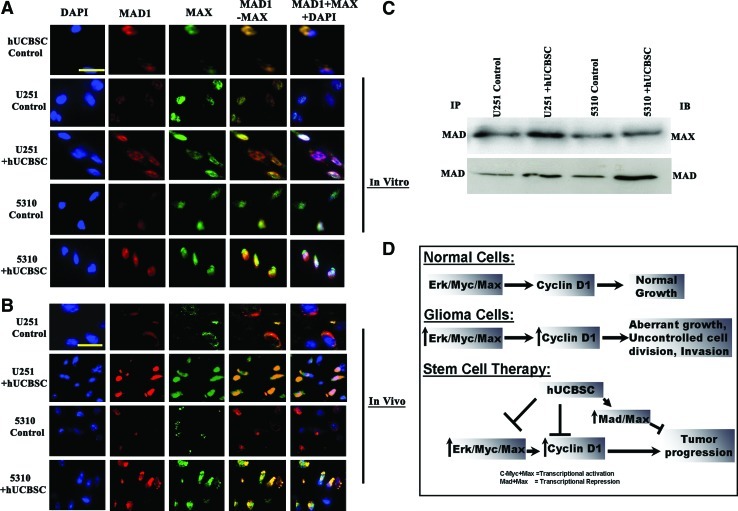
To evaluate whether the transcriptional control of ERK is mediated by Mad-Max interactions in coculture with hUCBSC and to promote the hypothesis that expression of Mad-Max complex represses ERK activity, we carried out co-localization experiments with Mad and Max in both control and hUCBSC-treated glioma samples. In hUCBSC-treated cocultures and also in vivo experiments, Mad1 was found to be associated with Max as shown by their high degree of co-localization in the co-localization experiments. However, no such association was observed in the controls, both in vitro and in vivo models (Fig. 8A, B). We also performed immunoprecipitation studies to identify physical interactions between Mad and Max. Western blot analysis on the immunoprecipitates showed that Mad-Max interaction was greater in the U251 and 5310 glioma cells cocultured with hUCBSC when compared with control glioma cells. The possible mechanism by which hUCBSC may regulate the glioblastoma progression was schematically represented in Fig. 8D.
FIG. 8.
Activation of Mad-Max complex in hUCBSC-treated cells in vitro and in vivo. (A) We carried out immunocytochemistry of U251 and 5310 cells either alone or in coculture with hUCBSC to study the expression of Mad-Max. Mad is conjugated with Alexa Fluor-594 (red), and Max is conjugated with Alexa Flour-488 (green). Bar=100 μm. (B) Similarly, dual immunohistochemical staining for co-localization was conducted with anti-Mad and anti-Max antibodies followed by the secondary antibodies conjugated with fluorophores for red and green fluorescence, respectively. Representative merged images show the cells expressing Mad and Max. Bar=100 μm. Each experiment was performed in triplicate with each sample (n=3). (C) Co-immunoprecipitation assay was carried out to determine the interaction of Mad with Max in control and hUCBSC-treated tissue lysates. Total tissue lysates were immunoprecipitated with Mad, and the immunoprecipitates of Mad were analyzed by immunoblotting with Max antibody. Immunoprecipitates of Mad were analyzed by western blotting with anti-Mad antibody. (D) Schematic representation showing the possible regulatory mechanism of Mad and Max in controlling glioblastoma progression. Color images available online at www.liebertonline.com/scd
Discussion
The dismal prognosis observed in glioblastoma treatment is mainly due to the highly invasive nature of glioblastoma cells. Knowledge of the mechanisms and factors involved in tumor cell invasion helps us to understand the mandatory need for the identification of new therapeutic targets [22].
In our earlier studies we demonstrated that hUCBSC control glioma cell cycle progression by downregulating the expression levels of cyclin D1 and its partner kinases Cdk4 and Cdk6 [11]. Based on our previous results, we explored the possibility that hUCBSC downregulate the expression of ERK, pERK, and c-Myc, thereby downregulating cyclin D1 and promoting cell cycle arrest. ERK translocation to the nucleus is essential for G1 phase progression [23]. Immunohistochemical staining of the tissue sections from control and hUCBSC-treated mice have shown that hUCBSC not only downregulates ERK but also its active form pERK. In line with our findings, U0126 treatment in embryonal rhabdomyosarcoma tumors downregulated phosphoactive ERK, c-Myc, and cyclin D1, suggesting the role of ERK in reducing c-Myc and cyclin D1 activity [24].
Accumulating evidence from various studies suggest the potential role of the ERK pathway in the migration of various cell types, and that the activation of ERK contributes to the progression of tumors through transcriptional regulation of metastasis-related genes [25,26]. The ERK inhibitor PD98059 suppressed the proliferation of glioma C6 cells indicating that activation of ERK was integral to the invasion/migration of human chondrosarcoma cells [27] and U87 glioma cells [28]. These findings provide support for the possibility that ERK activation is conceivably critical for the invasion/migration of glioma cells. In the present study, we showed suppression of glioma cell invasion with hUCBSC treatment. Transfection with shERK suppressed the invasion of glioma cells, testifying that activation of ERK is essential in the invasion of glioma cells. Addition of rERK increased invasion potential of the control glioblastoma cells. Surprisingly, rERK-induced invasion of the glioma cells was blocked by hUCBSC coculture treatment. Our results suggest that ERK activation is critical for the invasion of glioma cells and hUCBSC identifies ERK as a potential target molecule, eventually downregulating ERK to treat glioblastoma. In line with the results of the Matrigel invasion assay, immunoblotting showed that expression of ERK, pERK, c-Myc, and cyclin D1 was inhibited when cells were treated with hUCBSC or shERK, or rERK treated cells cocultured with hUCBSC. In addition to the aforementioned means, FACS analysis provides an alternative method to explain the effect of ERK on cell proliferation and survival. Results show that shERK decreased G0–G1 phase and retarded the cell cycle. Notably, the addition of rERK to shERK-transfected U251 and 5310 cells induced an obvious increase in the G0–G1 peak. These results support previous findings by Liu et al. [29] who demonstrated that inhibition of MEK1/2 in ERK-depleted cells arrest cells at the G1 phase. This suggests that downregulation of ERK by hUCBSC may be regarded as a key step for the regulation of glioblastoma progression.
Earlier studies propose that integrins directly regulate ERK by modulating growth factor-stimulated ERK activity [30]. Integrins are imperative for cell migration and invasion since they directly mediate adhesion to the extracellular matrix, regulate intracellular signaling pathways involving phosphorylation of FAK, and recruit adaptor proteins like GRB2 [31] along with subsequent downstream effector molecules like vincullin and paxillin. Activated FAK binds to ERK and is mediated by integrin activation [32], which in turn, promotes cell proliferation. Based on earlier findings, we hypothesize that integrins directly activate ERK through FAK-dependent pathways. In this study, we propose that α9β1 integrin is an active target of ERK in U251 and 5310 cells. α9β1 integrin is not found on normal astrocytes whereas it is highly expressed in glioblastoma cells, suggesting its importance in pathological tumor progression. Immunoblotting experiments with cocultured glioma cells with hUCBSC both in vitro and in vivo showed decreased expression levels of α9, β1, α9β1, FAK, GRB2, vincullin, and paxillin. These results are in accord with recent evidence by Brown et al. [33] that the expression of α9β1 in LN229 and LN18 cell lines was necessary for NGF-induced tumor progression through direct interaction and activation of MAPK ERK1/2 pathway. Young et al. [34] reported that the cytoplasmic domain of the α9 integrin subunit is important to augment the rate of cell migration, which is in agreement with α9β1 integrin-dependent pro-migratory activity of NGF. In another report, α9β1 integrin, after binding to its ligand NGF, induced activation of a focal adhesion adaptor protein, paxillin, thereby activating cell migration [35]. In the present study, for the first time, we suggest the possibility of regulating glioblastoma invasion by targeting α9β1 and ensuring its downregulation by reducing ERK levels. hUCBSC exerts its potential role in regulating invasion by downregulating ERK, thereby reducing its interactions with FAK, which in turn, controls the expression levels of α9β1 integrin.
To elucidate the role of ERK in functional regulation of glioma cell progression, we focused our studies on transcriptional regulation of c-Myc and its downstream molecule cyclin D1. c-Myc and Ras/MEK/ERK pathways play important roles in the progression of the G1-cell cycle phase [24,36,37] by enhancing cyclin expression and CDK/cyclin complex activities [38]. Use of U0126, a MEK/ERK pathway inhibitor, demonstrates the role of ERK in promoting G1 cell cycle arrest [39]. In the present study, we found that inhibition of ERK activity by decreased c-Myc expression and treatment with 10074-G5 decreased both c-Myc and pERK expression levels. Taken together, these data are in line with the notion that aberrant growth of glioblastoma cell lines can be impeded by targeting c-Myc following ERK inhibition.
It is understood that the ability of c-Myc to modulate the transcription of its target genes is regulated by stoichiometry of the Myc, Max, and Mad proteins [40]. The formation of the Myc-Max heterodimer promotes its binding to the E-box with subsequent interaction with transformation/transcription domain-associated protein and recruitment of histone acetylases to activate transcription [41]. In another study, it was found that Max functions as the central molecule forming transactivating complexes when it dimerizes with c-Myc or transrepressor complexes when it dimerizes with Mad. Therefore, the target gene activation or repression depends on the levels of Myc–Max–Mad interactions [42]. Mad expression reinforces the downregulation of c-Myc protein function by binding to Max and forming heterodimers, thereby making Max unavailable for c-Myc and also competes with Myc-Max complexes for binding to DNA target sites [43]. In addition, previous studies have shown that expression of Mad blocks the human hepatocellular carcinoma BEL-7404 cells at the G0–G1 phase. It has also been shown that an adenoviral Mad-expressing vector, when transfected in U373MG glioblastoma cells, led to increased accumulation of cells in G2–M phase [44]. Marampon et al. [36] reported that transfection of Mad-Myc chimera in the human muscle derived rhabdomyosarcoma cell line and in nonmuscle-derived human tumor cell lines SW403, IGR39 and PC3 repressed c-Myc activity and caused growth arrest in the G0-G1 phase. Our data prompted us to study the underlying mechanisms that possibly regulate ERK and c-Myc. In the present study, FACS analysis carried out to study the different phases of cell cycle after treatment with hUCBSC only shows a G0–G1 peak. Immunoblotting of the samples treated with hUCBSC demonstrated high expression levels of Mad and Max, suggesting that expression of Mad baits Max to form a complex. The high expression levels of either Mad or Max in the immunoblot cocultures, the presence of only G0/G1 phase in the FACS analysis, and the great degree of co-localization of Mad-Max in the hUCBSC-treated samples suggest a hypothetical mechanism in which hUCBSC express more of the transcriptional repressor protein Mad, which in turn, quenches Max from c-Myc, which reduces glioblastoma progression and invasion. In fact, we show increased expression levels of Myc and Mad in shERK-transfected U251 and 5310 cells whereas rERK treatment decreased expression levels of Mad and Max; these data support our hypothetical mechanism and are in line with our immunohistological data. Moreover, it is evident from our study that the interaction between Mad and Max ultimately downregulates c-Myc expression, thereby functionally causing downregulation of the ERK pathway. This functional correlation should be explored as a potential target in new therapeutic approaches for glioblastoma. In conclusion, the results of our study demonstrate that Mad and Max and their interactions serve as negative regulators of glioblastoma progression and promote tumor regression by virtue of their effect on c-Myc and ERK.
Supplementary Material
Acknowledgments
We thank Peggy Mankin and Noorjehan Ali for their technical assistance. We also thank Shellee Abraham for manuscript preparation and Diana Meister and Sushma Jasti for manuscript review. This project was supported by award number NS057529 (to J.S.R.) from the National Institute of Neurological Disorders and Stroke (NINDS). Contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Author Disclosure Statement
All authors declare no conflict of interest exists with this manuscript.
References
- 1.Lang FF. Bruner JM. Fuller GN. Aldape K. Prados MD. Chang S. Berger MS. McDermott MW. Kunwar SM, et al. Phase I trial of adenovirus-mediated p53 gene therapy for recurrent glioma: biological and clinical results. J Clin Oncol. 2003;21:2508–2518. doi: 10.1200/JCO.2003.21.13.2508. [DOI] [PubMed] [Google Scholar]
- 2.Steelman LS. Pohnert SC. Shelton JG. Franklin RA. Bertrand FE. McCubrey JA. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 3.Chang F. Steelman LS. Lee JT. Shelton JG. Navolanic PM. Blalock WL. Franklin RA. McCubrey JA. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia. 2003;17:1263–1293. doi: 10.1038/sj.leu.2402945. [DOI] [PubMed] [Google Scholar]
- 4.Cobb MH. Goldsmith EJ. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 5.McCubrey JA. Steelman LS. Franklin RA. Abrams SL. Chappell WH. Wong EW. Lehmann BD. Terrian DM. Basecke J, et al. Targeting the RAF/MEK/ERK, PI3K/AKT and p53 pathways in hematopoietic drug resistance. Adv Enzyme Regul. 2007;47:64–103. doi: 10.1016/j.advenzreg.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coller HA. Grandori C. Tamayo P. Colbert T. Lander ES. Eisenman RN. Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daksis JI. Lu RY. Facchini LM. Marhin WW. Penn LJ. Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene. 1994;9:3635–3645. [PubMed] [Google Scholar]
- 8.Jansen-Durr P. Meichle A. Steiner P. Pagano M. Finke K. Botz J. Wessbecher J. Draetta G. Eilers M. Differential modulation of cyclin gene expression by MYC. Proc Natl Acad Sci U S A. 1993;90:3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adhikary S. Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 10.Schreiber-Agus N. Stein D. Chen K. Goltz JS. Stevens L. DePinho RA. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proc Natl Acad Sci U S A. 1997;94:1235–1240. doi: 10.1073/pnas.94.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velpula KK. Dasari VR. Tsung AJ. Gondi CS. Klopfenstein JD. Mohanam S. Rao JS. Regulation of glioblastoma progression by cord blood stem cells is mediated by downregulation of cyclin D1. PLoS One. 2011;6:e18017. doi: 10.1371/journal.pone.0018017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao JS. Bhoopathi P. Chetty C. Gujrati M. Lakka SS. Matrix metalloproteinase-9 short interfering RNA induced senescence resulting in inhibition of medulloblastoma growth via p16INK4 and mitogen-activated protein kinase pathway. Cancer Res. 2007;67:4956–4964. doi: 10.1158/0008-5472.CAN-07-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenormand G. Henon S. Richert A. Simeon J. Gallet F. Elasticity of the human red blood cell skeleton. Biorheology. 2003;40:247–251. [PubMed] [Google Scholar]
- 14.Sato T. Miwa A. Ets-1 and integrin beta3 for lung metastasis from colorectal cancer. APMIS. 2002;110:347–353. doi: 10.1034/j.1600-0463.2002.100410.x. [DOI] [PubMed] [Google Scholar]
- 15.Wei J. Zhou S. Bachem MG. Debatin KM. Beltinger C. Infiltration of blood outgrowth endothelial cells into tumor spheroids: role of matrix metalloproteinases and irradiation. Anticancer Res. 2007;27:1415–1421. [PubMed] [Google Scholar]
- 16.Zhao J. Pestell R. Guan JL. Transcriptional activation of cyclin D1 promoter by FAK contributes to cell cycle progression. Mol Biol Cell. 2001;12:4066–4077. doi: 10.1091/mbc.12.12.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaller MD. Hildebrand JD. Parsons JT. Complex formation with focal adhesion kinase: A mechanism to regulate activity and subcellular localization of Src kinases. Mol Biol Cell. 1999;10:3489–3505. doi: 10.1091/mbc.10.10.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlaepfer DD. Jones KC. Hunter T. Multiple Grb2-mediated integrin-stimulated signaling pathways to ERK2/mitogen-activated protein kinase: summation of both c-Src- and focal adhesion kinase-initiated tyrosine phosphorylation events. Mol Cell Biol. 1998;18:2571–2585. doi: 10.1128/mcb.18.5.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivasan R. Zabuawala T. Huang H. Zhang J. Gulati P. Fernandez S. Karlo JC. Landreth GE. Leone G. Ostrowski MC. Erk1 and Erk2 regulate endothelial cell proliferation and migration during mouse embryonic angiogenesis. PLoS One. 2009;4:e8283. doi: 10.1371/journal.pone.0008283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee A. Hu J. Goss DJ. Thermodynamics of protein-protein interactions of cMyc, Max, and Mad: effect of polyions on protein dimerization. Biochemistry. 2006;45:2333–2338. doi: 10.1021/bi0522551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mauleon I. Lombard MN. Munoz-Alonso MJ. Canelles M. Leon J. Kinetics of myc-max-mad gene expression during hepatocyte proliferation in vivo: Differential regulation of mad family and stress-mediated induction of c-myc. Mol Carcinog. 2004;39:85–90. doi: 10.1002/mc.20000. [DOI] [PubMed] [Google Scholar]
- 22.Teodorczyk M. Martin-Villalba A. Sensing invasion: cell surface receptors driving spreading of glioblastoma. J Cell Physiol. 2010;222:1–10. doi: 10.1002/jcp.21901. [DOI] [PubMed] [Google Scholar]
- 23.Brunet A. Roux D. Lenormand P. Dowd S. Keyse S. Pouyssegur J. Nuclear translocation of p42/p44 mitogen-activated protein kinase is required for growth factor-induced gene expression and cell cycle entry. EMBO J. 1999;18:664–674. doi: 10.1093/emboj/18.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marampon F. Gravina GL. Di Rocco A. Bonfili P. Di SM. Fardella C. Polidoro L. Ciccarelli C. Festuccia C, et al. MEK/ERK inhibitor U0126 increases the radiosensitivity of rhabdomyosarcoma cells in vitro and in vivo by downregulating growth and DNA repair signals. Mol Cancer Ther. 2011;10:159–168. doi: 10.1158/1535-7163.MCT-10-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendes O. Kim HT. Lungu G. Stoica G. MMP2 role in breast cancer brain metastasis development and its regulation by TIMP2 and ERK1/2. Clin Exp Metastasis. 2007;24:341–351. doi: 10.1007/s10585-007-9071-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhou HY. Pon YL. Wong AS. Synergistic effects of epidermal growth factor and hepatocyte growth factor on human ovarian cancer cell invasion and migration: role of extracellular signal-regulated kinase 1/2 and p38 mitogen-activated protein kinase. Endocrinology. 2007;148:5195–5208. doi: 10.1210/en.2007-0361. [DOI] [PubMed] [Google Scholar]
- 27.Su CM. Lu DY. Hsu CJ. Chen HT. Huang CY. Yang WH. Su YC. Yang SN. Fong YC. Tseng WP. Tang CH. Glial cell-derived neurotrophic factor increases migration of human chondrosarcoma cells via ERK and NF-kappaB pathways. J Cell Physiol. 2009;220:499–507. doi: 10.1002/jcp.21802. [DOI] [PubMed] [Google Scholar]
- 28.Chiu WT. Shen SC. Chow JM. Lin CW. Shia LT. Chen YC. Contribution of reactive oxygen species to migration/invasion of human glioblastoma cells U87 via ERK-dependent COX-2/PGE(2) activation. Neurobiol Dis. 2010;37:118–129. doi: 10.1016/j.nbd.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 29.Liu X. Yan S. Zhou T. Terada Y. Erikson RL. The MAP kinase pathway is required for entry into mitosis and cell survival. Oncogene. 2004;23:763–776. doi: 10.1038/sj.onc.1207188. [DOI] [PubMed] [Google Scholar]
- 30.Keely P. Parise L. Juliano R. Integrins and GTPases in tumour cell growth, motility and invasion. Trends Cell Biol. 1998;8:101–106. doi: 10.1016/s0962-8924(97)01219-1. [DOI] [PubMed] [Google Scholar]
- 31.Schlaepfer DD. Hauck CR. Sieg DJ. Signaling through focal adhesion kinase. Prog Biophys Mol Biol. 1999;71:435–478. doi: 10.1016/s0079-6107(98)00052-2. [DOI] [PubMed] [Google Scholar]
- 32.Sieg DJ. Hauck CR. Schlaepfer DD. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci. 1999;112:2677–2691. doi: 10.1242/jcs.112.16.2677. [DOI] [PubMed] [Google Scholar]
- 33.Brown MC. Staniszewska I. Lazarovici P. Tuszynski GP. Del Valle L. Marcinkiewicz C. Regulatory effect of nerve growth factor in alpha9beta1 integrin-dependent progression of glioblastoma. Neuro Oncol. 2008;10:968–980. doi: 10.1215/15228517-2008-047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young BA. Taooka Y. Liu S. Askins KJ. Yokosaki Y. Thomas SM. Sheppard D. The cytoplasmic domain of the integrin alpha9 subunit requires the adaptor protein paxillin to inhibit cell spreading but promotes cell migration in a paxillin-independent manner. Mol Biol Cell. 2001;12:3214–3225. doi: 10.1091/mbc.12.10.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staniszewska I. Zaveri S. Del Valle L. Oliva I. Rothman VL. Croul SE. Roberts DD. Mosher DF. Tuszynski GP. Marcinkiewicz C. Interaction of alpha9beta1 integrin with thrombospondin-1 promotes angiogenesis. Circ Res. 2007;100:1308–1316. doi: 10.1161/01.RES.0000266662.98355.66. [DOI] [PubMed] [Google Scholar]
- 36.Marampon F. Ciccarelli C. Zani BM. Down-regulation of c-Myc following MEK/ERK inhibition halts the expression of malignant phenotype in rhabdomyosarcoma and in non muscle-derived human tumors. Mol Cancer. 2006;5:31. doi: 10.1186/1476-4598-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marampon F. Bossi G. Ciccarelli C. Di Rocco A. Sacchi A. Pestell RG. Zani BM. MEK/ERK inhibitor U0126 affects in vitro and in vivo growth of embryonal rhabdomyosarcoma. Mol Cancer Ther. 2009;8:543–551. doi: 10.1158/1535-7163.MCT-08-0570. [DOI] [PubMed] [Google Scholar]
- 38.Ussar S. Voss T. MEK1 and MEK2, different regulators of the G1/S transition. J Biol Chem. 2004;279:43861–43869. doi: 10.1074/jbc.M406240200. [DOI] [PubMed] [Google Scholar]
- 39.Mauro A. Ciccarelli C. De Cesaris P. Scoglio A. Bouche M. Molinaro M. Aquino A. Zani BM. PKCalpha-mediated ERK, JNK and p38 activation regulates the myogenic program in human rhabdomyosarcoma cells. J Cell Sci. 2002;115:3587–3599. doi: 10.1242/jcs.00037. [DOI] [PubMed] [Google Scholar]
- 40.Baudino TA. Cleveland JL. The Max network gone mad. Mol Cell Biol. 2001;21:691–702. doi: 10.1128/MCB.21.3.691-702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMahon SB. Wood MA. Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ayer DE. Kretzner L. Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 43.Hachem A. Gartenhaus RB. Oncogenes as molecular targets in lymphoma. Blood. 2005;106:1911–1923. doi: 10.1182/blood-2004-12-4621. [DOI] [PubMed] [Google Scholar]
- 44.Chen J. Willingham T. Margraf LR. Schreiber-Agus N. DePinho RA. Nisen PD. Effects of the MYC oncogene antagonist, MAD, on proliferation, cell cycling and the malignant phenotype of human brain tumour cells. Nat Med. 1995;1:638–643. doi: 10.1038/nm0795-638. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.