Abstract
Cellular pluripotency is associated with expression of the homeobox transcription factor genes NANOG, SOX2, and POU5F1 (OCT3/4 protein). Some reports suggest that mesenchymal progenitor cells (MPCs) may express increased quantities of these genes, creating the possibility that MPCs are more “pluripotent” than other adult cell types. The objective of this study was to determine whether equine bone marrow–derived MPCs had gene expression or DNA methylation patterns that differed from either early fetal-derived or terminally differentiated adult cells. Specifically, this study compared DNA methylation of the NANOG and SOX2 promoter regions and concurrent gene expression of NANOG, SOX2, and POU5F1 in equine induced pluripotent stem (iPS) cells, fetal fibroblasts, fetal brain cells, adult chondrocytes, and MPCs. Results indicate that NANOG and POU5F1 were not detectable in appreciable quantities in tissues other than the equine iPS cell lines. Equine iPS cells expressed large quantities of all three genes examined. Significantly increased quantities of SOX2 were noted in iPS cells and both fetal-derived cell types compared with adult cells. MPCs and adult chondrocytes expressed equivalent, low quantities of SOX2. Further, NANOG and SOX2 expression inversely correlated with the DNA methylation pattern in the promoter region, such that as gene expression increased, DNA methylation decreased. The equine iPS cell lines examined demonstrated DNA methylation and gene expression patterns that were consistent with pluripotency features described in other species. Results do not support previous reports that NANOG, SOX2, and POU5F1 are poised for increased activity in MPCs compared with other adult cells.
Introduction
Pluripotency is defined as the ability of a cell line to give rise to differentiated cells of all three primary germ layers. Pluripotency is associated with expression of the homeobox transcription factor genes NANOG, SOX2, and POU5F1 (the gene that codes for OCT3 and OCT4 proteins) [1–6]. These transcription factors are involved in sustaining pluripotency through transcriptional regulatory networks that function by repression of genes associated with differentiation [5–8]. Through complex signaling pathways, transcription factors repress or activate a subset of target genes to either maintain pluripotency or activate differentiation programs.
Analyses of SOX2, NANOG, and OCT3 and OCT4 indicate it is the relative quantities of these proteins which determine cell fate [1,9–15]. Although these factors play a role in maintenance of pluripotency, one of these factors alone cannot be regarded as the “Master” pluripotency regulator, as each cannot solely sustain self-renewal and prevent differentiation [16]. Pluripotency is dependent on nonlinear interactions, where molecular cues exert their effects dependent on the magnitude, combination, and duration of exposure to many different factors, such as leukemia inhibitory factor (LIF) and bone morphogenetic proteins (BMPs) [17].
Several epigenetic control mechanisms are used to regulate gene expression including DNA methylation and posttranslational modifications of histone proteins to affect remodeling of the chromatin structure. DNA methylation is one mechanism that controls DNA accessibility to transcriptional machinery. Previous work has demonstrated that increased gene expression is inversely correlated with DNA methylation [18–20]. Hypermethylation of cytosine preceding guanine (CpG)s in the promoter region recruits enzymes that downregulate transcription through inhibition of transcription machinery binding either directly or indirectly through modification of the chromatin structure. The exact mechanism is not fully understood but seems to be dependent on the density of CpG dinucleotides, the presence or absence of various histone modifications such as the addition or hydroxylation of a methyl group to the 5 position of the cytosine pyrimidine ring or the number 6 nitrogen of the adenine purine ring, and protein complex binding with polypeptides such as histone deacetylases and other chromatin remodeling proteins near CpGs [20–23].
DNA methylation can be utilized as a biomarker to determine whether cells are in an epigenetic state poised for activation of developmental regulatory genes [24–26]. For example, hypomethylation of the NANOG promoter has been demonstrated in fully validated pluripotent stem cells [27]. The epigenetic state of mouse and human embryonic stem (ES) and adult-derived cells have been investigated in several studies [28–31]. To date, no epigenetic studies have been reported using any type of equine somatic cells. An improved understanding of the basic biology of equine somatic cells is needed because the horse has emerged as an important species of interest in the field of regenerative and pluripotency research.
Clinically, equine “stem cell” therapies are being used extensively with very little understanding of the biologic properties of cells being implanted. Many companies (e.g., VetStem and VetCell) and universities (e.g., University of California, Davis) are commercializing the application of equine cells to regenerate muscle, tendon and ligament, and enhance cartilage healing in equine patients. Promotional promises are being made for great success based on little knowledge of how cells are contributing to the repair. Lack of information about the behavior of these cells has created both confusion and unrealistic expectations for horse owners who elect these unproven therapies for their animals. Due to similarities between equine and human athletes in the types and severity of injuries, the horse is an important model organism for musculoskeletal research. Further, the horse can provide large quantities of multiple tissue types in a nonlethal, minimally invasive manner for use in experimental studies, so repeated sampling from or multiple implantations into the same animal are possible. We sought to investigate the epigenetic state of an equine bone marrow–derived “stem cell” source frequently used for clinical applications to expand our knowledge about the pluripotency of these cells, with potential implications in validation of their use for both veterinary and comparable human therapies.
Although pluripotency has been attributed primarily to ES cell lines, some studies have suggested that mesenchymal progenitor cells (MPCs) may have increased gene expression of NANOG and POU5F1 [32] or potential for in vitro differentiation of MPCs into nonmesenchymal tissues [33]. The aim of this study was to evaluate the expression of the pluripotency genes (NANOG, SOX2, and POU5F1) using an epigenetic approach. We aimed to determine whether these genes were expressed in appreciable quantities in MPCs, with corresponding patterns of decreased DNA methylation compared with other equine cell types. Since there are no fully validated equine ES cell lines available, equine induced pluripotent stem (iPS) cells lines capable of forming teratomas were selected for use as control cells in this experiment [34]. For comparison, samples from equine iPS cells, fetal brain, fetal fibroblasts, adult bone marrow–derived MPCs, and adult chondrocytes were used. Our hypothesis was that since MPCs are of adult tissue origin, progenitor cells derived from adult equine bone marrow would have little to no expression of NANOG, SOX2, and POU5F1 and would be in an epigenetic state similar to terminally differentiated chondrocytes (another adult stromal tissue). In contrast, equine iPS and possibly other early fetal-derived cell types would have increased expression of these pluripotency markers with concomitant evidence of decreased DNA methylation.
Samples were evaluated using quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) with concurrent evaluation of DNA methylation patterns in the promoter regions of NANOG and SOX2. NANOG was selected for analysis of DNA methylation based on previous work suggesting it was likely to be highly methylated in adult tissues [35]. SOX2 was selected as a transcription factor that, while important in combination with other factors, is less specific for pluripotency maintenance, and therefore would likely have less promoter DNA methylation. Evaluation of the relationship between DNA methylation and gene expression of the pluripotency genes used in this study could improve our understanding of MPC biology and contribute to broader knowledge of differentiation-associated epigenetic changes.
Materials and Methods
Experimental overview
Samples of fetal brain, fetal fibroblasts, adult chondrocytes, and adult bone marrow–derived MPCs were collected. RNA and genomic DNA were isolated from each sample type. RNA and genomic DNA isolated from recently developed equine iPS cell lines generously donated by the Dr. Andras Nagy laboratory were used as a positive control tissue, since a fully validated equine ES cell line has not been reported [34]. Samples were assayed using qRT-PCR and MALDI-TOF mass spectrometry. All procedures were performed in compliance with institutional guidelines for research on animals.
Isolation of fetal brain
Brain tissue was harvested from a normal 180-day-gestation fetus (normal equine gestation is 340 days). The tissue was rinsed in phosphate buffered saline (PBS), rapidly frozen in liquid nitrogen, and stored as part of a tissue bank at −80°C until needed. A portion of the fetal brain tissue was used for genomic DNA and RNA isolation.
Isolation of equine fetal fibroblasts
An equine 34-day conceptus was collected by uterine lavage. A segment of body wall was harvested for fibroblast isolation. The tissue sample was gently homogenized in high-glucose Dulbecco's modified Eagle's medium (DMEM; 4,500 mg/L) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), streptomycin (100 U/mL), and 2 mM L-glutamine. The resultant mixture was pipetted to suspend the sample in media, transferred to 100-mm-diameter tissue culture plates, and incubated at 37°C, with 5% CO2 and a humidified atmosphere. At 24 h, the plates were rinsed with PBS and the culture media were exchanged. After 48 h of culture, the adherent fetal fibroblasts were trypsinized and the harvested cells were used for genomic DNA and RNA isolation.
Isolation of equine chondrocytes
Cartilage was aseptically harvested from the metacarpophalangeal joints from 3 horses (age range 5–6 years) euthanized for an unrelated study. Horses had no evidence of musculoskeletal abnormalities in the metacarpophalangeal joints. Chondrocytes were isolated using an overnight matrix digestion in 0.075% type II collagenase as previously described [36]. Chondrocytes were cryopreserved until needed. Prior to DNA and RNA isolation, chondrocytes were thawed and cultured overnight using Ham's F-12 medium supplemented with 10% FBS, penicillin (100 U/mL), streptomycin (100 U/mL), 25 mM HEPES, 2 mM L-glutamine, 50 mg/mL ascorbic acid, and 30 mg/mL α-ketoglutaric acid. Samples were cultured in 100-mm-diameter tissue culture plates, incubated at 37°C, with 5% CO2 and a humidified atmosphere. The following day, adherent chondrocytes were trypsinized and the harvested cells were used for genomic DNA and RNA isolation.
Collection of bone marrow aspirates and isolation of mononuclear cells
Bone marrow mononuclear cells were isolated from bone marrow aspirated from the sternum of 7 horses (age range 5–17 years). The aspirate (60 mL) from each horse was collected into a final concentration of 33 U/mL of preservative-free heparin (American Pharmaceutical Partners Inc.). Samples were diluted to 180 mL total volume using PBS and 0.5% bovine serum albumin. Samples were layered over Ficoll-Plaque Plus (GE Healthcare Biosciences) for density gradient centrifugation and collection of the enriched nucleated cell fraction as previously described [37,38].
MPC culture isolation and expansion
The nucleated cell fraction was cultured in a monolayer at a density of ∼300,000 cells/cm2 (∼20×106 cells/plate) on 100-mm-diameter tissue culture plates. Cells were propagated in low glucose (DMEM 1,000 mg/L) supplemented with 10% FBS, penicillin (100 U/mL), streptomycin (100 U/mL), 2 mM L-glutamine, and 1 ng/mL basic fibroblast growth factor (bFGF). This media is subsequently referred to as BM-media. The BM-media was exchanged every 72–96 h. Samples were cultured in a humidified atmosphere of 37°C and 5% CO2. The cells reached subconfluence of 70%–90% as determined by microscopic evaluation after 10–12 days and were passaged using Accumax® cell dissociation solution (Innovative Cell Technologies Inc.) to avoid damage to the cell surface proteins. The cells were replated at a density of 4,000–8,000 cells/cm2.
Genomic DNA isolation
The DNeasy® blood and tissue kit (Qiagen GmbH) was used for isolation and purification of genomic DNA from equine iPS cells, fetal fibroblasts, fetal brain tissue, adult bone marrow–derived MPCs, and adult chondrocytes according to the manufacturer's recommendations. Quality and quantity of DNA was determined using a Nanodrop® ND-1000 Spectrophotometer (NanoDrop Technologies Inc.). DNA purity was verified using gel electrophoresis. Isolated genomic DNA was sheared by passage through a 27-gauge needle in preparation for subsequent treatments.
DNA methylation primer design
Candidate primers for NANOG and SOX2 were designed using the EpiDesigner-Program™ software (www.epidesigner.com) with DNA sequences published in Genbank. Primers were selected based on the density of CpG dinucleotides and their proximity to the promoter region (Table 1). By searching the DNA coding sequence and the sequence 2,000 kb upstream, 4 CpG islands were identified in both NANOG and SOX2 that fit the definition of CpG islands as described by Takai and Jones [39]. One primer set was designed for each transcription factor to cover as many CpG dinucleotides as possible. Primers were designed to bind just outside CpG-rich areas to avoid bias due to the existence of both methylated and nonmethylated cytosines in CpG islands. For NANOG, all 13 CpG dinucleotides detected in the promoter region were covered by the primer set. For SOX2, the resultant primer set covered 27 of the 37 CpG dinucleotides identified in the promoter region. Primer sets were validated for specificity using fetal brain tissue, since no other positive control tissue was available at the time of initial primer selection. When genomic DNA from equine iPS cells was obtained, primer sets underwent further validation to confirm specificity for the gene of interest. Following PCR amplification (GeneAmp® PCR System 9600; Applied Biosystems), products were run on a 0.8% agarose gel. UV transillumination confirmed the correct size of PCR product for each gene.
Table 1.
Sequence of Primers Employed for the DNA Methylation Assay
| Gene name | NANOG | SOX2 |
|---|---|---|
| Genomic regiona | EquCab2-6-35481584-35487923 | EquCab2-19-20356019-20359070 |
| Analyzed sequence | Coding sequence and 2 kb upstream size: 6344 bp | Coding sequence and 2 kb upstream size: 3053 bp |
| Primer design | Product size: 478 bp, 1.4 kb upstream of transcription start site; CpG dinucleotides included: 13 | Product size: 497 bp, 0.88 kb upstream of transcription start site; CpG dinucleotides included: 27 |
| Primerb Sequence | F:5′-TAATTTAGGGTAAGTTAGGATGGGG-3′ | F:5′-AATTTTTTTTGGAGGGAGGTTTAG-3′ |
| R:5′-AACACCTAAACTAACAACTTACCAATTC-3′ | R:5′-TATCCTACTAAAATTTCAAAAACCC-3′ |
UCSC genome browser on horse September 2007 (Broad/EquCab2) assembly.
Primers were ordered with the standard Sequenom MassCLEAVE tails (F: AGGAAGAGAG; R: CAGTAATACGACTCATAGGGAGAAGGCT).
bp, base pairs; F, forward; R, reverse.
DNA methylation analysis
Quantitative determination of DNA methylation in the CpG promoter regions of NANOG and SOX2 in the samples required several steps prior to mass spectrometry analysis. Bisulfite treatment was performed to convert nonmethylated cytosine nucleotides to uracil, while leaving the 5-methylcytosine residues of the sample undisturbed. Bisulfite treatment was performed using the EZ-96 DNA Methylation Kit (Zymo Research) according to manufacturer's recommendations. Next, gene specific primers were added to amplify the template, while preserving the bisulfite sequence changes using PCR. All PCRs were carried out in the GeneAmp PCR System 9600 (Applied Biosystems) according to EpiTYPER's recommendations using AccuPrime™ Taq Polymerase and 10× AccuPrime buffer II (Invitrogen). The PCR products were run on a 2% agarose gel to verify specific amplification of the respective SOX2 and NANOG sequences. Following amplification of the target region, samples were cleaved in a base-specific manner to create fragments of differing sizes dependent on the sequence changes generated through bisulfite treatment. Samples were then assayed using MALDI-TOF mass spectrometry to determine what CpGs in the promoter region of the sample were originally methylated.
SssI methylase treatment
Prior to bisulfite treatment, DNA from a portion of the cell lysates was treated with SssI methylase (Cat. #M0226; New England BioLabs® Inc.) according to the manufacturer's recommendations. This enzyme induces DNA hypermethylation and helps confirm the bisulfite treatment reaction is complete in later analysis. SssI methylase–treated samples were utilized as positive controls, as the resultant samples should be fully methylated and can be used for comparison with untreated samples.
RNA extraction and one-step reverse transcription and qRT-PCR
Gene expression analysis was performed to evaluate the quantity of NANOG, SOX2, and POU5F1 in cells derived from equine iPS, fetal, and adult tissue sources. RNA was extracted from approximately 1 to 3×106 cells of the corresponding samples collected for DNA methylation assays using the 5 Prime Perfect Pure RNA® extraction kit (5 Prime Inc.) according to the manufacturer's recommendations. RNA quantity and quality were determined using a Nanodrop spectrophotometer (NanoDrop Technologies, Inc.) and visualization of the 18 and 28S bands on 0.8% agarose gels.
RNA samples were diluted to a concentration of 100 ng/well; two replicate wells were used for quantitative gene expression assays. Total RNA was reverse transcribed and amplified using the one-step RT-PCR technique and the ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). The primers and dual-labeled fluorescent probe [6-FAM as the 5′ label (reporter dye) and TAMRA as the 3′ label (quenching dye)] were designed using Primer Express Software Version 2.0b8a (Applied Biosystems). The NANOG, SOX2, and POU5F1 probes and primers were designed using predicted equine-specific sequences published in Genbank and the sequences obtained in our laboratory. Portions of the genes were cloned and gene sequences agreed with previously reported data (NANOG: XM_001498808; SOX2: NM_001143799; POU5F1: XM_001490108).
NANOG had the following primers and probe:
Forward 5′-ACAGCCCCGATTCATCCA-3′
Probe FAM (5′)-CAGTCCCAGAGTAAAACCGCTGCCC-TAMRA (3′)
Reverse 5′-TCTTTGCCTCGCTCGTCTCT-3′
SOX2 had the following primers and probe:
Forward 5′-TGCGAGCGCTGCACAT-3′
Probe FAM (5′)-ATAAATACCGTCCTCGGCGGAAAACCAA-TAMRA (3′)
Reverse 5′-AGCGTGTACTTATCCTTCTTCATGAG-3′
POU5F1 had the following primers and probe:
Forward 5′-CGGGCACTGCAGGAACAT-3′
Probe FAM (5′)-TTCTCCAGGTTGCCTCTCACTCGGTTC-TAMRA (3′)
Reverse 5′-CCGAAAGAGAAAGCGAACTAGTATTG-3′
Statistical analysis
Gene expression data and DNA methylation data were categorized into four tissue types: (1) equine iPS cells; (2) fetal brain or fibroblasts; (3) bone marrow–derived MPCs; (4) adult chondrocytes. Groups were compared using a one-way ANOVA with a Tukey all-pairwise comparisons post hoc test to determine whether there were differences in gene expression or DNA methylation of pluripotency genes between the different tissue types. A P-value of <0.05 was considered significant.
Results
Validation of bisulfite treatment
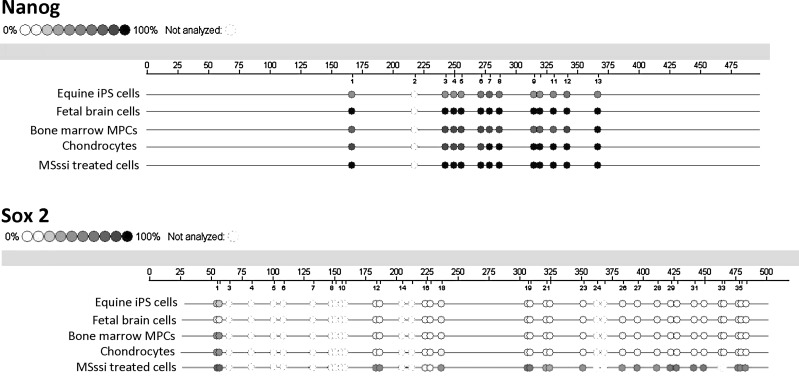
Treatment of genomic DNA with SssI methylase resulted in 90%–100% methylation of all CpG dinucleotides for NANOG and up to 80% methylation of all CpG dinucleotides of SOX2 (Fig. 1). SssI methylase treatment resulted in increased DNA methylation compared with untreated samples but did not cause full methylation in all samples.
FIG. 1.
Epigrams of the overall DNA methylation of NANOG and SOX2 promoter regions. Circles represent the examined CpG dinucleotides upstream of the coding sequences of NANOG and SOX2. The degree of filling of the circles represents the degree of DNA methylation from 0% to 100% in the various sample types. Incomplete, unfilled circles represent dinucleotide sites that did not have results.
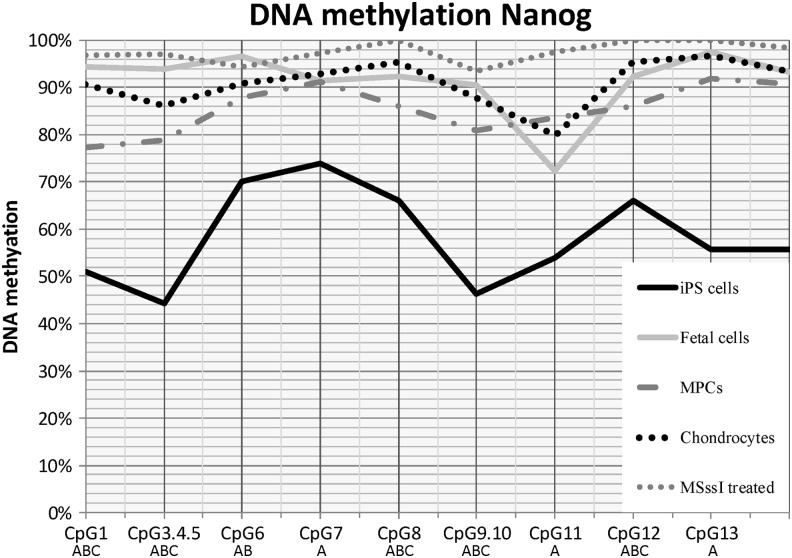
DNA methylation analysis of NANOG
At least 75% of the CpG dinucleotides in the promoter region of NANOG were methylated in most samples examined (Fig. 2). The exceptions were the equine iPS cell lines, where there was a significant (P≤0.005) decrease in DNA methylation at all NANOG CpG dinucleotide sites analyzed compared with the other sample types. The high degree of DNA methylation across all of the CpG dinucleotides examined did not allow for clear distinction between the remaining tissue types. Although several CpG dinucleotides had statistical differences detected between the non-iPS groups, there was no apparent correlation to NANOG expression in these samples.
FIG. 2.
NANOG DNA methylation pattern at individual CpG sites. Lines reflect sample type. MSssi-treated samples are included for comparison. Letters (A, B, C) denote statistical differences (P≤0.05) between (A) iPS cells, fetal cells, MPCs, or chondrocytes; (B) fetal cells, MPCs, or chondrocytes; (C) MPCs versus chondrocytes. iPS, induced pluripotent stem.
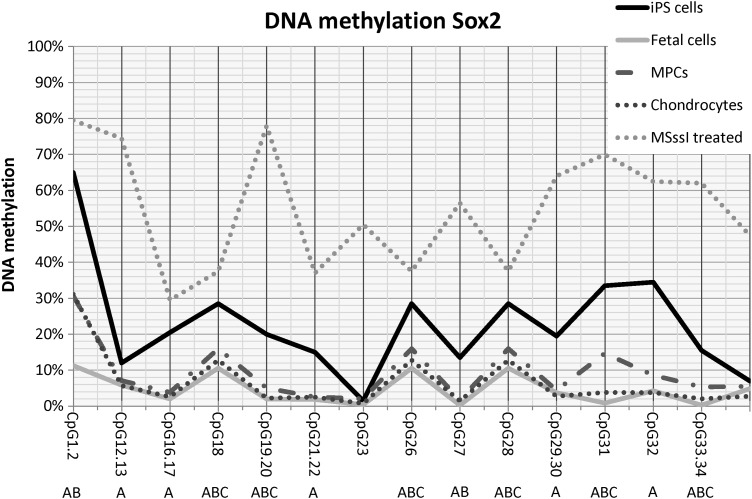
DNA CpG methylation analysis of SOX2
The DNA methylation of SOX2 across the promoter region was very low for all samples (Fig. 3). Although the overall percentage of DNA methylation was low, differences in DNA methylation in the promoter region of SOX2 between tissue types were evident. Significant differences (P≤0.02) were noted between tissue types for the majority of CpG dinucleotides. Interestingly, the DNA methylation pattern of equine iPS samples was the highest of all of the groups examined. Fetal tissues had significantly less DNA methylation than MPCs at most CpG dinucleotides and chondrocytes had less methylation than MPCs when a statistical difference was noted.
FIG. 3.
SOX2 DNA methylation pattern at individual CpG sites. Lines reflect sample type. MSssi-treated samples are included for comparison. Letters (A, B, C) denote statistical differences (P≤0.05) between (A) iPS cells, fetal cells, MPCs, or chondrocytes; (B) fetal cells, MPCs, or chondrocytes; (C) MPCs versus chondrocytes.
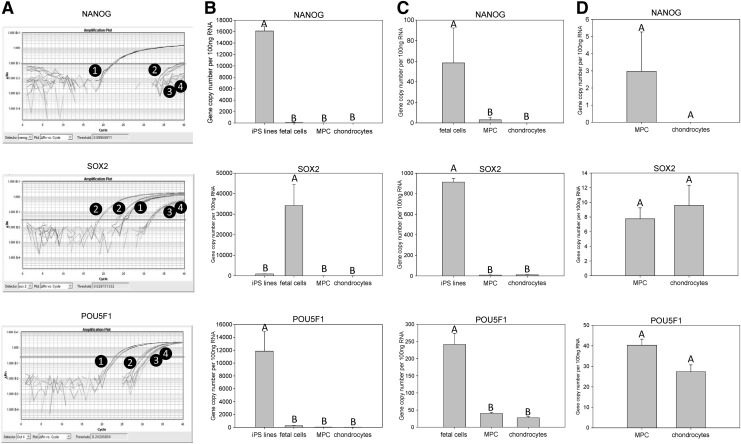
Gene expression analysis of NANOG
NANOG expression was significantly (P≤0.005) increased in equine iPS cell lines with over 160 copies/ng of RNA compared with the other groups examined. The expression of NANOG was very low in all remaining samples analyzed, with less than 1 gene copies/ng of RNA observed in all samples. Significant differences (P≤0.005) in NANOG expression were noted between tissue types (Fig. 4). Fetal cells had significantly (P≤0.005) more NANOG expression than either adult MPCs or chondrocytes. There was no difference (P=0.50) in NANOG expression between adult MPCs and chondrocytes.
FIG. 4.
Quantitative gene expression of NANOG, SOX2, and POU5F1. (A) Representative amplification plots for (1) equine iPS cell lines, (2) fetal cells, (3) MPC samples, and (4) chondrocytes. (B) Gene expression comparison between the four sample types. (C) Gene expression comparison between three remaining sample types when the group expressing the highest quantity of the gene of interest was omitted from the analysis. (D) Gene expression comparison between MPCs and chondrocytes. The Y-axis represents mean normalized gene copy numbers in 100 ng of RNA; bars on the X-axis represent sample types (±SEM). Letters (A, B) denote statistical differences (P≤0.05) between groups. SEM, standard error of the mean.
Gene expression analysis of SOX2
In contrast to the very low copy numbers of NANOG, gene expression of SOX2 was much higher in some tissues (Fig. 4). For example, over 500 gene copies/ng of RNA were detected in the fetal brain sample. There was a significant increase in SOX2 expression in fetal cells compared with all other tissue types (P≤0.005). Equine iPS cells had significantly higher (P≤0.005) amounts of SOX2 expression than adult tissues. There was no significant difference noted when MPCs were compared with adult chondrocytes (P=0.56).
Gene expression analysis of POU5F1
Similar to the pattern seen for NANOG, the gene expression of POU5F1 was very low in all samples aside from the equine iPS cells (Fig. 4). Equine iPS cells had significantly (P≤0.005) increased quantities of POU5F1, with over 120 copies/ng of RNA. Less than 3 gene copies/ng of RNA were observed in all remaining samples. Significant differences (P≤0.005) were noted between the remaining tissue types with fetal cells having significantly more POU5F1 expression than either adult MPCs or chondrocytes. There was no difference (P=0.25) in POU5F1 expression between MPCs and chondrocytes.
Discussion
In this study, DNA methylation analysis suggests that NANOG is a more specific marker of pluripotency than SOX2, as NANOG was far less methylated in equine iPS cells than any other cell type examined. The remaining samples were not in an epigenetic state poised for transcriptional activation in any of the cell types analyzed, including tissues from early development. Our data support previous reports that the NANOG promoter region becomes hypermethylated very early during development to avoid overexpression in neonatal cells [40–42]. SOX2 was less specific for pluripotent cells, as all tissues examined had low levels of DNA methylation and expressed variable quantities of SOX2 dependent on the tissue of origin.
Gene expression data provided additional information in at least one part of the mechanism of transcriptional regulation to support the DNA methylation results. Based solely on the DNA methylation results, we anticipated that NANOG would have very little gene expression in all tissues other than the iPS cell lines. This was supported by our results. For both NANOG and POU5F1, it is likely the low copy numbers (less than 3 gene copies/ng of RNA) detected in the adult and fetal samples had an insignificant, if any, biological role. These low gene quantities more likely represented detection of background noise. This is consistent with results of NANOG expression in cultured MPCs from other species, where gene copies have been detected via RT-PCR, but not by Northern blot [43] or microarray [44–47]. In addition, NANOG and POU5F1 have pseudogenes that may further complicate accurate detection of gene expression [35,43].
The molecular regulation of mammalian development, lineage commitment, and cellular specification involves transcriptional mechanisms governed through environmental signaling with molecules such as LIF, BMP, and Wnt signaling pathways, combined with epigenetic and translational mechanisms [40,48–53]. It is likely that epigenetic modifications play a dual role by stabilizing cellular lineage commitment and providing cues to promote differentiation [50]. Our results suggest that fetal-derived cells, MPCs, and chondrocytes are not comparable with equine iPS cells. The non-iPS cell types examined in this study appear to be in an epigenetic state committed to an established cellular lineage. Human iPS cell lines have shown significant reprogramming variability, aberrant reprogramming, incomplete reprogramming, and other differences from human ES cell lines [54]. Had it been available, the ideal control cell to demonstrate pluripotency in this study would have been a fully validated equine ES cell line. The choice to use equine iPS cell lines capable of forming teratomas as the positive control, while imperfect, was the most stringent control cell type currently available. In support of our findings, results from a previous study demonstrated that reprogrammed cells hold an epigenetic memory reflecting their tissue of origin [55].
A few studies have suggested that established MPCs from other species hold differentiation abilities comparable to ES cells and express transcription factors associated with pluripotency such as NANOG, SOX2, and OCT3 and OCT4 [33,56–58]. We had no evidence of increased expression of these transcription factors in adult MPCs compared with either adult-derived or fetal-derived cell types and significantly decreased expression of these genes compared with validated iPS cells. It is possible that the increased gene expression reported previously in bone marrow–derived MPCs represents a response to the in vitro culture environment. Several studies that reported pluripotent-like characteristics in MPCs used LIF as a culture media supplement [33,56–58]. This is in contrast to our study, where MPC cultures were derived and expanded in medium that had been enriched with only bFGF. Other studies verified that LIF in the presence of serum is capable of changing cell fate without genetic manipulation [41]. In addition, a recent publication demonstrated that adding trypsin to cell culture medium mimics a stress situation, as occurs during injury and disease [59]. Trypsin-treated cells undergoing subsequent suspension culture conditions could be induced to mimic cell types from all germ layers in vitro and in vivo but were still unable to form teratomas in testes in immunodeficient mice [59]. From these studies we can conclude that evaluation of potency of established MPC cultures necessitates uniform culture conditions.
Highly conserved regulatory regions have been identified in the 5′ flanking region upstream of the transcription start site of NANOG in mice, humans, and nonhuman primates. Octamer elements, Sox elements, and other transcription factor binding motifs with both repressive and activating effects on NANOG have been documented [51–53]. Epigenetic studies using undifferentiated human ES cells identified a hypomethylated CpG island near the transcription start site that extends as far as 1.7 kb upstream of the 5′ flanking region [25]. This region was included in the primers designed for NANOG in this study. DNA methylation of the upstream 5′ flanking region of NANOG is a well-conserved epigenetic regulatory mechanism among species that contributes to gene repression and lineage restriction.
In this study, we have reported the DNA methylation pattern in the NANOG or SOX2 promoter regions and gene expression of NANOG, SOX2, and POU5F1 in a number of equine iPS, fetal-derived, and adult-derived cell types. Several differences in gene expression and DNA methylation patterns were demonstrated between the cell types. The data did not support that equine bone marrow–derived MPCs were in an epigenetic state different from adult chondrocytes, confirming our hypothesis that MPCs derived from equine bone marrow would have little to no expression of NANOG, SOX2, and POU5F1 and would be in an epigenetic state similar to terminally differentiated chondrocytes. Further, gene expression correlated with DNA methylation pattern in the promoter regions of both transcription factors examined, with increased methylation correlating with decreased gene expression. Our results did not support that equine bone marrow–derived MPCs are in an epigenetic state poised for activation of NANOG, SOX2, and POU5F1.
Acknowledgments
The authors would like to express gratitude to Peter Schweitzer and Liz Paronett for providing invaluable guidance in the use of the EpiTyper™ assay. We would also like to thank Taralyn McCarrel for her contribution to the plasmid preparation for the pluripotency genes. Finally, we are very grateful to Dr. Andras Nagy and his research group for the generous contribution of RNA and genomic DNA from recently developed equine iPS cell lines to be used as positive controls in this study. This work was supported by grant number C023050 from New York State Stem Cell Science and grant number F32 AR056187-02 from NIAMS/NIH (Catherine H. Hackett).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Wang J. Rao S. Chu J. Shen X. Levasseur DN. Theunissen TW. Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 2.Keenen B. de la Serna IL. Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation. J Cell Physiol. 2009;219:1–7. doi: 10.1002/jcp.21654. [DOI] [PubMed] [Google Scholar]
- 3.Kalmar T. Lim C. Hayward P. Munoz-Descalzo S. Nichols J. Garcia-Ojalvo J. Martinez A. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JB. Sebastiano V. Wu G. Arauzo-Bravo MJ. Sasse P. Gentile L. Ko K. Ruau D. Ehrich M, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 5.Eiges R. Schuldiner M. Drukker M. Yanuka O. Itskovitz-Eldor J. Benvenisty N. Establishment of human embryonic stem cell-transfected clones carrying a marker for undifferentiated cells. Curr Biol. 2001;11:514–518. doi: 10.1016/s0960-9822(01)00144-0. [DOI] [PubMed] [Google Scholar]
- 6.Judson RL. Babiarz JE. Venere M. Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adachi K. Suemori H. Yasuda SY. Nakatsuji N. Kawase E. Role of SOX2 in maintaining pluripotency of human embryonic stem cells. Genes Cells. 2010;15:455–470. doi: 10.1111/j.1365-2443.2010.01400.x. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki A. Raya A. Kawakami Y. Morita M. Matsui T. Nakashima K. Gage FH. Rodriguez-Esteban C. Izpisua JC. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niwa H. Ogawa K. Shimosato D. Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 10.Rosner MH. Vigano MA. Ozato K. Timmons PM. Poirier F. Rigby PW. Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- 11.Silva J. Chambers I. Pollard S. Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 12.Meissner A. Wernig M. Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 13.Lin T. Chao C. Saito S. Mazur SJ. Murphy ME. Appella E. Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 14.Masui S. Nakatake Y. Toyooka Y. Shimosato D. Yagi R. Takahashi K. Okochi H. Okuda A. Matoba R, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 15.Loh YH. Wu Q. Chew JL. Vega VB. Zhang W. Chen X. Bourque G. George J. Leong B, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 16.Chickarmane V. Troein C. Nuber UA. Sauro HM. Peterson C. Transcriptional dynamics of the embryonic stem cell switch. PLoS Comput Biol. 2006;2:e123. doi: 10.1371/journal.pcbi.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zandstra PW. Le HV. Daley GQ. Griffith LG. Lauffenburger DA. Leukemia inhibitory factor (LIF) concentration modulates embryonic stem cell self-renewal and differentiation independently of proliferation. Biotechnol Bioeng. 2000;69:607–617. [PubMed] [Google Scholar]
- 18.Docherty SJ. Davis OS. Haworth CM. Plomin R. Mill J. Bisulfite-based epityping on pooled genomic DNA provides an accurate estimate of average group DNA methylation. Epigenetics Chromatin. 2009;2:3. doi: 10.1186/1756-8935-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaenisch R. Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 20.Weber M. Hellmann I. Stadler MB. Ramos L. Paabo S. Rebhan M. Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 21.Meissner A. Mikkelsen TS. Gu H. Wernig M. Hanna J. Sivachenko A. Zhang X. Bernstein BE. Nusbaum C, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ku M. Koche RP. Rheinbay E. Mendenhall EM. Endoh M. Mikkelsen TS. Presser A. Nusbaum C. Xie X, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikkelsen TS. Ku M. Jaffe DB. Issac B. Lieberman E. Giannoukos G. Alvarez P. Brockman W. Kim TK, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitelaw NC. Whitelaw E. How lifetimes shape epigenotype within and across generations. Hum Mol Genet. 2006;15:R131–R137. doi: 10.1093/hmg/ddl200. [DOI] [PubMed] [Google Scholar]
- 25.Yeo S. Jeong S. Kim J. Han JS. Han YM. Kang YK. Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2007;359:536–542. doi: 10.1016/j.bbrc.2007.05.120. [DOI] [PubMed] [Google Scholar]
- 26.Yu XF. Kim JH. Jung EJ. Jeon JT. Kong IK. Cloning and characterization of cat POU5F1 and NANOG for identification of embryonic stem-like cells. J Reprod Dev. 2009;55:361–366. doi: 10.1262/jrd.20035. [DOI] [PubMed] [Google Scholar]
- 27.Fouse SD. Shen Y. Pellegrini M. Cole S. Meissner A. Van L. Jaenisch R. Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Go MJ. Takenaka C. Ohgushi H. Forced expression of Sox2 or Nanog in human bone marrow derived mesenchymal stem cells maintains their expansion and differentiation capabilities. Exp Cell Res. 2008;314:1147–1154. doi: 10.1016/j.yexcr.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 29.Boquest AC. Noer A. Sorensen AL. Vekterud K. Collas P. CpG methylation profiles of endothelial cell-specific gene promoter regions in adipose tissue stem cells suggest limited differentiation potential toward the endothelial cell lineage. Stem Cells. 2007;25:852–861. doi: 10.1634/stemcells.2006-0428. [DOI] [PubMed] [Google Scholar]
- 30.Boquest AC. Noer A. Collas P. Epigenetic programming of mesenchymal stem cells from human adipose tissue. Stem Cell Rev. 2006;2:319–329. doi: 10.1007/BF02698059. [DOI] [PubMed] [Google Scholar]
- 31.Noer A. Sorensen AL. Boquest AC. Collas P. Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue. Mol Biol Cell. 2006;17:3543–3556. doi: 10.1091/mbc.E06-04-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guillot PV. Gotherstrom C. Chan J. Kurata H. Fisk NM. Human first-trimester fetal MSC express pluripotency markers and grow faster and have longer telomeres than adult MSC. Stem Cells. 2007;25:646–654. doi: 10.1634/stemcells.2006-0208. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y. Jahagirdar BN. Reinhardt RL. Schwartz RE. Keene CD. Ortiz-Gonzalez XR. Reyes M. Lenvik T. Lund T, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 34.Nagy K. Sung HK. Zhang P. Laflamme S. Vincent P. Agha-Mohammadi S. Woltjen K. Monetti C. Michael IP. Smith LC. Nagy A. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. 2011;7:693–702. doi: 10.1007/s12015-011-9239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatano SY. Tada M. Kimura H. Yamaguchi S. Kono T. Nakano T. Suemori H. Nakatsuji N. Tada T. Pluripotential competence of cells associated with Nanog activity. Mech Dev. 2005;122:67–79. doi: 10.1016/j.mod.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Nixon AJ. Lillich JT. Burton-Wurster N. Lust G. Mohammed HO. Differentiated cellular function in fetal chondrocytes cultured with insulin-like growth factor-I and transforming growth factor-beta. J Orthop Res. 1998;16:531–541. doi: 10.1002/jor.1100160503. [DOI] [PubMed] [Google Scholar]
- 37.Radcliffe CH. Flaminio MJ. Fortier LA. Temporal analysis of equine bone marrow aspirate during establishment of putative mesenchymal progenitor cell populations. Stem Cells Dev. 2009;19:269–282. doi: 10.1089/scd.2009.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hackett CH. Flaminio MJ. Fortier LA. Analysis of CD14 expression levels in putative mesenchymal progenitor cells isolated from equine bone marrow. Stem Cells Dev. 2010;20:721–735. doi: 10.1089/scd.2010.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takai D. Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hattori N. Imao Y. Nishino K. Hattori N. Ohgane J. Yagi S. Tanaka S. Shiota K. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells. 2007;12:387–396. doi: 10.1111/j.1365-2443.2007.01058.x. [DOI] [PubMed] [Google Scholar]
- 41.Boiani M. Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 42.Chambers I. Colby D. Robertson M. Nichols J. Lee S. Tweedie S. Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 43.Hart AH. Hartley L. Ibrahim M. Robb L. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev Dyn. 2004;230:187–198. doi: 10.1002/dvdy.20034. [DOI] [PubMed] [Google Scholar]
- 44.Evsikov AV. Solter D. Comment on “ ‘Stemness': transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature”. Science. 2003;302:393. doi: 10.1126/science.1082380. author reply 393. [DOI] [PubMed] [Google Scholar]
- 45.Fortunel NO. Otu HH. Ng HH. Chen J. Mu X. Chevassut T. Li X. Joseph M. Bailey C, et al. Comment on “‘Stemness': transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature”. Science. 2003;302:393. doi: 10.1126/science.1086384. author reply 393. [DOI] [PubMed] [Google Scholar]
- 46.Schepers U. A stem cell molecular signature: are there hallmark properties that are shared by all stem cells? Chembiochem. 2003;4:716–720. doi: 10.1002/cbic.200300661. [DOI] [PubMed] [Google Scholar]
- 47.Ivanova NB. Dimos JT. Schaniel C. Hackney JA. Moore KA. Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 48.Lu R. Markowetz F. Unwin RD. Leek JT. Airoldi EM. MacArthur BD. Lachmann A. Rozov R. Ma'ayan A, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu da Y. Yao Z. Isolation and characterization of the murine Nanog gene promoter. Cell Res. 2005;15:317–324. doi: 10.1038/sj.cr.7290300. [DOI] [PubMed] [Google Scholar]
- 50.Zhao X. Ruan Y. Wei CL. Tackling the epigenome in the pluripotent stem cells. J Genet Genomics. 2008;35:403–412. doi: 10.1016/S1673-8527(08)60058-2. [DOI] [PubMed] [Google Scholar]
- 51.Liang J. Wan M. Zhang Y. Gu P. Xin H. Jung SY. Qin J. Wong J. Cooney AJ. Liu D. Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 52.Yuri S. Fujimura S. Nimura K. Takeda N. Toyooka Y. Fujimura Y. Aburatani H. Ura K. Koseki H. Niwa H. Nishinakamura R. Sall4 is essential for stabilization, but not for pluripotency, of embryonic stem cells by repressing aberrant trophectoderm gene expression. Stem Cells. 2009;27:796–805. doi: 10.1002/stem.14. [DOI] [PubMed] [Google Scholar]
- 53.Pan G. Li J. Zhou Y. Zheng H. Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 2006;20:1730–1732. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- 54.Lister R. Pelizzola M. Kida YS. Hawkins RD. Nery JR. Hon G. Antosiewicz-Bourget J. O'Malley R. Castanon R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim K. Doi A. Wen B. Ng K. Zhao R. Cahan P. Kim J. Aryee MJ. Ji H, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang Y. Vaessen B. Lenvik T. Blackstad M. Reyes M. Verfaillie CM. Multipotent progenitor cells can be isolated from postnatal murine bone marrow, muscle, and brain. Exp Hematol. 2002;30:896–904. doi: 10.1016/s0301-472x(02)00869-x. [DOI] [PubMed] [Google Scholar]
- 57.Breyer A. Estharabadi N. Oki M. Ulloa F. Nelson-Holte M. Lien L. Jiang Y. Multipotent adult progenitor cell isolation and culture procedures. Exp Hematol. 2006;34:1596–1601. doi: 10.1016/j.exphem.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 58.Ulloa-Montoya F. Kidder BL. Pauwelyn KA. Chase LG. Luttun A. Crabbe A. Geraerts M. Sharov AA. Piao Y, et al. Comparative transcriptome analysis of embryonic and adult stem cells with extended and limited differentiation capacity. Genome Biol. 2007;8:R163. doi: 10.1186/gb-2007-8-8-r163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuroda Y. Kitada M. Wakao S. Nishikawa K. Tanimura Y. Makinoshima H. Goda M. Akashi H. Inutsuka A, et al. Unique multipotent cells in adult human mesenchymal cell populations. Proc Natl Acad Sci U S A. 2010;107:8639–8643. doi: 10.1073/pnas.0911647107. [DOI] [PMC free article] [PubMed] [Google Scholar]