Abstract
We have shown that hematopoietic stem/progenitor cell phenotype and differentiative potential change throughout cell cycle. Lung-derived microvesicles (LDMVs) also change marrow cell phenotype by inducing them to express pulmonary epithelial cell-specific mRNA and protein. These changes are accentuated when microvesicles isolated from injured lung. We wish to determine if microvesicle-treated stem/progenitor cell phenotype is linked to cell cycle and to the injury status of the lung providing microvesicles. Lineage depleted, Sca-1+ (Lin-/Sca-1+) marrow isolated from mice were cultured with interleukin 3 (IL-3), IL-6, IL-11, and stem cell factor (cytokine-cultured cells), removed at hours zero (cell cycle phase G0/G1), 24 (late G1/early S), and 48 (late S/early G2/M), and cocultured with lung tissue, lung conditioned media (LCM), or LDMV from irradiated or nonirradiated mice. Alternatively, Lin-/Sca-1+ cells not exposed to exogenous cytokines were separated into G0/G1 and S/G2/M cell cycle phase populations by fluorescence-activated cell sorting (FACS) and used in coculture. Separately, LDMV from irradiated and nonirradiated mice were analyzed for the presence of adhesion proteins. Peak pulmonary epithelial cell-specific mRNA expression was seen in G0/G1 cytokine-cultured cells cocultured with irradiated lung and in late G1/early S cells cocultured with nonirradiated lung. The same pattern was seen in cytokine-cultured Lin-/Sca-1 cells cocultured with LCM and LDMV and when FACS-separated Lin-/Sca-1 cells unexposed to exogenous cytokines were used in coculture. Cells and LDMV expressed adhesion proteins whose levels differed based on cycle status (cells) or radiation injury (LDMV), suggesting a mechanism for microvesicle entry. These data demonstrate that microvesicle modification of progenitor/stem cells is influenced by cell cycle and the treatment of the originator lung tissue.
Introduction
For over 40 years, the “hierarchy model” has been widely accepted to describe the process by which differentiated hematopoietic cells are produced from bone marrow-derived hematopoietic stem cells (HSCs). In this model, a stem cell enters cell cycle, divides, and 1 or both of the daughter cells become a more differentiated progenitor cell. With subsequent divisions, daughter cells obtain more differentiated characteristics and lose self-renewal potential. Contrary to this model, our group has shown that HSC are capable of reversibly changing their functional phenotype as they progress though cell cycle [1–13]. We have used cocktails of cytokines including interleukins (ILs)-3, 6, and 11 and stem cell factor (SCF) or SCF, thrombopoietin, and FLK-2, to induce HSC to progress though cell cycle in a synchronous fashion. Previous work has shown that the majority of lineage depleted (Lin-), stem cell antigen-1 positive (Sca-1+) cells, a marrow population enriched with stem and progenitor cells, are in G0/G1 phase of cell cycle at isolation and for up to 16 h in cytokine culture (80%–90%) then enter into S phase after 20–24 h in cytokine culture. By 48 h, 90% of cells are found to be in late S/G2/M phase of cell cycle [6]. Gene expression profiles of highly purified murine HSC change dramatically, as “stem cell” genes are highly expressed at G0/G1 phase and turned off at S/G2/M phase, while “cell division” genes are turned on at S/G2/M phase [14]. Surface expression of adhesion proteins are also linked to cell cycle, altering the ability of these cells to bind extracellular matrix in vitro [6,7]. Differential adhesion protein expression may explain the engraftment nadir that we have observed of HSC, as cells in late S/early G2 phase prior to transplantation into myeloablated mice are defective at engrafting the host bone marrow relative to cells in other points of cycle [14]. As engraftment potential is significantly better in cells prior to and after late S/early G2 phase, then nadirs again at the next late S/early G2 phase, these changes appear to be reversible. Their fluctuating differentiative potential results in the production of populations of specific lineages of differentiated hematopoietic cells depending on where they are in cell cycle as we have shown that HSCs at early S phase and mid S phase give rise to megakaryocytic and nonproliferative granulocytic-predominant populations (respectively) of differentiated cells in secondary culture [3]. These observations have led to the continuum model of stem cell biology, in which the differentiation potential of HSCs is linked to cell cycle [15–23]. In addition to cell cycle, the potential of HSCs likely influenced other factors. Our group has shown that cellular vesicles derived from a variety of sources are capable of altering the gene and protein expression profile of cells of the bone marrow [24,25].
Cellular vesicles were first described to be present in the human circulatory system over 40 years ago [26]. These intriguing spherical structures are bound by a lipid bilayer which is similar in composition to the cell membrane from which the vesicle was derived. They contain elements that are a reflection of their cell of origin, including mRNA, microRNA, DNA, and protein, and are capable of selectively entering different cell populations [24,25]. Consequently, as vesicles are released into the extracellular compartment, other cells are exposed to these membranes and cytoplasmic elements. Cellular vesicles can be characterized by their size, density, appearance by electron microscopy, lipid and protein composition, mechanism of genesis, and intracellular origins [27]. This characterization has led to the description of distinct populations of cellular vesicles, including exosomes [28], ectosomes [29], microvesicles [30], membrane particles [30], and apoptotic vesicles [31]. Exosomes (50–80 nm in size) are released with the fusion of multivesicular bodies to plasma membrane. Microvesicles (100 nm–1 μm in size), on the other hand, are released by cell surface membrane blebbing. In the literature, generic terms have often been used to describe cellular vesicles, including “microparticles.” However, it is clear that most populations that have been studied by investigators represent a mixture of discrete entities. In practice, most vesicle preparations are inherently heterogeneous with different approaches giving enrichment of 1 type over another [27]. In this article, we will employ microvesicles as an all-encompassing term, as the population that we have studied is enriched with microvesicles but contains other cellular vesicles.
Although their biological significance has been largely overlooked for years, there is growing evidence that microvesicles may be important mediators of cell-to-cell communication. Earlier reports have described microvesicles as mediators of thrombosis [32] or as a potential tumor vaccine [33], but more recently focus has been on their capacity to modify cell fate. Cells that interact with them can display altered phenotypic and functional characteristics via a variety of different mechanisms. This may occur by the direct stimulation of cells with microvesicle-based growth factors or bioactive lipids, by the transfer of a variety of membrane surface receptors, or by epigenetic reprogramming by transfer of transcription factors or other factors which alter the cellular transcriptome [34]. They have been shown to transfer CD41, integrins, and C-X-C chemokine receptor type 4 (CXCR4) between cells [31,35–37] and deliver the human immunodeficiency virus and prions to cells [38,39]. Descriptions of functional changes induced by the transfer of vesicle-derived elements are becoming more abundant in the literature. Messenger RNA and protein contained within embryonic stem cell-derived microvesicles has been shown to reprogram hematopoietic progenitor cells [40]. DNA can be transferred by apoptotic bodies from irradiated Epstein-Barr virus (EBV)-carrying cell lines to cocultured cells, leading to integration of EBV copies and expression of EBV genes [41]. T lymphocyte extracts have also been shown to induce fibroblasts to express lymphoid genes [42]. In a similar fashion, endothelial cells exposed to endothelial progenitor cell-derived microvesicles had improved survival in culture and formed capillary-like structures both in vitro and after transplantation into nonobese diabetic severe combined immunodeficient mice [43]. Microvesicle-cell interactions may play an important role in the transfer of the “malignant” phenotype to nonmalignant cells as tumor-derived microvesicles can transfer determinants to monocytes [35] and human prostate cancer tissue can induce tissue-specific mRNA expression in cocultured human marrow cells [44].
Our previous work has focused on microvesicles derived from murine lung and their ability to induce a pulmonary epithelial cell phenotype on marrow cells in coculture [24,25]. In these studies, we have found that although our microvesicle preparation is vastly heterogeneous, it appears to be enriched with microvesicles originating from pulmonary epithelial cells. We have shown that lung-derived microvesicles (LDMVs) enter marrow cells in coculture and induce them to express high levels of pulmonary epithelial cell-specific mRNA and protein. Conversely, marrow cells exposed to LDMVs in coculture that did not internalize them have minimal to no expression of these genes, indicating that the microvesicle internalization is vital for these changes to occur. This effect was enhanced in marrow cells cocultured with lungs from previously-irradiated mice. LDMVs change the differentiation potential of cocultured marrow cells. When these cocultured cells are transplanted into lethally-irradiated mice, they preferentially engraft the recipient lung as functioning type II pneumocytes relative to transplanted marrow cells not exposed to microvesicles. Elements transferred by microvesicles, which include mRNA, miRNA, and lung-specific proteins, are responsible for these phenotypic changes. Our data indicate that among these transferred elements are those which impact the transcriptome of the target cell in a stable fashion. Supporting this statement are observations that marrow cells cocultured with LDMVs express pulmonary epithelial cell-specific mRNA for up to 12 weeks in vitro. These stable changes in gene expression are also seen in nonpulmonary tissue of mice transplanted with marrow cells that have been cocultured with LDMVs. In these experiments, myeloablated mice transplanted with microvesicle-modified marrow cells express pulmonary epithelial cell-specific mRNA in cells of their bone marrow, liver, spleen, and thymus 6 weeks after transplantation (unpublished data). Additionally, there are 3 times more bone marrow-derived pulmonary epithelial cells (type II pneumocytes) in the lungs of mice transplanted with microvesicle-modified marrow cells compared with mice transplanted with unmanipulated marrow cells. These findings establish that the influence of microvesicles on cells is not a transient phenomenon but rather results in a persistent change in cellular phenotype.
As LDMVs influence marrow cell fate and as the injury status of the microvesicle source contributes to these changes, we speculate that the cell cycle status of the cocultured marrow cell is another important variable in marrow cell receptivity to microvesicle-induced phenotypic change. In present studies, we show that Lin-/Sca-1+ marrow cell cycle status, both dependent and independent of exogenous cell cycle-inducing cytokines, and microvesicle source are important determinants in the ability of these cells to express pulmonary epithelial cell-specific genes.
Materials and Methods
Experimental animals
All studies used 6-to-8 week-old male C57BL/6 mice (Jackson Laboratories) and were approved by the Institutional Animal Care and Use Committee at Rhode Island Hospital. Animals had ad libitum access to food and water and were euthanized by CO2 inhalation or isofluorane inhalation followed by cervical dislocation.
Radiation injury
Mice were exposed to a single dose of 500 centigray (cGy) total body irradiation (TBI) using a Gammacell 40 Exactor Irradiator at 110 cGy/min (MDS Nordion) and euthanized 5 days after irradiation.
Tissue collection
For lung harvest, after euthanasia, blood was flushed from the vasculature using ice-cold 1× Dulbecco's Phosphate-Buffered saline (1× PBS; Invitrogen) infused thought the right ventricle. Lungs were placed in ice-cold 1× PBS supplemented with 5% heat-inactivated fetal calf serum (HIFCS; Hyclone) and 1% penicillin–streptomycin (PS; Invitrogen). For whole bone marrow (WBM) cell harvest, tibiae, femurs, iliac crests and spines were collected and all surrounding muscle removed with sterile gauze. Bones were placed in ice-cold 1× PBS/5% HIFCS/1% PS and crushed using mortar and pestle. Cells were strained through 40 μm cell strainer (Allegiance) placed over 50 mL conical tube (Fisher) then centrifuged at 300g for 10 min at 4°C.
Lineage depletion
Mononuclear cells were isolated from WBM by discontinuous density centrifugation at 1,000g for 30 min at room temperature using OptiPrep (Accurate Chemical). Mononuclear cells were then lineage depleted (Lin-) by adding the following rat-anti mouse antibodies: anti-Ter119, B220, Mac-1, Gr-1, CD4, and CD8 (BD Biosciences). After 15 min of incubation on ice, Dynabead M450 anti-rat IgG (Dynal) was added and lineage positive cells were removed by a magnetic column. Remaining Lin- cells were counted and percent viability determined was using Trypan Blue stain (Gibco).
Isolation of Lin-Sca-1+ cells
Allophycocyanin (APC)-conjugated anti-mouse LY-6AE (Sca-1; BD Biosciences) was added to a final concentration of 1 mg/106 Lin- cells, then incubated for 30 min on ice. Cells were washed with PBS, centrifuged at 300g for 10 min then passed through a 40 μm filter. Propidium iodide (0.05 mg/mL) was added (1:1,000 dilution) and Lin-/Sca-1+ cells were then separated using a 5 laser Becton Dickenson/Cytopeia Influx High Speed Cell Sorter.
Cell culture and coculture experiments
Cytokine-stimulated Lin-/Sca-1+ cell culture
Lin-/Sca-1+ marrow cells were cultured (1×105 cells/mL) in Teflon-coated flasks with Dulbecco's modified Eagle medium (DMEM glutamax; Invitrogen) supplemented with 15% HIFCS, 1% PS, 1% l-glutamine (Invitrogen), and the following cytokines: recombinant mouse (rm) IL-3, 50 U/mL, rm IL-11, 50 ng/mL, rm IL-6, 50 U/mL, and rm Steel factor 50 ng/mL. Cultures were harvested at 0, 20, 24, 28, 36, 44, 48, 52, and 72 h after initiation of culture. Time 0 cells were exposed to cytokine-supplemented media and immediately washed. Cells were washed with 1× PBS by centrifugation at 300g for 10 min then used in culture or coculture or for RNA extraction (Table 1, experiment 1).
Table 1.
Experimental Design: Cell Culture and Coculture Experiments
| Experiment number | Marrow cells | Cell cycle separation technique | Coculture established | Analysis performed |
|---|---|---|---|---|
| 1 | Lin-/Sca-1+ | Cultured with IL-3, IL-6, IL-11, SCF for 0–72 h (various time points) | No coculture established | RT-PCR on cytokine-cultured marrow cells after various time points in cytokine culture |
| 2 | Lin-/Sca-1+ | Cultured with IL-3, IL-6, IL-11, SCF for 0, 24 or 48 h | Cocultured with: Irradiated lung Nonirradiated lung No lung (control) | RT-PCR on marrow cells after 7 days of coculture with lung |
| 3 | Lin-/Sca-1+ | Hoechst 33258 labeled, separated by FACS into G0/G1 and S/G2/M cells (no exogenous cytokines) | Cocultured with: Irradiated lung No lung (control) | RT-PCR on marrow cells after 7 days of coculture with lung |
| 4 | Lin-/Sca-1+ | Cultured with IL-3, IL-6, IL-11, SCF for 0 or 48 h | Cocultured with: LCM, irradiated lung Unconditioned media (control) | RT-PCR on marrow cells after 7 days of coculture with LCM |
| 5 | Lin-/Sca-1+ | Cultured with IL-3, IL-6, IL-11, SCF for 0 or 48 h | Cocultured with: MVs, irradiated lung Media, MVs (control) | RT-PCR on marrow cells after 7 days of coculture with MVs |
In experiment 1, three experiments were performed (n=6–10 total per time point).
In experiment 2, five experiments were performed (n=8–12 per condition).
In experiment 3, two experiments were performed (n=4–6 per condition).
In experiment 4, two experiments were performed (n=4–8 per condition).
In experiment 5, two experiments were performed (n=4–8 per condition).
IL, interleukin; SCF, stem cell factor; RT-PCR, reverse transcription–polymerase chain reaction; FACS, fluorescence-activated cell sorting; LCM, lung conditioned media; MVs, microvesicles.
Coculture experiments of lung with cytokine-stimulated Lin-/Sca-1+ cells
About 1–1.5×105 Lin-/Sca-1+ cells that had been cultured with cytokines (as described above) for 0, 24, or 48 h were washed and placed in culture wells containing Dexter culture medium, consisting of Fischer medium supplemented with 1% PS, 0.0125 g/mL fungizone, 10−7 M hydrocortisone sodium succinate (all from Invitrogen), and 20% horse serum (HyClone). Cell-impermeable well inserts were placed in each well and the following cocultures were established: 0, 24, or 48 h cytokine-stimulated Lin-/Sca-1+ cells cocultured opposite nonirradiated lung, 500 cGy irradiated lung or Dexter media containing no lung (control). One half of a minced murine lung was used in each coculture well. Cocultures were incubated at 33°C/5% CO2 for 7 days. Well inserts were then removed, Lin-/Sca-1+ cells were washed with 1× PBS and centrifuged at 300g for 10 min at 4°C and lysed for total RNA extraction (Table 1, experiment 2).
Hoechst-labeled fluorescence-activated cell sorting-separated Lin-/Sca-1+ cells
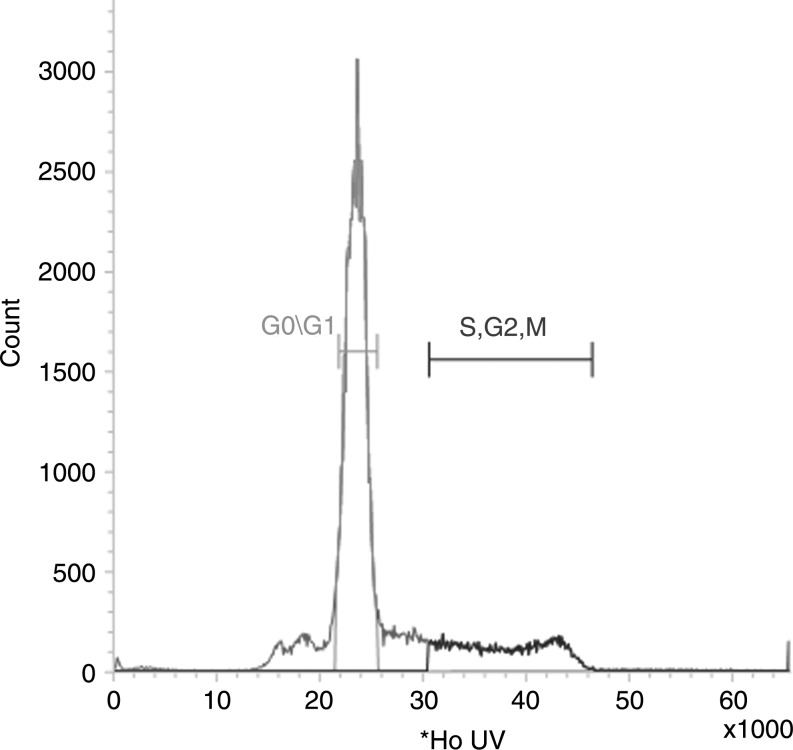
Hoechst 33258 dye (Sigma) was added (final concentration 10 μM/mL) to Lin-/Sca-1+ cells and incubated at 37°C for 90 min. Cells were washed with 1× PBS/5% HIFCS/1% PS and centrifuged at 300g for 10 min at 4°C. Cells were filtered through a 40 μm filter and separated using a 5 laser Becton Dickenson/Cytopeia Influx High Speed Cell Sorter based on their signal intensity of Hoechst dye. Cells with low signal intensities were considered to be in G0/G1 phase of cell cycle. Cells with signal intensities 1.25–1.75× that of G0/G1 cells were considered to be in S phase of cell cycle. Cells with signal intensities 1.75–2× that of G0/G1 cells were considered to be in G2/M phase of cell cycle. Doublets will be excluded by pulse width. G0/G1 cells and S/G2/M cells were separately collected and used in culture or coculture (Fig. 1).
FIG. 1.
Cell cycle status separation of Lin-/Sca-1+ cells using Hoechst 33258 dye. Lin-/Sca-1+ cells were labeled with Hoechst 33258 dye and separated into G0/G1 and S/G2/M cell cycle populations based on their Hoechst content.
Coculture experiments of lung with Hoechst-labeled FACS-separated Lin-/Sca-1+ cells
About 7.5 × 104–1.5 × 105 Lin-/Sca-1+ G0/G1 or S/G2/M cells were placed in cell-impermeable well-separated 6-well culture plates and cocultured with 500 cGy irradiated minced lung (one half lung per coculture well) or Dexter media containing no lung (control). Cocultures were incubated at 33°C/5% CO2 for 7 days. Well inserts were then removed, Lin-/Sca-1+ cells were washed with 1× PBS and centrifuged at 300g for 10 min at 4°C and lysed for total RNA extraction (Table 1, experiment 3).
Coculture experiments of cell-free lung conditioned media with cytokine-stimulated Lin-/Sca-1+ cells
Minced lungs isolated from 500 cGy irradiated mice were placed on top of cell-impermeable well insets (0.4 μm pore size; Millipore) in 6-well culture plates filled with DMEM-glutamax supplemented with 15% fetal calf serum (Hyclone) and 1% PS. Plates were incubated at 37°C/5% CO2 for 7 days then lung-containing well inserts were removed and the media collected. Media was centrifuged at 300g for 10 min to ensure that it was truly cell-free. Cell-free lung conditioned media (LCM) or DMEM-glutamax containing no lung tissue (control) were then used for coculture with 0 or 48 h cytokine-stimulated Lin/Sca-1+ cells. Cocultures were incubated at 33°C/5% CO2 for 7 days. Lin-/Sca-1+ cells were then washed with 1× PBS and centrifuged at 300g for 10 min at 4°C and lysed for total RNA extraction (Table 1, experiment 4).
Coculture experiments of LDMVs with cytokine-stimulated Lin-/Sca-1+ cells
LCM made from 500 cGy irradiated mice was ultracentrifuged at 10,000g for 1 h at 4°C in a Thermo Scientific Sorval WX Ultra series ultracentrifuge to remove apoptotic debris. The supernatant was collected and ultracentrifuged at 100,000g for 1 h at 4°C. The supernatant was discarded and the pellet resuspended in 1× PBS. The pelleted material was ultracentrifuged again at 100,000g for 1 h at 4°C and resuspended in 1× PBS. The ultracentrifuged (UCF) pellet derived from LCM was resuspended in DMEM-glutamax and used for coculture with Lin-Sca-1+ cells. The supernatant was discarded and the UCF pellet, containing LDMVs, was resuspended in DMEM-glutamax and used for coculture. LDMVs or DMEM-glutamax containing no lung tissue (control) was then used for coculture with 0 or 48 h cytokine-stimulated Lin/Sca-1+ cells. Cocultures were incubated at 33°C/5% CO2 for 7 days. Lin-/Sca-1+ cells were then washed with 1× PBS and centrifuged at 300g for 10 min at 4°C and lysed for total RNA extraction (Table 1, experiment 5).
RNA extraction, cDNA amplification, and real-time reverse transcription–polymerase chain reaction analysis
Total RNA from cultured or cocultured Lin-/Sca-1+ cells or uncultured minced lung was isolated using the RNeasy Mini or Micro Kit (Qiagen). RNA was measured for quantity and quality using a Nanodrop ND/1000 spectrophotometer (Thermo Scientific). Isolated RNA was used to amplify cDNA using the High Capacity cDNA transcription kit (Applied Biosystems) in a final volume of 20ul, per manufacturer's recommendations. The amount of RNA used ranged from 10 to 500 ng, depending on the yield from any given sample; however, equal amounts of RNA were used for all samples of any given experiment. Amplification reactions consisted of 1 cycle for 10 min at 25°C, 1 cycle for 120 min at 37°C, and 1 cycle for 5 min at 85°C. Gene expression was analyzed by reverse transcription–polymerase chain reaction (RT-PCR) using a 9800 Fast Thermal Cycler (Applied Biosystems). All 20× assay mixes were purchased from Applied Biosystems. Murine assays used were as follows: Beta 2 microglobulin (Mm00437762_m1), surfactant A (Mm00499170_m1), surfactant B (Mm00455681_m1), surfactant C (Mm00488144_m1), surfactant D (Mm00486060_m1), CCSP (Mm00442046_m1), aquaporin-5 (Mm00437578_m1), BMP4 (Mm00432087_m1), PDGFA (Mm00833533_m1), PGE2 (Mm00460181_m1), VEGFR-1 (Mm00438980_m1), VEGFR-2 (Mm00440099_m1), FGF10 (Mm00433275_m1), Nkx2.1 (Mm00447558_m1), Shh (Mm00436527_m1), PTHrP (Mm00436057_m1), integrin alpha 2 (Mm00434371_m1), integrin alpha 4 (Mm00439770_m1), integrin alpha 5 (Mm00439797_m1), integrin alpha L (Mm00801807_m1), integrin beta 1 (Mm01253227_m1), integrin beta 2 (Mm00434513_m1), L-selectin (Mm00441291_m1), P-selectin (Mm00441295_m1), VCAM-1 (Mm00449197_m1), PECAM-1 (Mm00476702_m1). cDNA preamplification reactions was performed with the following reagents in a final volume of 50: 12.5 μL of a pooled mixture for all assays (made by combining equal volumes all 20× TaqMan gene expression assays and diluting to a final concentration of 0.2×), 25 μL of TaqMan Preamp Master mix (Applied Biosystems), and 12.5 μL of cDNA. The reaction was performed on the 9800 Fast Thermal Cycler (Applied Biosystems) consisting of a 10 min cycle at 95°C followed by 14 cycles at 95°C for 15 s, then 60°C for 4 min. The final product was diluted with TE buffer [1 mM Tris (Invitrogen), 0.1 mM ethylenediaminetetraacetic acid (Gibco) pH 8.0] and then kept at −20°C. All Real Time RT-PCRs were performed in 96-well plates on a 7900HT Fast RT-PCR System with the following reagents in a final volume of 25 μL: 20× assay mix (for either β2 microglobulin or one of the target genes) and 2× TaqMan PCR Master Mix. Predetermined amounts of cDNA were added to this mixture. Duplicate reactions of the target and housekeeping genes were performed simultaneously for each cDNA template analyzed. The PCR consisted of an initial enzyme activation step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. A cycle threshold (CT) value was obtained for each sample, and duplicate sample values were averaged. The 2-ΔΔCT method was used to calculate relative expression of each target gene [45]. Briefly, mean CT value of target genes in each sample were normalized to its averaged housekeeping gene CT value to give a ΔCT value. This was then normalized to control samples (ΔΔCT), and the 2-ΔΔCT value was obtained. To calculate 2-ΔΔCT for target genes with no expression in the control group, a CT value of 40 was assigned to the control group so that a relative quantity of the target gene could be reported. In some instances, the ΔCT value (CT value of the endogenous gene Beta 2 Microglobulin subtracted from the CT value of the target gene) was used as a means of reporting gene expression. The control group used for all comparisons was 0 h Lin-/Sca-1+ cells cultured without lung, LCM or LDMVs or fluorescence-activated cell sorting (FACS)-separated G0/G1 or S/G2/M cells that were cocultured without lung. All controls were cultured for the same duration as the experimental groups.
Uptake of carboxyfluorescein N-succinimidyl ester-labeled microvesicles by Lin-/Sca-1+ cells
LDMVs were labeled with the cell cytoplasm dye carboxyfluorescein N-succinimidyl ester (CFSE; Molecular Probes), final concentration of 0.02 μM, for 15 min at 37°C. An equal volume of 10% fetal bovine serum solution in 1× PBS was then added and the samples were ultracentrifuged again at 100,000g for 1 h at 4°C. Lin-/Sca-1+ cells were cocultured with CFSE-labeled LDMVs in DMEM-glutamax at 37°C for 48 h. Cells were then harvested, washed with 1× PBS, and centrifuged at 300g for 10 min at 4°C then passed through a 40 μm filter. Lin-/Sca-1+ cells were then separated using a 5 laser Becton Dickenson/Cytopeia Influx High Speed Cell Sorter based on their presence or absence of CFSE. CFSE was excited at 488 nm and detected through a 528/38 bandpass filter. CFSE+ events were defined as cells that had internalized microvesicles in culture whereas CFSE- events were defined as cells exposed to microvesicles in culture that did not internalize them. Five micro liters of each sorted cell populations were placed on slides and Vectashield (Vector) with 0.4 mmol 4,6-Diamidino-2-phenyindole (Sigma) was added to counterstain nuclei. Samples were visualized using conventional and deconvolution fluorescence microscopy (Zeiss Axioplan 2 microscope; Carl Zeiss) at room temperature. No photosubtraction or processing of artifact was performed.
Microvesicle surface protein determination
LCM made from nonirradiated and irradiated lung was ultracentrifuged and the UCF pellet was resuspended in 1× PBS at a concentration of 1 × 107 microvesicles/mL (estimating 1 × 106 microvesicles per lung). Equal aliquots were labeled with of one of the following antibodies (final concentration: 0.5 μg antibody/1 × 106 microvesicles): anti-mouse CD49e (α5 integrin) AlexaFluor 647 (Biolegend), anti-human/mouse CD49f (α6 integrin) AlexaFluor 647 (Biolegend), rat IgG2aκ AlexaFluor 647 (isotype control for α5 and α6 integrin; Biolegend), anti-mouse/rat CD29 (β1 integrin) APC (eBioscience), armenian hampster IgG APC (isotype control for β1 integrin; eBioscience), rat anti-mouse CD184 (CXCR4) APC (BD Pharmingen), rat IgG2b APC (isotype control for CXCR4; BD Pharmingen), anti-mouse CD107a (LAMP-1) AlexaFluor 647 (eBioscience), rat IgG2a AlexaFluor 647 (isotype control for LAMP-1; eBioscience), rat anti-mouse VEGFR2/Ly-73 APC (BD Pharmingen), rat IgG 2a kappa APC (isotype control for VEGFR2; BD Pharmingen), anti-mouse CD54 (ICAM-1) fluorescein isothiocyanate (FITC; BD Pharmingen), Armenian Hamster IgG 1, kappa FITC (isotype control for ICAM-1; BD Pharmingen), anti-mouse CD81 (TAPA-1) phycoerythrin (PE; BD Pharmingen), Hampster IgG1 kappa PE (isotype control for TAPA-1; BD Pharmingen) anti-mouse CD154 (CD40 Ligand) APC (BD Pharmingen), Armenian Hamster IgG3 kappa APC (isotype control for CD40 Ligand; BD Pharmingen). Samples were incubated on ice for 15 min then washed by ultracentrifugation. Antibody-positive microvesicles were detected by FACS relative to a relevant isotype control. The quantity of antibody-positive microvesicles, expressed as a percentage of all sorted microvesicles, was determined.
Statistical analysis
Data were analyzed using a 2-sided Student's t-test in cases where there were fewer than 6 measurements within 2 parent groups. Alternatively, Wilcoxon rank sum test was performed in cases where there were 6 or more measurements within 2 parent groups. When more than 2 parent groups were compared, a 1-way ANOVA analysis was performed. We considered results to be statistically significant when P<0.05. Data were presented as mean+standard error.
Results
Baseline gene expression of Lin-/Sca-1+ cells in cytokine-stimulated cell cycle
Real Time RT-PCR analysis was performed on Lin-/Sca-1+ cells cultured in the presence of recombinant murine IL-3, IL-6, IL-11, and SCF (cytokines). Cells were removed from culture immediately (0 h), corresponding to G0/G1 phase of cell cycle, after 24 h, corresponding to late G1/early S phase of cell cycle, after 48 h, corresponding to late S/early G2 phase of cell cycle, and at 4 h intervals clustered around the 24 h and 48 h time points. Analysis focused on the expression on various pulmonary epithelial cell-specific genes and adhesion proteins. Expression of pulmonary epithelial cell-specific genes was extremely low or absent (data not shown). Expression for most other genes varied with time in cytokine culture but these changes were not statistically significant. However, changes in expression of α5, αL, β1, and β2 integrins and P-selectin were statistically significant (Table 2). Data are reported as ΔCT values (CT value of the housekeeping gene Beta 2 microglobulin subtracted from the CT value of the target gene), with lower values representing higher gene expression. These data indicate that the expression of certain adhesion proteins in Lin-/Sca-1+ cells significantly vary with cytokine-induced cell cycle.
Table 2.
Adhesion Protein Gene Expression of Lin-/Sca-1+ Cells in Cytokine-Induced Cell Cycle
| |
|
Time in cytokine culture |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 h | 20 h | 24 h | 28 h | 36 h | 44 h | 48 h | 52 h | ||
| α2 integrin | 7.96 (1.26) | 7.61 (0.17) | 8.79 (0.91) | 7.78 (0.53) | 8.97 (0.42) | 8.31 (0.02) | 8.26 (0.82) | 9.93 (0.68) | |
| α4 integrin* | 6.76 (0.38) | 8.07 (0.16) | 4.38 (0.54) | 3.33 (0.07) | 3.63 (0.28) | 3.17 (0.57) | 4.05 (0.29) | 2.68 (0.11) | |
| α5 integrin* | 5.73 (0.33) | 6.03 (0.30) | 7.66 (0.29) | 6.64 (0.32) | 7.29 (0.23) | 6.47 (0.31) | 9.49 (0.42) | 8.22 (0.22) | |
| ΔCT values, Lin-/Sca-1+ cells mean (SE) | αLintegrin* | 2.26 (0.28) | 2.29 (0.26) | 1.58 (0.18) | 3.88 (0.36) | 4.95 (0.59) | 6.54 (0.31) | 8.23 (0.87) | 8.06 (1.10) |
| β1 integrin* | 2.83 (0.38) | 2.81 (0.28) | 2.51 (0.25) | 2.21 (0.06) | 4.63 (0.24) | 4.90 (0.34) | 5.56 (0.36) | 5.76 (0.38) | |
| β2 integrin* | 4.10 (0.17) | 3.10 (0.21) | 3.02 (0.37) | 3.27 (0.16) | 4.78 (0.22) | 7.12 (0.23) | 6.98 (0.34) | 9.79 (0.89) | |
| L-selectin | 2.25 (0.34) | 2.86 (0.19) | 2.90 (0.44) | 3.64 (0.27) | 4.05 (0.13) | 3.56 (0.62) | 3.89 (0.91) | 3.56 (0.35) | |
| P-selectin* | 5.57 (0.31) | 6.67 (0.16) | 8.76 (0.21) | 8.77 (0.36) | 9.54 (0.22) | 10.38 (0.21) | 9.75 (0.99) | 7.74 (0.71) | |
| VCAM-1 | 8.40 (1.27) | 7.97 (1.14) | 8.70 (1.88) | 8.48 (2.00) | 9.09 (1.88) | 9.25 (1.06) | 8.99 (0.59) | 8.20 (0.28) | |
| PECAM-1 | 3.04 (0.87) | 3.44 (1.45) | 3.59 (1.56) | 3.33 (1.17) | 4.45 (1.45) | 3.56 (1.65) | 2.19 (0.72) | 3.51 (1.17) | |
P≤0.05, 1-way ANOVA.
VCAM-1, vascular cell adhesion protein 1; PECAM-1, platelet/endothelial cell adhesion molecule-1; ANOVA, analysis of variance; SE, standard error; CT, cycle threshold.
Gene expression of Lin-/Sca-1+ cells in cytokine-stimulated cell cycle cocultured with lung
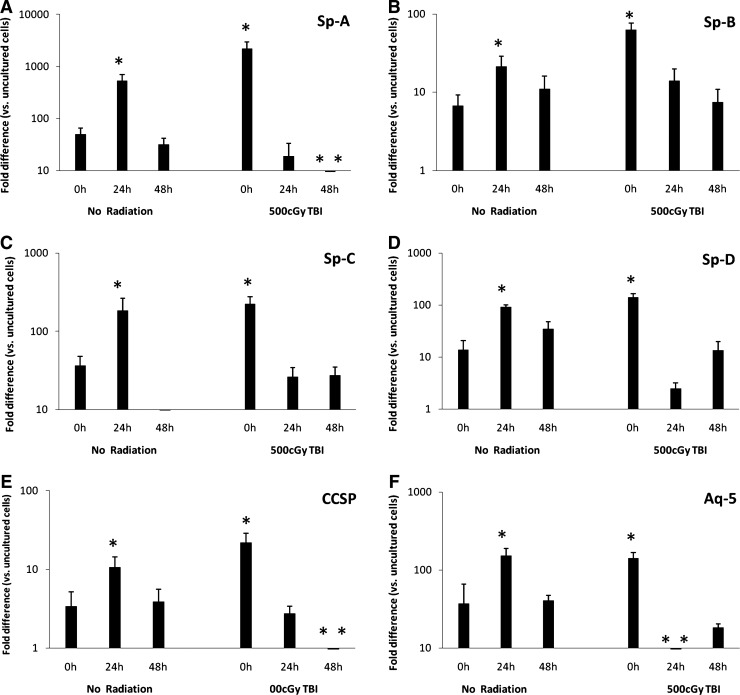
Lin-/Sca-1+ cells were cultured with cytokines for 0, 24, or 48 h then placed into coculture with lungs from mice exposed to 500 cGy of TBI, lungs from nonirradiated mice or no lung (control). During coculture, lung and marrow cells were separated by a cell-impermeable membrane (0.4 μm pore size). Lin-/Sca-1+ cell expression of adhesion proteins did not vary significantly with respect to time of Lin-/Sca-1+ cells in cytokine culture or radiation status of cocultured lung (data not shown). However, Lin-/Sca-1+ cell expression of all 6 pulmonary epithelial cell genes analyzed did significantly vary with respect to time of Lin-/Sca-1+ cells in cytokine culture and radiation status of cocultured lung (Fig. 2). When cocultured with nonirradiated lung, pulmonary epithelial cell gene expression was significantly elevated in 24 h cytokine-cultured Lin-/Sca-1+ cells compared with control cells and 0 and 48 h cytokine-cultured Lin-/Sca-1+ cells. Alternatively, when cocultured with irradiated lung, pulmonary epithelial cell gene expression was significantly elevated in 0 h cytokine-cultured Lin-/Sca-1+ cells compared with control cells and 24 and 48 h cytokine-cultured Lin-/Sca-1+ cells. These data indicate that Lin-/Sca-1+ marrow cells express a variety of pulmonary epithelial cell genes when cocultured with lung and that the magnitude of expression varies depending on the cytokine-induced cell cycle status and radiation status of the cocultured lung.
FIG. 2.
Cytokine-cultured Lin-/Sca-1+ cells cocultured with lung. (A) Sp-A, (B) Sp-B, (C) Sp-C, (D) Sp-D, (E) CCSP, (F) Aq-5 expression in cytokine-cultured Lin-/Sca-1+ cells that were then cocultured with lung. *P<0.05, Wilcoxon, 24 h versus 0 and 48 h or 0 h versus 24 and 48 h. **P<0.05, Wilcoxon, versus control. Sp-A, surfactant protein A; CCSP, clara cell-specific protein.
Gene expression of Lin-/Sca-1+ cells in cytokine-independent cell cycle cocultured with lung
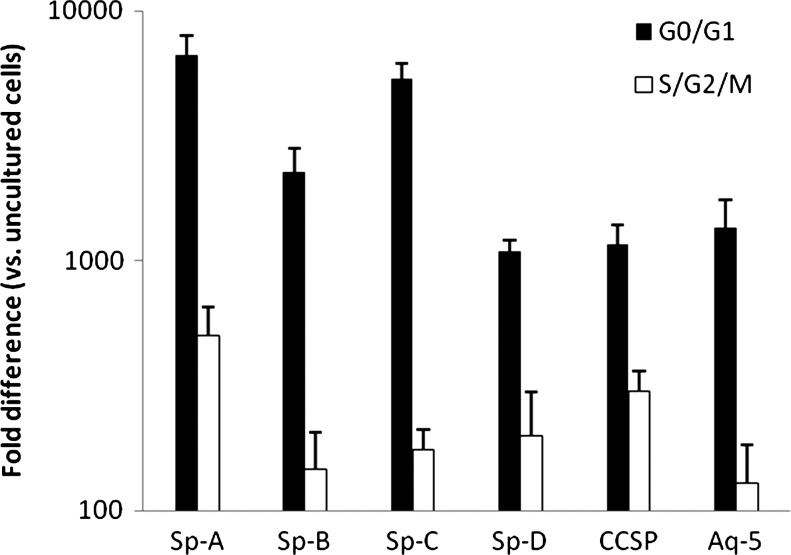
Lin-/Sca-1+ cells labeled with Hoechst 33258 dye were FACS-separated into G0/G1 (corresponding to 0 h in cytokine culture) and S/G2/M (corresponding to time points of up to 48 h in cytokine culture) then cocultured with irradiated lung or no lung (control). Lin-/Sca-1+ cell expression of adhesion proteins and growth factors were not significantly different in G0/G1 or S/G2/M Lin-/Sca-1+ cells cocultured with irradiated lung (data not shown). G0/G1 and S/G2/M Lin-/Sca-1+ cells cocultured with irradiated lung expressed significantly elevated levels of all 6 pulmonary epithelial cell genes analyzed compared with control cells (Fig. 3). In addition, G0/G1 Lin-/Sca-1+ cells cocultured with irradiated lung expressed significantly higher levels of pulmonary epithelial cell genes compared with S/G2/M cells cocultured with irradiated lung. These data indicate that pulmonary epithelial cell gene expression in Lin-/Sca-1+ cells exposed to lung in coculture vary according to Lin-/Sca-1+ cell cycle and that this variability is independent of exogenous cytokines.
FIG. 3.
Lin-/Sca-1+ cells in cytokine-independent cell cycle cocultured with lung. For all genes, G0/G1 versus S/G2/M cocultured Lin-/Sca-1+ cells and G0/G1 and S/G2/M cocultured Lin-/Sca-1+ cells versus control, P<0.05, Wilcoxon.
Gene expression of Lin-/Sca-1+ cells cocultured with LCM and LDMVs
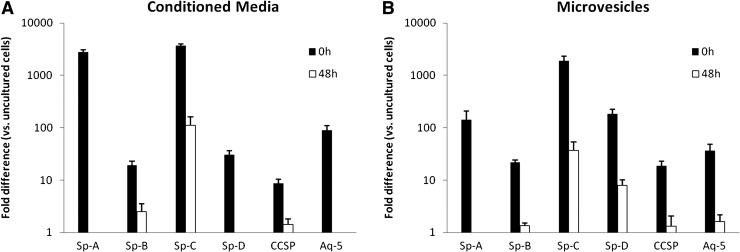
Lin-/Sca-1+ cells cultured for 0 or 48 h in cytokines were cocultured with either LCM or LDMVs isolated from irradiated mice or no lung (control). Expression of all of the 6 pulmonary epithelial cell genes analyzed was significantly elevated in 0 h cytokine-cultured Lin-/Sca-1+ cells cocultured either with LCM or LDMVs compared to 48 h cytokine-cultured Lin-/Sca-1+ cells (Fig. 4A, B). These data indicate that LDMVs induce expression of pulmonary epithelial cell genes in cocultured Lin-/Sca-1+ cells and that the magnitude of these changes is dependent on the cell cycle status of the cocultured Lin-/Sca-1+ cell.
FIG. 4.
Lin-/Sca-1+ cells cocultured with LCM, lung-derived microvesicles. Cytokine cultured Lin-/Sca-1+ cells (0 or 48 h) that were then cocultured with (A) LCM or (B) microvesicles from irradiated lung. For all genes, 0 versus 48 h and 0 and 48 h versus control, P<0.05, Student's t-test. LCM, lung conditioned media.
LDMVs are internalized by Lin-/Sca-1+ cells in culture
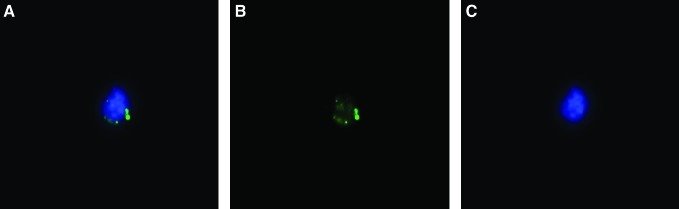
LDMVs labeled with the cell cytoplasmic dye, CFSE, were incubated with Lin-/Sca-1+ for 48 h then FACS-separated into 2 cell populations: cells that had internalized microvesicles (CFSE+ cells) and cells exposed to microvesicles that had not internalized microvesicles (CFSE− cells). Internalization of CFSE+ microvesicles into FACS-separated cells was confirmed by fluorescence microscopy (Fig. 5).
FIG. 5.
Lin-/Sca-1+ cells internalize lung-derived microvesicles. CFSE-labeled, lung-derived microvesicles internalized by a Lin-/Sca-1+ cell in culture. (A) Merged image, (B) FITC filter, (C) DAPI filter. 63 ×, room temperature. CFSE, carboxyfluorescein N-succinimidyl ester; FITC, fluorescein isothiocyanate; DAPI, 4,6-diamidino-2-phenyindole.
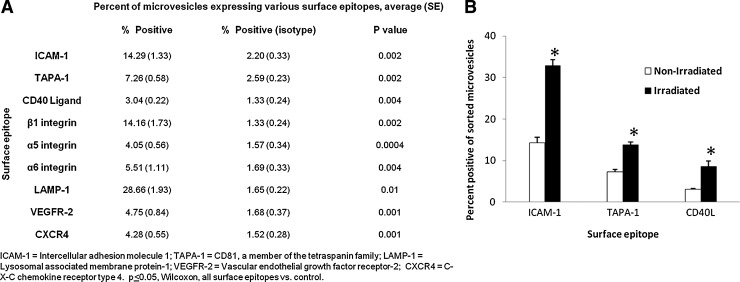
Adhesion protein profile of LDMVs
LDMVs were labeled with directly-conjugated antibodies to a variety of adhesion proteins or their isotype control antibodies. Antibody positive microvesicles were identified by FACS and quantified relative to their isotype controls (Fig. 6A). Adhesion proteins expressed in excess to their isotype controls on the surface of LDMVs included α5 integrin (CD49e), α6 integrin (CD49f), β1 integrin (CD29), CXCR4 (CD184), LAMP-1 (lysosomal associated membrane protein, CD107a), VEGFR2, ICAM-1 (CD54), TAPA-1 (CD81), and CD40 Ligand (CD154). Microvesicles isolated from irradiated lung contained a significantly higher percentage of microvesicles positive for ICAM-1, TAPA-1, and CD40 Ligand compared with microvesicles isolated from nonirradiated lung (Fig. 6B).
FIG. 6.
Surface epitopes of microvesicles isolated from irradiated, nonirradiated lung. (A) Percentage of microvesicles isolated from nonirradiated lung positive adhesion proteins, P<0.05, Wilcoxon, versus isotype control. (B) Percentage of ICAM-1, TAPA-1, CD40L positive microvesicles isolated from irradiated and nonirradiated lung. *P<0.05, Wilcoxon, irradiated versus nonirradiated. ICAM-1, intercellular adhesion molecule 1; TAPA-1, CD81, a member of the tetraspanin family.
Discussion
Our group has established that microvesicles isolated from murine lung induce pulmonary epithelial cell-specific gene expression in bone marrow cells which internalize them. This expression is accentuated in marrow cells that have been exposed to microvesicles from radiation-injured lung. We elected to use lung tissue as the source of microvesicles as means of studying cell cycle-related differences in microvesicle internalization due to our extensive experience with this population of microvesicles. These data presented here can be viewed from 2 separate but related perspectives. One is the variable influence of microvesicles on bone marrow-derived Lin-/Sca-1+ cells at different points in cell cycle, either cytokine-stimulated or independent of exogenous cytokines. The other is the impact of originator lung tissue injury by irradiation on the capacity of evolved microvesicles to influence Lin-/Sca-1+ cell gene expression at different points in cell cycle. The influence of microvesicles on cytokine-stimulated cells at different points in cell cycle represents another example of cycle-related phenotype alteration of marrow-derived stem/progenitor cells. We have shown that global gene expression, the expression profile of adhesion proteins and cytokine receptors, homing to marrow after transplantation and short- and long-term multi-lineage engraftment are all linked to changes in cell cycle [1–14]. Differentiation potential of marrow-derived stem-progenitor cells are also linked to cell cycle as evidenced by their ability to preferentially form megakaryocytes and granulocytes in vitro and convert into pulmonary epithelial cells after transplantation into irradiated mice [15–23]. The present work adds microvesicle modulation of cell phenotype to the cycle related changes in bone marrow-derived stem/progenitor cells.
The mechanism for the differing cycle specificity of microvesicles derived from normal or irradiated mice is as yet unclear. Our group has found that LDMVs contain hundreds of different miRNA and proteins, but the specific element (or elements) responsible for these phenotypic changes has yet to be clarified. Microvesicles isolated from radiation-injured and nonirradiated lung contain several miRNA and proteins species which are distinct from one another and the miRNA and protein profile of marrow cells exposed to them also differ (unpublished findings). Although the mechanism of these transformative changes is not known, their differential influence on Lin-/Sca-1+ cells may be related to the adhesion protein profile expressed on their surface. There are a variety of mechanisms that govern the uptake of microvesicles into target cells, including receptor-mediated and nonreceptor-mediated processes. The mechanism by which microvesicles are incorporated also depends on the source of microvesicles and the type of target cell; therefore, findings from a specific experimental condition shouldn't be generalized to all possible experimental conditions. Deregibus et al. [43] demonstrated that entry of endothelial progenitor cell-derived microvesicles into human microvascular endothelial cells can be greatly attenuated by exposure of microvesicles to anti-α4 integrin and anti-β1 integrin blocking antibodies. These findings suggest that in the context of these experimental conditions, receptor-mediated internalization is an important mechanism. In the present studies, we report that CD40 Ligand, ICAM-1, and TAPA-1 positive populations of microvesicles appeared to be enhanced when derived from radiation-injured lung cells. Additionally, Lin-/Sca-1+ cells expressed a multitude of adhesion protein genes, some of which (α4, α5, αL, β1 and β2 integrins, and P-selectin) had levels which changed significantly with cell cycle. Thus, fluctuating adhesion protein expression might explain the observed differences between cycle phase and originator tissue. For example, interactions between differentially expressed microvesicle-based ICAM-1 or CD40L and Lin-/Sca-1+ -based LFA-1 (αL/β2 integrins) or α5/β1 integrins, respectively, may account for these differences. However, to establish this, antibody blocking experiments are necessary. In this instance, we have been unable to determine microvesicle entry specificity since, in preliminary experiments, the isotope controls for blocking antibodies studied blocked the genetic conversions as effectively as the blocking antibodies themselves. This is of interest in itself and will be separately studied.
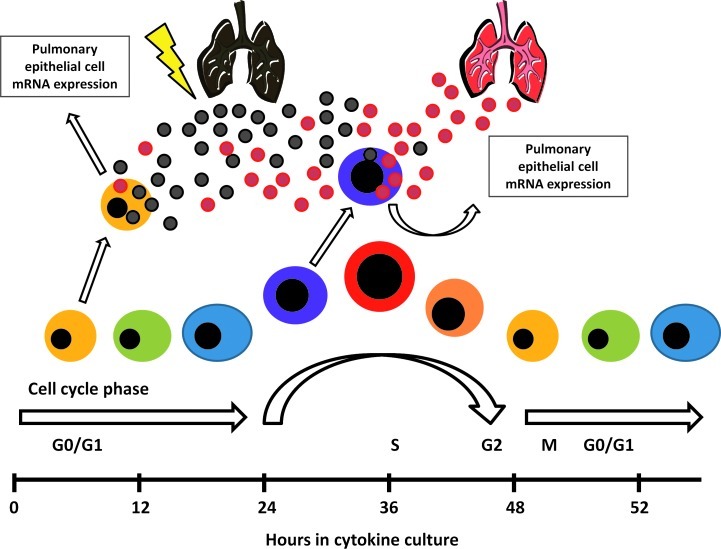
Our present results indicate that the influence of LDMVs on Lin-/Sca-1+ cell gene expression is strongly influenced by the cycle phase of the target cell and the treatment of the originator lung tissue (Fig. 7). We speculate that in the setting of lung injuries where the pulmonary capillary endothelial barrier is compromised, microvesicles originating from the pulmonary epithelium are able to gain access to the circulation. These microvesicles may then interact with bone marrow-derived stem/progenitor cells within the circulation, influencing phenotypic changes of these cells and may allow them to contribute to the cellular component of the injured lung. The influence of cell cycle phase on the ability of target cells to be modified by microvesicles further highlights the complexity of microvesicle cell fate modulation. The clinical relevance of these findings pertains to the development of cell-based therapies for lung disease. Microvesicle-modification may help to prime transplanted cells, enhancing their ability to engraft injured tissue. Optimal conditions, including transplanted cell population and its cell cycle status, would have to be defined. In summary, microvesicle modulation of cell phenotype is an evolving biology which is changing our views on the stability of cellular systems.
FIG. 7.
Progenitor/stem cell fate determination: a proposed model. The phenotype of progenitor/stem cells changes reversibly throughout cell cycle. Lung-derived microvesicles preferentially influence progenitor/stem cell gene expression at different points of cell cycle depending on the injury status of the lung tissue.
Acknowledgments
This work is supported by the following NIH grants: 5K08 HL086868-04, 1P20 RR025179-01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of this article.
Author Disclosure Statement
None of the authors have any commercial associations that might create a conflict of interest in connection with this manuscript.
References
- 1.Habibian HK. Peters SO. Hsieh CC. Wuu J. Vergilis K. Grimaldi CI. Reilly J. Carlson JE. Frimberger AE, et al. The fluctuating phenotype of the lymphohematopoietic stem cell with cell cycle transit. J Exp Med. 1998;188:393–398. doi: 10.1084/jem.188.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerny J. Dooner M. McAuliffe C. Habibian H. Stencil K. Berrios V. Reilly J. Carlson J. Cerny AM, et al. Homing of purified murine lymphohematopoietic stem cells: a cytokine-induced defect. J Hematother Stem Cell Res. 2002;11:913–922. doi: 10.1089/152581602321080574. [DOI] [PubMed] [Google Scholar]
- 3.Colvin GA. Dooner MS. Dooner GJ. Sanchez-Guijo FM. Demers DA. Abedi M. Ramanathan M. Chung S. Pascual S. Quesenberry PJ. Stem cell continuum: directed differentiation hotspots. Exp Hematol. 2007;35:96–107. doi: 10.1016/j.exphem.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Colvin GA. Lambert JF. Carlson JE. McAuliffe CI. Abedi M. Quesenberry PJ. Rhythmicity of engraftment and altered cell cycle kinetics of cytokine-cultured murine marrow in simulated microgravity compared with static cultures. In Vitro Cell Dev Biol Anim. 2002;38:343–351. doi: 10.1290/1071-2690(2002)038<0343:ROEAAC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 5.Colvin GA. Lambert JF. Moore BE. Carlson JE. Dooner MS. Abedi M. Cerny J. Quesenberry PJ. Intrinsic hematopoietic stem cell/progenitor plasticity: inversions. J Cell Physiol. 2004;199:20–31. doi: 10.1002/jcp.10436. [DOI] [PubMed] [Google Scholar]
- 6.Dooner GJ. Colvin GA. Dooner MS. Johnson KW. Quesenberry PJ. Gene expression fluctuations in murine hematopoietic stem cells with cell cycle progression. J Cell Physiol. 2008;214:786–795. doi: 10.1002/jcp.21273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker PS. Nilsson SK. Li Z. Berrios VM. Dooner MS. Cooper CL. Hsieh CC. Quesenberry PJ. Adhesion receptor expression by hematopoietic cell lines and murine pro genitors: modulation by cytokines and cell cycle status. Exp Hematol. 2007;27:533–541. doi: 10.1016/s0301-472x(98)00037-x. [DOI] [PubMed] [Google Scholar]
- 8.Berrios VM. Dooner GJ. Nowakowski G. Frimberger A. Valinski H. Quesenberry PJ. Becker PS. The molecular basis for the cytokine-induced defect in homing and engraftment of hematopoietic stem cells. Exp Hematol. 2001;29:1326–1335. doi: 10.1016/s0301-472x(01)00734-2. [DOI] [PubMed] [Google Scholar]
- 9.Reddy GP. McAuliffe CI. Pang L. Quesenberry PJ. Bertoncello I. Cytokine receptor repertoire and cytokine responsiveness of Ho dull/Rh dull stem cells with differing potentials for G1/S phase progression. Exp Hematol. 2002;30:792–800. doi: 10.1016/s0301-472x(02)00814-7. [DOI] [PubMed] [Google Scholar]
- 10.Quesenberry PJ. Dooner GJ. DelTatto M. Colvin GA. Johnson K. Dooner MS. Expression of cell cycle related genes with cytokine-induced cell cycle progression of primitive hematopoietic stem cells. Stem Cells Dev. 2010;19:453–460. doi: 10.1089/scd.2009.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colvin GA. Berz D. Liu L. Dooner MS. Dooner G. Pascual S. Chung S. Sui Y. Quesenberry PJ. Heterogeneity of non-cycling and cycling synchronized murine hematopoietic stem/progenitor cells. Cell Physiol. 2010;222:57–65. doi: 10.1002/jcp.21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dooner MS. Aliotta JM. Pimentel J. Dooner GJ. Abedi M. Colvin G. Liu Q. Weier HU. Johnson KW. Quesenberry PJ. Conversion potential of marrow cells into lung cells fluctuates with cytokine-induced cell cycle. Stem Cells Dev. 2008;17:207–219. doi: 10.1089/scd.2007.0195. [DOI] [PubMed] [Google Scholar]
- 13.Quesenberry PJ. Colvin GA. Lambert JF. The chiaroscuro stem cell: a unified stem cell theory. Blood. 2002;100:4266–4271. doi: 10.1182/blood-2002-04-1246. [DOI] [PubMed] [Google Scholar]
- 14.Lambert JF. Liu M. Colvin GA. Dooner M. McAuliffe CI. Becker PS. Forget BG. Weissman SM. Quesenberry PJ. Marrow stem cells shift gene expression and engraftment phenotype with cell cycle transit. J Exp Med. 2003;197:1563–1572. doi: 10.1084/jem.20030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quesenberry P. Colvin G. Lambert JF. Abedi M. Cerny J. Dooner M. Moore B. McAuliffe C. Demers D, et al. Marrow stem cell potential within a continuum. Ann N Y Acad Sci. 2003;996:209–221. doi: 10.1111/j.1749-6632.2003.tb03248.x. [DOI] [PubMed] [Google Scholar]
- 16.Quesenberry PJ. Colvin GA. Abedi M. Lambert JF. Moore B. Demers D. Greer D. McAuliffe C. Dooner M, et al. The marrow stem cell: the continuum. Bone Marrow Transplant. 2003;32(Suppl. 1):S19–S22. doi: 10.1038/sj.bmt.1703938. [DOI] [PubMed] [Google Scholar]
- 17.Quesenberry PJ. Dooner G. Colvin G. Abedi M. Stem cell biology and the plasticity polemic. Exp Hematol. 2005;33:389–394. doi: 10.1016/j.exphem.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Quesenberry P. Abedi M. Dooner M. Colvin G. Sanchez-Guijo FM. Aliotta J. Pimentel J. Dooner G. Greer D, et al. The marrow cell continuum: stochastic determinism. Folia Histochem Cytobiol. 2005;43:187–190. [PubMed] [Google Scholar]
- 19.Quesenberry PJ. The continuum model of marrow stem cell regulation. Curr Opin Hematol. 2006;13:216–221. doi: 10.1097/01.moh.0000231417.08031.ac. [DOI] [PubMed] [Google Scholar]
- 20.Quesenberry PJ. Dooner G. Dooner M. Colvin G. The stem cell continuum: considerations on the heterogeneity and plasticity of marrow stem cells. Stem Cell Rev. 2005;1:29–36. doi: 10.1385/SCR:1:1:029. [DOI] [PubMed] [Google Scholar]
- 21.Quesenberry PJ. Colvin G. Dooner G. Dooner M. Aliotta JM. Johnson K. The stem cell continuum: cell cycle, injury, and phenotype lability. Ann N Y Acad Sci. 2007;106:20–29. doi: 10.1196/annals.1392.016. [DOI] [PubMed] [Google Scholar]
- 22.Quesenberry PJ. Aliotta JM. The paradoxical dynamism of marrow stem cells: considerations of stem cells, niches, and microvesicles. Stem Cell Rev. 2008;4:137–147. doi: 10.1007/s12015-008-9036-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quesenberry PJ. Dooner GJ. Dooner MS. Problems in the promised land: status of adult marrow stem cell biology. Exp Hematol. 2009;37:775–783. doi: 10.1016/j.exphem.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Aliotta JM. Sanchez-Guijo FM. Dooner GJ. Johnson KW. Dooner MS. Greer KA. Greer D. Pimentel J. Kolankiewicz LM, et al. Alteration of marrow gene expression, protein production and engraftment into lung by lung-derived microvesicles. Stem Cells. 2007;9:2245–2256. doi: 10.1634/stemcells.2007-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aliotta JM. Pereira M. Johnson KW. de Paz N. Dooner MS. Puente N. Ayala C. Brilliant K. Berz D, et al. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp Hematol. 2010;38:233–245. doi: 10.1016/j.exphem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269–288. doi: 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- 27.Théry C. Ostrowski M. Segura E. Membrane vesicles as conveyors of immune responses. Nature Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 28.Keller S. Sanderson MP. Stoeck A. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Morel O. Toti F. Hugel B. Freyssinet JM. Cellular microparticles: a disseminated storage pool of bioactive vascular effectors. Curr Opin Hematol. 2004;11:156–164. doi: 10.1097/01.moh.0000131441.10020.87. [DOI] [PubMed] [Google Scholar]
- 30.Heijnen HF. Schiel AE. Fijnheer R. Geuze HJ. Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- 31.Janowska-Wieczorek A. Majka M. Kijowski J. Baj-Krzyworzeka M. Reca R. Turner AR. Ratajczak J. Emareson SG. Kowalska MA. Ratajczak MZ. Platelet-derived microparticles bind to hematopoietic stem/progenitor cells and enhance their engraftment. Blood. 2001;98:3143–3149. doi: 10.1182/blood.v98.10.3143. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert GE. Sims PJ. Wiedmer T. Furie B. Furie BC. Shattil SJ. Platelet-derived microparticles express high affinity receptors for factor VIII. J Biol Chem. 1991;266:17261–17268. [PubMed] [Google Scholar]
- 33.Little SR. Lynn DM. Ge Q. Anderson DG. Puram SV. Chen J. Eisen HN. Langer R. Poly-beta amino ester-containing microparticles enhance the activity of nonviral genetic vaccines. Proc Natl Acad Sci U S A. 2004;101:9534–9539. doi: 10.1073/pnas.0403549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratajczak MZ. Microvesicles: from “dust to crown.”. Blood. 2006;108:2885. [Google Scholar]
- 35.Baj-Krzyworzeka M. Szatanek R. Weglarczyk K. Baran J. Urbanowicz B. Branski P. Ratajczak MZ. Zembala M. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Imunother. 2006;55:808–818. doi: 10.1007/s00262-005-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozmyslowicz T. Majka M. Kijowski J. Murphy SL. Conover DO. Poncz M. Ratajczak J. Gaulton GN. Ratajczak MZ. Platelet and megakaryocyte-derived microparticles transfer CXCR4 receptor to CXCR4-null cells and make them susceptible to infection by×4-HIV. AIDS. 2003;17:33–42. doi: 10.1097/00002030-200301030-00006. [DOI] [PubMed] [Google Scholar]
- 37.Graves LE. Ariztia EV. Navari JR. Matzel HJ. Stack MS. Fishman DA. Proinvasive properties of ovarian cancer ascites-derived membrane vesicles. Cancer Res. 2004;64:7045–7049. doi: 10.1158/0008-5472.CAN-04-1800. [DOI] [PubMed] [Google Scholar]
- 38.Fackler OT. Peterlin BM. Endocytic entry of HIV-1. Curr Biol. 2000;10:1005–1008. doi: 10.1016/s0960-9822(00)00654-0. [DOI] [PubMed] [Google Scholar]
- 39.Fevrier B. Vilette D. Archer F. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004;100:10592–10597. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratajczak J. Miekus K. Kucia M. Zhang J. Reca R. Dvorak P. Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 41.Holmgren L. Szeles A. Rajnavolgyi R. Folkman J. Klein G. Ernberg I. Falk KI. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood. 1999;93:3956–3963. [PubMed] [Google Scholar]
- 42.Hakelien AM. Landsverk HB. Rob JM. Skalhegg BS. Collas P. Reprogramming fibroblasts to express T-cell functions using cell extracts. Nat Biotech. 2002;20:460–466. doi: 10.1038/nbt0502-460. [DOI] [PubMed] [Google Scholar]
- 43.Deregibus MC. Cantaluppi V. Calogero R. Lo Iacono M. Tetta C. Biancone L. Bruno S. Bussolati B. Camussi G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 44.Renzulli JF., 2nd DelTatto M. Dooner G. Aliotta J. Goldstein L. Dooner M. Colvin G. Chatterjee D. Quesenberry PJ. Microvesicle induction of prostate specific gene expression in normal human bone marrow cells. J Urol. 2010;184:2165–2171. doi: 10.1016/j.juro.2010.06.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ. Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]