Abstract
Tumor recurrence after chemotherapy is a major cause of patient morbidity and mortality. Recurrences are thought to be secondary to small subsets of cancer cells that are better able to survive traditional forms of chemotherapy and thus drive tumor regrowth. The ability to isolate and better characterize these therapy-resistant cells is critical for the future development of targeted therapies aimed at achieving more robust and long-lasting responses. Using a novel application for the proliferation marker carboxyfluorescein diacetate, succinimidyl ester (CFSE), we have identified a population of slow-cycling, label-retaining tumor cells in both in vitro sphere cultures and in vivo xenograft models. Strikingly, label-retaining cells exhibit a multifold increase in ability to survive traditional forms of chemotherapy and reenter the cell cycle. Further, we demonstrate the innovative application of CFSE to live sort slow-cycling tumor cells and validate their chemoresistance and tumorigenic potential.
Introduction
The incidence of recurrence after treatment in patients with epithelial tumors is a major obstacle in developing truly curative treatments. Although many stage II colon cancer patients show initial responses to standard chemotherapies, 5-year recurrence rates can be as high as 25% [1]. In breast cancer patients, 15-year recurrence rates are as high as 20% [2]. While factors associated with recurrence (sizes, grade, etc.) can suggest which tumors are likely to recur, the inability to accurately predict recurrence risk can lead to both unnecessary and insufficient treatment. It appears likely that subsets of tumor cells evade initial chemotherapy and survive to repropagate the tumor [3,4]. Traditional chemotherapies like 5-fluorouracil (5-FU) and Oxaliplatin require active cycling cells to trigger cell death [4,5]. Cells that are quiescent or cycling slowly are therefore less likely to be susceptible to these drugs, suggesting an inherent recurrence mechanism in which slow-cycling cells evade therapeutic agents and repropagate tumors. Evidence for such chemoresistance abilities are observed in normal skin tissue where more slowly dividing cells in the bulge survive chemotherapy to regenerate the hair follicle [6]. In the mouse forebrain, high doses of tritiated thymidine (3H-TdR) kill constitutively proliferating cells, but have no effect on quiescent cells [7].
Similar to adult tissues, slow-cycling populations of cells have been identified in cancer tissues. Roesch et al. demonstrated that primary melanoma cell lines contain a PKH26 label–retaining population that has a doubling time of 4 weeks [8]. Using the same PKH dye, Kusumbe and Bapat demonstrated the existence of a slow-cycling population of cells in ovarian cancer [9]. Dembinski and Krauss used Vybrant® DiI to demonstrate a pancreatic adenocarcinoma slow-cycling cell population [10]. Even cell lines grown for years in vitro, like MDA.MB.231, have been found to contain label-retaining cell (LRC) populations [11]. The contribution that slow-cycling populations play in chemotherapy resistance is not well studied and it is unclear whether this characteristic may be a significant factor in tumor recurrence.
In this study, we use an innovative application for the cell tracing dye carboxyfluorescein diacetate, succinimidyl ester (CFSE) to identify and isolate slow-cycling LRCs in both commonly used colon and breast tumor cell lines, as well as a primary human breast tumor. We demonstrate that these slow-cycling cells are tumorigenic and more resistant to traditional chemotherapies than rapidly dividing cells. Importantly, slow-cycling cells survive treatment and demonstrate active DNA synthesis after the removal of chemotherapy drugs, suggesting that they may drive recurrence in the clinical setting.
Materials and Methods
Cell lines and CFSE labeling
Adherent HCT116 (ATCC CCL-247) and MDA-MB-231 (ATCC HTB-26) cultures were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco 11995) or Roswell Park Memorial Institute (RPMI) medium (Gibco 22400), respectively, with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin.
Sphere cultures were grown in Sphere media consisting of DMEM/F12 (Gibco 11320) with 1× B-27 (Gibco 12587), 15 mM HEPES (Gibco 15630), 1% penicillin/streptomycin, 20 ng/mL basic fibroblast growth factor (Invitrogen 13256-029), and 10 ng/mL epithelial growth factor (EGF) (Sigma E9644). Spheres were digested in alkaline solution (Sphere media with NaOH, pH 11.6) and quenched with acidic solution (Sphere media with HCl, pH 1.7) then filtered through a 40 μM mesh.
CFSE labeling was conducted with 10 μM CFSE stock according to manufacturer's protocol for cells in suspension (Molecular Probes C34554).
Mice and tumor xenografts
NOD.CB17-Prkdc scid/J mice were purchased from Jackson Laboratories and housed in the UMass Animal Medicine Facilities.
Adherent HCT116 (1×107) or MDA-MB-231 (7×106) CFSE-labeled cells were suspended in Matrigel (BD Biosciences 354234) and injected subcutaneously into the flank or #3 mammary fat pad, respectively. After 2 weeks, two to four tumor digests were combined to obtain adequate cell numbers and live sorted.
Primary patient breast tumor tissue (designated 2597T) was obtained from the UMass Cancer Center Tissue and Tumor Bank with IRB approval and was exclusively passaged in NOD/SCID mice.
Tumor digests were mechanically and enzymatically (2 mg/mL collagenase) digested, dissociated on a gentleMACs Dissociator (Miltenyi Biotech), and filtered through a 40 μM mesh.
To determine CFSE+ cell growth potential, 1.0×104 HCT116 or 2.5×104 2597T live sorted CFSE+ were resuspended in Matrigel and regrafted either into flanks or mammary fat pads.
Chemotherapy enrichment assays
Cells were cultured in Sphere media for at least 1 week and passaged once before use. Single-cell suspensions of CFSE-labeled HCT116 cells were plated at 2.0×104 cells/mL. One-week-chased single-cell suspensions were replated at 2.0×104 cells/mL in Sphere media containing dimethyl sulfoxide (DMSO) vehicle control, 2 μM Oxaliplatin (Sigma 9512), 250 μM 5-FU (Sigma F6627), or FOX (Oxaliplatin and 5-FU) for 3 days before being analyzed for CFSE content. Alternatively, FOX-treated cultures were replated in fresh Sphere media containing 10 μM BrdU (BD Pharmingen 550891) for 3 days. A range of drug concentrations was initially used to determine the above concentrations that would give 75%–90% kill curves.
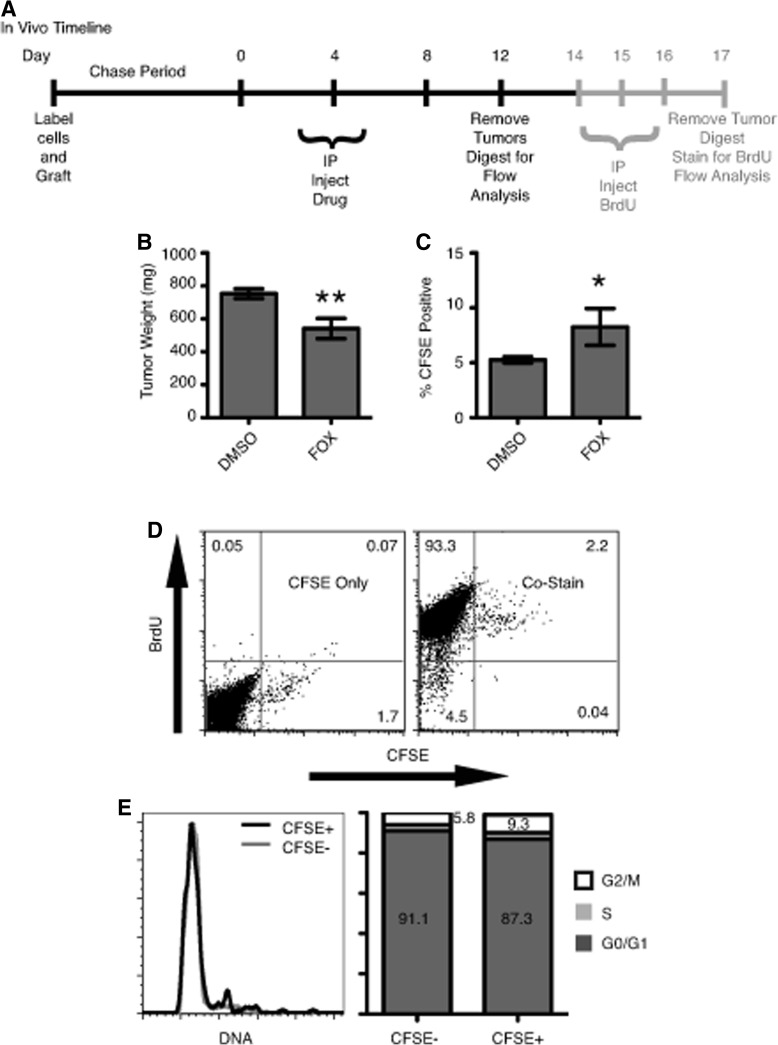
Mice were injected with 5×106 HCT116 CFSE-labeled cells into the #3 and #8 mammary fat pads. Twelve days after engraftment (day 0) mice were injected IP with 40 mg/kg 5-FU and 10 mg/kg Oxaliplatin diluted in phosphate-buffered saline (PBS). Injections were repeated on days 4 and 8. On day 12, tumors were collected and processed as described. BrdU pulsed tumors received 1 mg BrdU by IP injection on days 14, 15, and 16, and were processed on day 17.
Flow cytometry
Single-cell suspensions derived from sphere cultures were washed in PBS+ (PBS with 1% 1M HEPES and 2% FBS). Single-cell in vivo tumor digests were washed in PBS+ and stained with the epithelial-positive selection marker EpCAM (eBiosciences 50-9326) and the negative selection markers TER119, CD31, and CD45 (BD Pharmingen 553673, 553373, and 553089). In vitro and in vivo samples were suspended in PBS and 20% Sphere media with 7AAD (BD Pharmingen 559925). A maximum of 5% of the greatest CFSE intense-Live/Lineage negative cells were collected.
Chemotherapy-treated samples and BrdU-treated samples were labeled with the viability discriminator Live/Dead Blue (Invitrogen L23105) and fixed in Cytofix buffer (BD Biosciences 51-2090KZ). BrdU-treated samples were stained with anti-BrdU antibody (Roche 11170376001, APC secondary Santa Cruz sc-3818). Final samples were suspended in FACS buffer (1× PBS, 1 mM sodium azide, and 0.05 g/mL bovine serum albumin).
Cell cycle analysis was performed on cytofixed cells in 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) (Roche 236276).
All samples were analyzed and sorted by the UMass Medical School Flow Cytometry Facility. The proliferation wizard from the Modfit analysis package was used to generate generation peaks from HCT116 in vivo xenografts at 2 weeks.
Staining
Tumor samples were frozen in OCT compound, sectioned at 6 μm thickness, fixed in 4% paraformaldehyde, and stained with hematoxylin and eosin stains. Fluorescent stains were mounted with Vectashield containing DAPI (Vector H-1200).
Statistics
Statistical analysis and P values were calculated using the t-test functions of the GraphPad Prism software.
Results
Detection of in vitro and in vivo LRCs
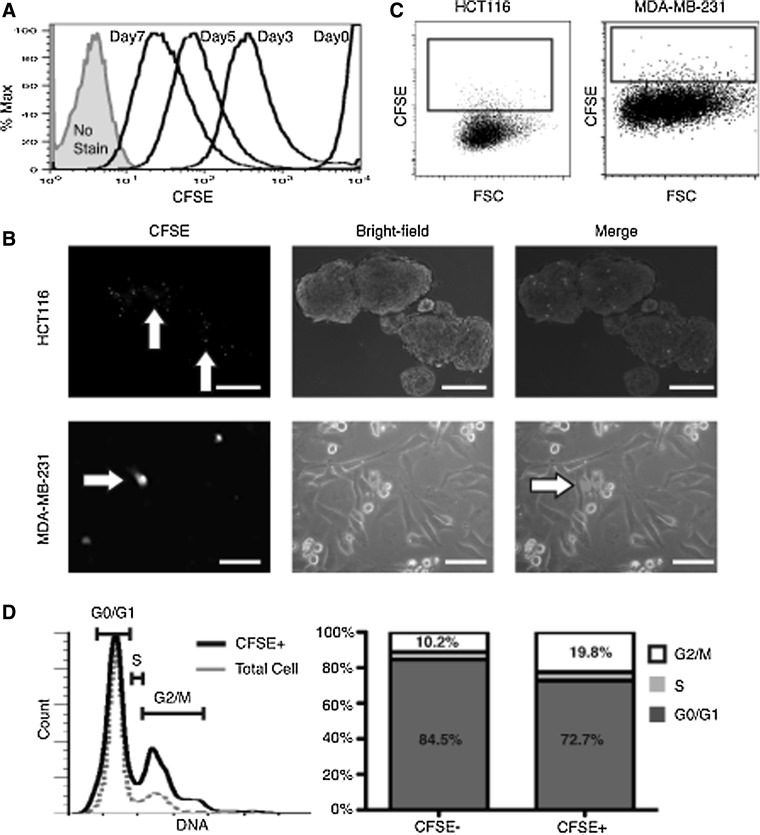
Sphere cultures are considered to better represent properties of tumor cells; we, therefore, used this system to determine whether slow-cycling cells were present in the colon tumor line HCT116. Similar to other groups, we found that in sphere media, MDA-MB-231 (MDA231) cells formed poorly proliferative, loosely associated clumps of cells that made pulse/chase growth assays impractical [12]. Therefore, MDA231 cells were grown as adherent cultures during evaluation for in vitro LRCs. MDA231 cultures or HCT116 spheres were digested into single cells and labeled with the fluorescent cell tracing dye CFSE, then plated back into RPMI or Sphere media, respectively. Over the course of 1 week, dividing cells progressively dilute out label and decrease in fluorescence intensity (Fig. 1A). At 1 week, a CFSE-intense cell population was distinguishable from non/low-labeled bulk cells by microscopy under both conditions (Fig. 1B). When single cells were analyzed by flow cytometry, an average CFSE+ bright LRC population of 4.2%±2.1% was detected in HCT116 spheres and 0.99%±0.14% in MDA231 adherent cultures (Fig. 1C, representative plots). Using the mean CFSE intensity to compare cells with the lowest CFSE intensity (mean CFSE=26.8) to the highest CFSE intensity (black box, mean CFSE=227), HCT116 CFSE bright cells were ∼3 cell divisions behind non/low-labeled bulk cells (Fig. 1C), indicating a distinct population of slowly dividing cells. Cell cycle analysis of HCT116 CFSE+ spheres revealed a 2-fold increase in G2/M phase cells when compared with total cells (Fig. 1D), suggesting an extended G2/M phase or arrest.
FIG. 1.
In vitro identification of LRCs. (A) Flow cytometry plot demonstrating HCT116 sphere dilution of CFSE over 7 days. All HCT116 cells are intensely positive directly after labeling. Over time, label is lost in rapidly proliferating cells, with a tail of more slowly cycling LRCs visible only after 3 days. (B) Florescent and bright-field image overlay of HCT116 sphere cultures and MDA231 adherent cultures 1 week after labeling (scale bar=100 μm). CFSE+ cells (white arrows) are visible under a fluorescent microscope. (C) Representative flow cytometry plot of digested HCT116 sphere cultures and MDA231 adherent cultures after 7 days. Black box depicts the approximate collection gate around the highest CFSE-intense cells. (D) Representative cell cycle profile of total HCT116 sphere cultures. Total cells (gray) and CFSE+ LRCs (black) after 1 week and adjacent quantification bar graphs. CFSE, carboxyfluorescein diacetate, succinimidyl ester; LRC, label-retaining cell.
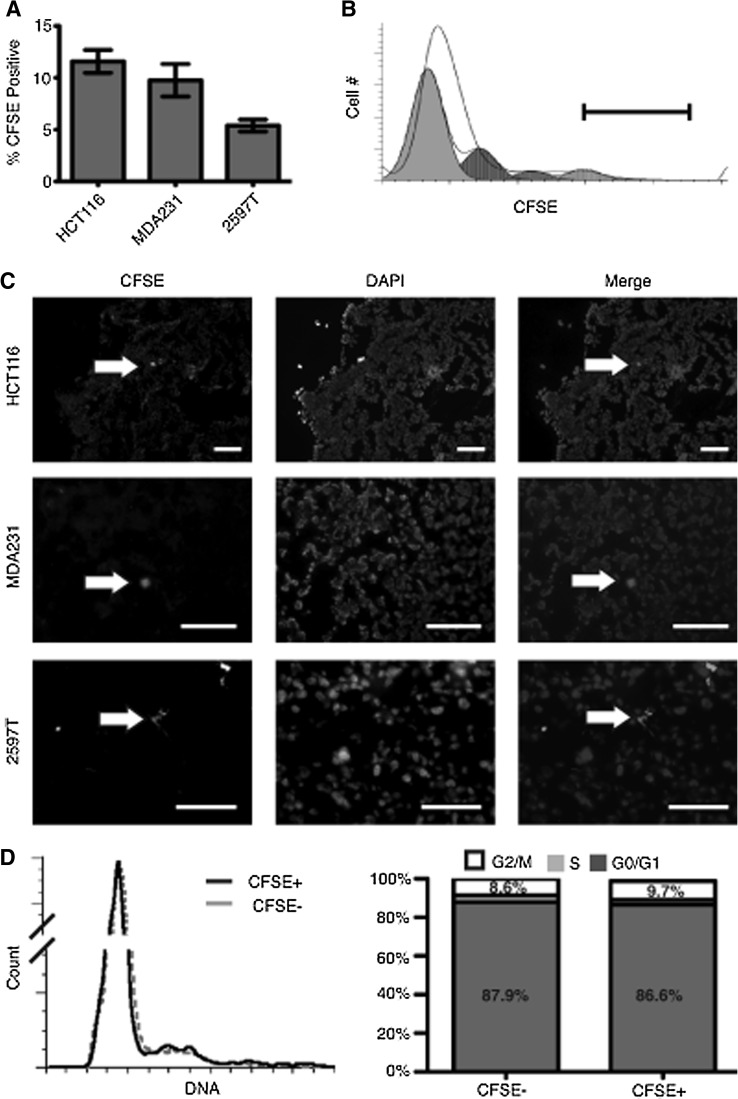
We next used CFSE labeling to determine whether slow-cycling cells were detectable in an in vivo system. HCT116 and MDA231 cells were labeled with CFSE and grafted subcutaneously or into the mammary fat pads, respectively, of NOD/SCID mice. After 2 weeks of growth, flow cytometry indicated a population of CFSE+ cells distinguishable from the bulk tumor cells: HCT116 with 11.6%±2.9% and MDA231 with 9.7%±3.5% CFSE+ cells (Fig. 2). To demonstrate that this was not limited to cell lines, we performed similar in vivo studies with a patient-derived primary breast tumor (2597T) that demonstrated an LRC content of 5.3%±0.8% (Fig. 2A). Using the proliferation wizard from the Modfit software package, we calculated that the top 5% CFSE-intense cells have undergone at least 3–5 fewer cell divisions than the bulk tumors cells in HCT116 xenografts (Fig. 2B). On frozen tissue sections of tumor xenografts, only a small number of scattered CFSE+ cells were detectable (Fig. 2C), demonstrating the sensitivity of flow cytometric analysis over immunofluorescence microscopy for this system. When analyzed for cell cycle stage, in vivo CFSE+ cell cycle profiles were similar to bulk tumor cells with only a slight enrichment for the G2/M phase (Fig. 2D).
FIG. 2.
In vivo identification of LRCs. (A) Composition of CFSE-positive cells after 2 weeks of tumor growth in HCT116 colon tumors, MDA231 breast tumors, and the primary breast sample 2597T tumors. (B) Representative flow cytometry plot of CFSE cell intensities for HCT116 tumor xenografts after 2 weeks of growth. White peak is the unanalyzed data according to CFSE intensity and cell number. Gray peaks represent predicted dilution populations calculated by the proliferation wizard of the Modfit software package. Six separate division peaks were calculated, with two small peaks of greatest CFSE intensity not visible. Black bar approximates the gate used during live sorting to collect the highest 5% intense cells. (C) Fluorescent staining of HCT116, MDA231, and 2597T xenograft tumors after 2 weeks of growth. Only rare cells retain a visible intensity of CFSE (white arrows) while less intense cells are still detectable by flow cytometry (scale bar=100 μm). (D) Cell cycle profiles for HCT116 xenografts. CFSE− cells (gray) and CFSE+ LRCs (black) after 2 weeks and adjacent quantification bar graphs.
LRCs are tumorigenic and not senescent
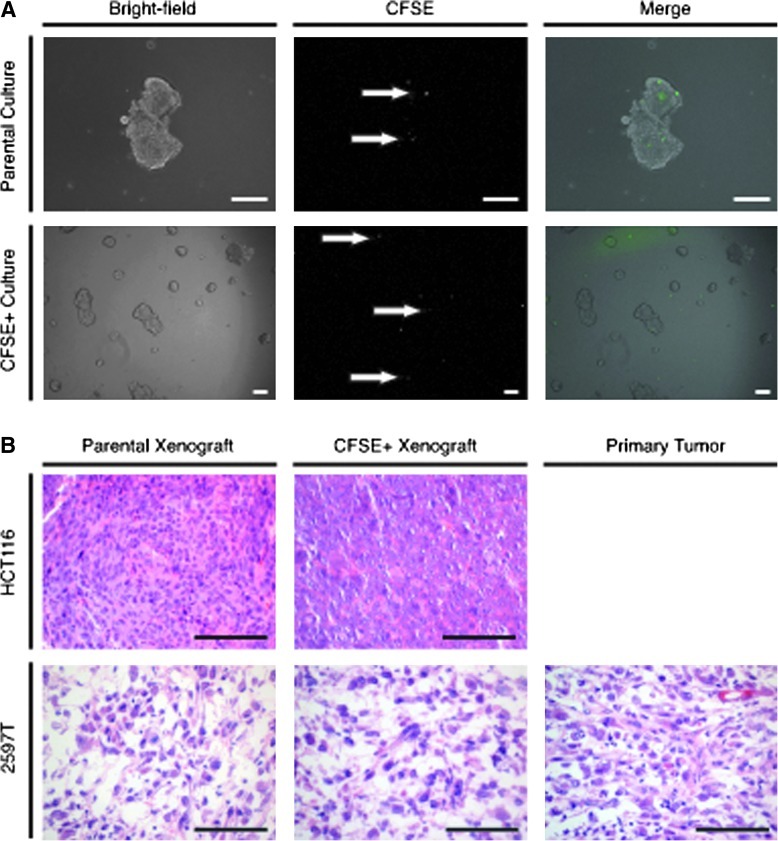
To assess the capacity of LRCs to contribute to tumor growth, we live sorted pulse-chased HCT116 spheres (Fig. 3A, Row 1) and replated CFSE+ cells. After 1 week, ∼75% of CFSE+ cells had divided to form spheres (Row 2). It is interesting to note that 25% of cells did not appear to divide and retained high levels of the CFSE label, suggesting that a population of LRCs may exhibit long-term cell cycle arrests.
FIG. 3.
LRCs are colony forming and tumorigenic. (A) HCT116-labeled spheres were digested after 1-week growth (row 1) and live sorted for CFSE+ cells (white arrows). Fluorescent and bright-field imaging of representative HCT116 CFSE+ cell growth after 1-week (row 2). (B) HCT116 or 2597T CFSE-labeled tumors (column 1) were digested into single cells and live sorted for CFSE+ cells (EpCAM+ and Lin−). Hematoxylin and eosin images of CFSE+ xenograft growth (column 2). CFSE+ tumors are histologically similar to both parental tumor xenografts, and in the case of 2597T, similar to the primary patient tumor as well (column 3) (scale bar=100 μm).
To determine whether in vivo–derived LRCs are tumorigenic, pulse-chased HCT116 and 2597T primary tumors were digested to single cells and live sorted for a maximum of 5% of the top CFSE+ cells. When reinjected into mice, large tumors were formed within 2 months. Histologically, CFSE+ tumors were similar to tumors formed from the established parental cell line (Fig. 3B). In the primary 2597T breast line, CFSE+ tumors were also similar to the histology of the primary patient sample.
LRCs are enriched following chemotherapy
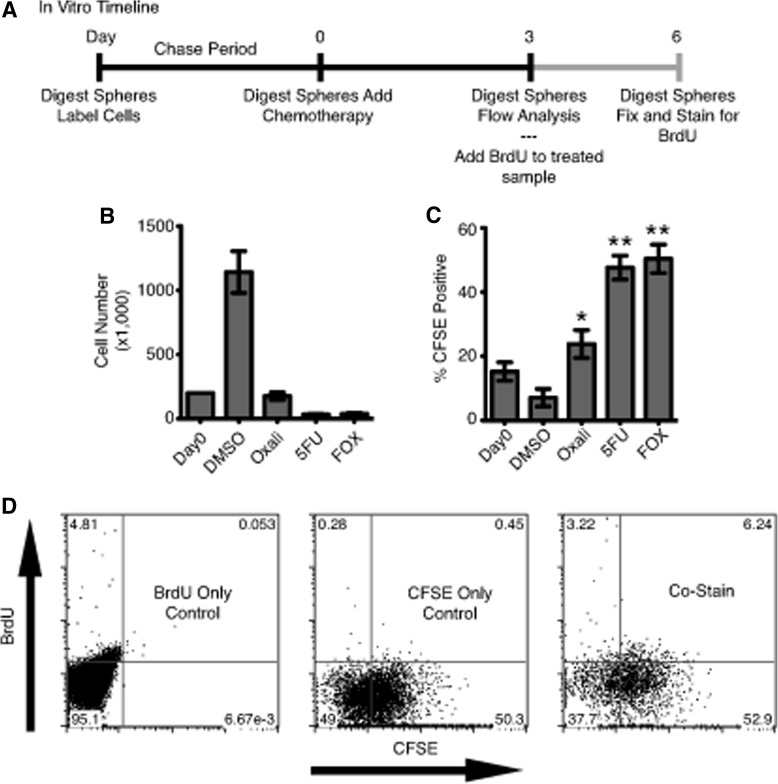
Slow-cycling LRCs in adult tissues have been demonstrated to survive chemotherapy better than other cells within the tissue [6,13,14]. To investigate whether chemotherapy resistance is also a characteristic of our slow-cycling tumor cells, we labeled HCT116 cells, and chased for 1 week in sphere media. Single-cell digests were then either plated in DMSO vehicle control or a chemotherapy reagent at a combined concentration to kill between 75% and 90% of cells and discern more resistant cells (Fig. 4A). After 3 days in culture, DMSO-treated cells had expanded by 5-fold, while Oxaliplatin- (15%), 5-FU– (2.8%), and FOX-treated (3.3%) cultures expectedly contained only a fraction of viable cells compared with DMSO controls (Fig. 4B). When analyzed for CFSE, DMSO control samples demonstrated an expected decrease in CFSE content (15.2% to 7.1%) as these cells continued to proliferate. Strikingly, Oxaliplatin-, 5-FU–, and FOX-treated cells demonstrated 3.4-, 6.7-, and 7.1-fold increase in CFSE+ cell content, respectively (Fig. 4C). To determine whether LRCs are capable of dividing after treatment, HCT116 FOX-treated cultures were replated in sphere media containing BrdU. After 3 days, 6.2% of total cells were BrdU+/CFSE+ cells, suggesting ∼10% of CFSE+ cells (6.2% of 59.1%) had undergone DNA replication and reentered the cell cycle during this early postchemotherapy recovery period (Fig. 4D).
FIG. 4.
In vitro LRCs demonstrate increased therapy resistance. (A) Timeline for in vitro HCT116 chemotherapy treatment experiments. (B) Number of surviving cells after 3 days in the presence of DMSO, 2 μM Oxaliplatin (Oxali), 250 μM 5-fluorouracil (5-FU), or the combination of Oxali and 5-FU “FOX.” (C) Percent CFSE-positive cells after 3 days in the presence of drug. Oxali-, 5-FU–, and FOX-treated samples are significantly enriched over DMSO-treated samples (n=3, *P=0.01, **P<0.0001). (D) Representative BrdU incorporation flow cytometry plots for FOX-treated sphere cultures with single color controls. DMSO, dimethyl sulfoxide.
To elucidate whether slow-cycling LRCs are chemoresistant in vivo, CFSE-labeled HCT116 cells were engrafted into NOD/SCID mice and allowed a 12-day chase before being placed on a FOX treatment regime (Fig. 5A). At the end of treatment, FOX-treated tumors were significantly smaller in size (541 mg) compared with DMSO-treated controls (753 mg) (Fig. 5B). When analyzed for CFSE content, DMSO-treated tumors averaged 5.3% CFSE+ cells while FOX-treated tumors had a statistically significant increase in CFSE+ cells at 8.3% (Fig. 5C). When FOX-treated tumors were pulsed with BrdU for 4 days, LRCs demonstrated the ability to cycle at least once over the time period through BrdU incorporation (Fig. 5D) and a cell fraction within all stages of the cell cycle via DAPI stain (Fig. 5E). Combined, these data demonstrate the ability for LRCs to reenter the cell cycle and actively proliferate shortly after chemotherapy treatment.
FIG. 5.
CFSE LRCs and chemotherapy resistance model. (A) Timeline for in vivo HCT116 xenograft treatment regime. (B) Tumor masses at the end of treatment for DMSO (n=12) and FOX (n=8) (**P<0.01). (C) CFSE composition for DMSO- and FOX-treated tumors (*P=0.045). (D) Representative BrdU incorporation flow cytometry plots for FOX-treated tumors pulsed with BrdU. CFSE+ cells incorporated approximately equal amounts of BrdU as CFSE−. (E) Representative 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI)-derived cell cycle profiles for FOX-treated tumors pulsed with BrdU with adjacent quantification bar graphs.
Discussion
The use of CFSE to demonstrate the existence of slow-cycling LRC populations in cancer is divergent from the dye's general use of short-term lineage tracing. The CFSE offers the distinctive ability to isolate and further characterize live, slow-cycling cells apart from other label-retaining methods, like BrdU, that require cell permeabilization. Further, CFSE dilution and retention relies on a functional cellular phenotype and is independent of protein markers with poorly understood functional contribution to cell dynamics. We demonstrate the existence of a CFSE LRC population in two epithelial tumors lines and one primary patient-derived tumor xenograft. Slow-cycling populations of cells have been found in the normal colon [15] and normal breast tissues [16–18], suggesting that this characteristic maybe carried over from their tissues of origin and serve an important functional role in tissue longevity.
Importantly, the populations of LRCs that we identified both in vitro and in vivo are capable of reactivation. While it is conceivable that a population of cells that enter a senescent state postlabeling would retain label, the identified LRCs are capable of division when live sorted and replated in Sphere media and can regenerate tumors in vivo. Taken together, these data suggest that CFSE+ LRCs are capable of driving tumor formation and regrowth in a clinical setting.
The observed enrichment of LRCs after chemotherapy requires either the preferential expansion of the LRC pool or a reduction of the nonlabel-retaining bulk cells. Back calculations of the in vitro–treated DMSO population suggest an increase in absolute LRC number from ∼30,000 to ∼80,000, due to LRC proliferation that generates partially labeled daughters. However, this expansion alone cannot account for the significant increase in overall LRC composition. In addition, the significant difference in survival between Oxaliplatin-treated samples and 5-FU/FOX–treated samples further supports the concept of LRC enrichment due to increased chemotherapy resistance. Oxaliplatin had insignificant overall kill and only modest LRC enrichment, while 5-FU killed a majority of cells and demonstrated a more profound LRC enrichment (Fig. 4). These data indicate that LRCs are better able to survive standard chemotherapy treatments and their enrichment is related to chemotherapeutic efficiency of killing nonlabel-retaining bulk cells.
In vivo we see a similar pattern where treatment with clinically relevant concentrations of FOX resulted in modest debulking of tumors, corresponding with only modest LRC enrichment (Fig. 5). Unfortunately, increasing drug concentrations or decreasing time between injections proved fatal for a high percentage of mice and hampered our ability to determine whether a more profound debulking of tumor cells would lead to greater fold LRC enrichment.
In in vitro sphere assays, HCT116 LRCs were enriched for cells in the G2/M phases of the cell cycle (Fig. 1C). Other groups have associated a G2/M phase arrest with increased resistance to multiple forms of chemotherapy and a propensity to evade apoptosis [19,20]. It is likely therefore that the G2/M arrest observed in our LRCs is a contributing factor to the observed enrichment, perhaps by extending the time for repair or providing the means to evade apoptotic signals. It is also possible that simply arresting in the G2/M state primes LRCs to better respond to cellular stresses and apoptotic signaling that results from therapeutic treatment.
While the ability of cancer cells to survive chemotherapy is important, clinically significant cells must also be capable of proliferation to stimulate tumor recurrence. Therefore, it is important to establish that CFSE+ LRCs are not avoiding the cytotoxic effects of FOX treatment by entering a permanently nondividing state. Both in vitro and in vivo, a subset of FOX-treated LRCs was able to incorporate BrdU shortly after removal of FOX (Figs. 4D and 5D), demonstrating the capacity of LRCs to actively proliferate and conceivably contribute to tumor recurrence after treatment. This suggests that slow-cycling cells are activated after chemotherapy withdrawal to repropagate the tumor.
Given the therapy-resistant and tumorigenic properties of LRCs, it is conceivable to conclude an association between slow-cycling cells and cancer stem cells (CSCs).
The concept that subsets of tumor cells have increased capacity for tumor propagation in mice is still evolving. For instance, in some models of cancer stem cells (such as melanoma), most if not all cells are tumorigenic [21], while in other system (such as in colon cancer), surface markers have been shown to be nonspecific, with marker-positive and negative cells showing tumorigenicity [22,23]. Evidence from other groups in ovarian tumors, mammary tumors, and melanoma cell lines has demonstrated a connection between the slow-cycling phenotype and identified CSC populations [8,18,24]. However, we believe our findings to be biologically and clinically significant, independent of a cancer stem cell model. Given the possibility that most or all cancer cells could be tumorigenic, we identify a subset of slow-cycling cells that is better able to survive treatment, and thus more capable of leading to tumor recurrence.
In summary, we propose a model in which a subset of slow-cycling tumor cells has enhanced chemoresistance and proliferate after chemotherapy withdrawal to allow for tumor recurrence in a clinical setting. Further, this study details an innovative method to isolate, enrich, and better characterize a population of live therapy-resistant cells in both in vitro cultures and in vivo xenografts. This work should allow for future live enrichment and better characterization of chemoresistant populations to develop more targeted therapies.
Acknowledgments
The authors thank the University of Massachusetts Medical School Cancer Center Tissue and Tumor Bank (www.umassmed.edu/cancercenter/tissuebank/index.aspx) for samples and staining.
W.M. Keck Foundation Award and Department of Defense (BC074714) (Stephen Lyle); R01-CA119061 (JeanMarie Houghton).
Author Disclosure Statement
There are no conflicts of interests in connection with this manuscript.
References
- 1.Andre T. Boni C. Navarro M. Tabernero J. Hickish T. Topham C. Bonetti A. Clingan P. Bridgewater J. Rivera F. de Gramont A. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27:3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 2.Brewster AM. Hortobagyi GN. Broglio KR. Kau SW. Santa-Maria CA. Arun B. Buzdar AU. Booser DJ. Valero V. Bondy M. Esteva FJ. Residual risk of breast cancer recurrence 5 years after adjuvant therapy. J Natl Cancer Inst. 2008;100:1179–1183. doi: 10.1093/jnci/djn233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao S. Wu Q. McLendon RE. Hao Y. Shi Q. Hjelmeland AB. Dewhirst MW. Bigner DD. Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 4.Jung Y. Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 5.Papamichael D. The use of thymidylate synthase inhibitors in the treatment of advanced colorectal cancer: current status. Oncologist. 1999;4:478–487. [PubMed] [Google Scholar]
- 6.Selleri S. Arnaboldi F. Palazzo M. Hussein U. Balsari A. Rumio C. Caveolin-1 is expressed on multipotent cells of hair follicles and might be involved in their resistance to chemotherapy. Br J Dermatol. 2005;153:506–513. doi: 10.1111/j.1365-2133.2005.06746.x. [DOI] [PubMed] [Google Scholar]
- 7.Morshead CM. Reynolds BA. Craig CG. McBurney MW. Staines WA. Morassutti D. Weiss S. van der Kooy D. Neural stem cells in the adult mammalian forebrain: a relatively quiescent subpopulation of subependymal cells. Neuron. 1994;13:1071–1082. doi: 10.1016/0896-6273(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 8.Roesch A. Fukunaga-Kalabis M. Schmidt EC. Zabierowski SE. Brafford PA. Vultur A. Basu D. Gimotty P. Vogt T. Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusumbe AP. Bapat SA. Cancer stem cells and aneuploid populations within developing tumors are the major determinants of tumor dormancy. Cancer Res. 2009;69:9245–9253. doi: 10.1158/0008-5472.CAN-09-2802. [DOI] [PubMed] [Google Scholar]
- 10.Dembinski JL. Krauss S. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin Exp Metastasis. 2009;26:611–623. doi: 10.1007/s10585-009-9260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fillmore CM. Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimshaw MJ. Cooper L. Papazisis K. Coleman JA. Bohnenkamp HR. Chiapero-Stanke L. Taylor-Papadimitriou J. Burchell JM. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10:R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng T. Rodrigues N. Shen H. Yang Y. Dombkowski D. Sykes M. Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 14.Dekaney CM. Gulati AS. Garrison AP. Helmrath MA. Henning SJ. Regeneration of intestinal stem/progenitor cells following doxorubicin treatment of mice. Am J Physiol Gastrointest Liver Physiol. 2009;297:G461–G470. doi: 10.1152/ajpgi.90446.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potten CS. Stem cells in gastrointestinal epithelium: numbers, characteristics and death. Philos Trans R Soc Lond B Biol Sci. 1998;353:821–830. doi: 10.1098/rstb.1998.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welm BE. Tepera SB. Venezia T. Graubert TA. Rosen JM. Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 17.Shackleton M. Vaillant F. Simpson KJ. Stingl J. Smyth GK. Asselin-Labat ML. Wu L. Lindeman GJ. Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 18.Pece S. Tosoni D. Confalonieri S. Mazzarol G. Vecchi M. Ronzoni S. Bernard L. Viale G. Pelicci PG. Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Chikamatsu K. Ishii H. Takahashi G. Okamoto A. Moriyama M. Sakakura K. Masuyama K. Resistance to apoptosis-inducing stimuli in CD44+ head and neck squamous cell carcinoma cells. Head Neck. 2011 doi: 10.1002/hed.21732. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 20.Harper LJ. Costea DE. Gammon L. Fazil B. Biddle A. Mackenzie IC. Normal and malignant epithelial cells with stem-like properties have an extended G2 cell cycle phase that is associated with apoptotic resistance. BMC Cancer. 2010;10:166. doi: 10.1186/1471-2407-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quintana E. Shackleton M. Sabel MS. Fullen DR. Johnson TM. Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shmelkov SV. Butler JM. Hooper AT. Hormigo A. Kushner J. Milde T. St Clair R. Baljevic M. White I, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ricci-Vitiani L. Lombardi DG. Pilozzi E. Biffoni M. Todaro M. Peschle C. De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 24.Gao MQ. Choi YP. Kang S. Youn JH. Cho NH. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene. 2010;29:2672–2680. doi: 10.1038/onc.2010.35. [DOI] [PubMed] [Google Scholar]