Abstract
Recent studies have highlighted the involvement of rare (<1% frequency) copy-number variations and point mutations in the genetic etiology of autism spectrum disorder (ASD); these variants particularly affect genes involved in the neuronal synaptic complex. The SHANK gene family consists of three members (SHANK1, SHANK2, and SHANK3), which encode scaffolding proteins required for the proper formation and function of neuronal synapses. Although SHANK2 and SHANK3 mutations have been implicated in ASD and intellectual disability, the involvement of SHANK1 is unknown. Here, we assess microarray data from 1,158 Canadian and 456 European individuals with ASD to discover microdeletions at the SHANK1 locus on chromosome 19. We identify a hemizygous SHANK1 deletion that segregates in a four-generation family in which male carriers—but not female carriers—have ASD with higher functioning. A de novo SHANK1 deletion was also detected in an unrelated male individual with ASD with higher functioning, and no equivalent SHANK1 mutations were found in >15,000 controls (p = 0.009). The discovery of apparent reduced penetrance of ASD in females bearing inherited autosomal SHANK1 deletions provides a possible contributory model for the male gender bias in autism. The data are also informative for clinical-genetics interpretations of both inherited and sporadic forms of ASD involving SHANK1.
Main Text
Autism is the prototypic form of a group of conditions, also known as “autism spectrum disorders” (ASDs [MIM 209850]), that share common characteristics (impairments in socialization and communication and a pattern of repetitive interests and behaviors) but differ in developmental course, symptom pattern, and cognitive and language abilities. Other ASD subtypes include Asperger disorder (which has less severe language and cognitive deficits) and pervasive developmental disorder not otherwise specified (PDD-NOS; subthreshold symptoms and/or later onset). Subclinical forms of ASD are often characterized as the broader autism phenotype (BAP).1 Twin and family studies provide evidence of the importance of complex genetic factors in the development of both sporadic and inherited forms of idiopathic autism. An enigma in ASD is the 4:1 male to female gender bias, which might rise to 11:1 when Asperger disorder is considered.2
Rare copy-number variations (CNVs) and sequence-level mutations have been identified as etiologic factors in ASD.3–5 De novo CNVs are observed in 5%–10% of ASD cases.6–10 A relative enrichment of CNVs disrupting synaptic complex genes11,12 is observed, and NLGN3 (MIM 300336), NLGN4 (MIM 300427),13 NRXN1 (MIM 600565),14 NRXN3 (MIM 600567),15 SHANK2 (MIM 603290),9,16,17 and SHANK3 (MIM 606230)18–20 have been identified as highly penetrant susceptibility loci for ASD and intellectual disability (ID). The SHANK gene family,21,22 which also includes SHANK1 (MIM 604999), encodes scaffolding proteins that localize to postsynaptic sites of excitatory synapses in the brain.
We describe seven individuals with deletions involving SHANK1. Four male cases have ASD with higher functioning or the BAP and are from a multigenerational family (see family 1 in Figure 1) that carries inherited gene deletions, an unrelated fifth ASD male case has a de novo deletion at the same locus (see family 2 in Figure S1, available online), and two female individuals from family 1 (Figure 1) have the SHANK1 deletion but no ASD or BAP.
Figure 1.

Pedigree of Family 1
Pedigree of a multigenerational family carrying a rare CNV that deletes one copy of SHANK1. Individuals with ASD and BAP are indicated by filled symbols and striped symbols, respectively. The proband is indicated by an arrow. “WT” indicates individuals that have the typical copy number of two at the SHANK1 locus, and “NA” indicates the unavailability of DNA.
The ASD dataset examined in this study was composed of 1,158 unrelated Canadian individuals (898 males and 260 females) and 456 unrelated European individuals (362 males and 94 females). All individuals with ASD were diagnosed by expert clinicians on the basis of the Autism Diagnostic Interview-Revised (ADI-R) and/or the Autism Diagnostic Observation Schedule (ADOS).23 Canadian cases were recruited from five different sites: The Hospital for Sick Children, Toronto, Ontario; McMaster University, Hamilton, Ontario; Memorial University of Newfoundland, St. John's, Newfoundland; University of Alberta, Edmonton, Alberta; and the Montreal Children's Hospital of the McGill University Health Centre, Montreal, Quebec. The European ASD subjects were recruited by the PARIS (Paris Autism Research International Sibpair) study and several other sites at specialized clinical centers dispersed in France, Sweden, Germany, Finland, and the UK. In Sweden, for some cases, the Diagnostic Interview for Social and Communication Disorders (DISCO-10) was applied instead of the ADI-R. The ID dataset consisted of 185 mostly French Canadians (98 males and 87 females) and 155 German nonsyndromic ID cases (93 males and 62 females). Further descriptions of these datasets and the assessment procedures used are available elsewhere.16,24 Approval was obtained from the research ethics boards at The Hospital for Sick Children (Toronto) and McMaster University for the study, and informed written consent was obtained from all participants.
For the assessment of the presence of CNVs on a genome-wide scale, DNA from the Canadian ASD dataset was genotyped at The Centre for Applied Genomics, Toronto with one of three high-resolution microarray platforms: Affymetrix GeneChip SNP 6.0, Illumina Infinium 1M single SNP, or Agilent SurePrint G3 Human CGH (comparative genomic hybridization) 1x1M. The probe coverage of these platforms at the SHANK1 region is shown in Figure S2. CNVs were analyzed with published methods.9,25,26 Briefly, CNV calling was performed with a multialgorithm approach that used at least two different algorithms for the three array platforms: Affymetrix 6.0 (Birdsuite, iPattern, and Genotyping Console), Illumina 1M (PennCNV, QuantiSNP, and iPattern), and Agilent CGH 1x1M (DNA Analytics and CBS from DNAcopy package). Subsequent analyses focused on those CNVs spanning five or more array probes and detected by at least two algorithms. Independent validation of the deletion at the SHANK1 locus in family 1 was performed with SYBR-Green-based real-time quantitative PCR (qPCR), for which two independent primer pairs were placed at the SHANK1 locus and the FOXP2 (MIM 605317) locus as a negative (diploid) control (Figure S3). All primers used in qPCR validation and breakpoint mapping are listed in Table S1.
Using the Illumina Human 1M-Duo BeadChip, we genotyped DNA from the European ASD dataset at the Centre National de Génotypage at the Institut Pasteur. CNVs were analyzed with previously published methods.9 Validation of the array CNV calls was performed with qPCR in a similar way as described above; two independent primer pairs were placed at the SHANK1 locus and at the exon 18 locus of SHANK1 as a negative (diploid) control (Figure S3).
We initially identified a hemizygous microdeletion in chromosomal region 19q13.33 in ASD proband III-5 from the Canadian cohort (see family 1 in Table 1 and Figure 1). The deletion, which was determined to be 63.8 kb, eliminated exons 1–20 of SHANK1 and the neighboring CLEC11A (MIM 604713), which codes for a growth factor for primitive hematopoietic progenitor cells (Figure 2). Subsequent genotyping in family 1 revealed that the deletion was also present in males I-1, IV-1, and IV-3 as well as in females II-4 and III-2 (Figure 1).
Table 1.
Clinical Description of Individuals Carrying SHANK1 Deletion
|
Clinical Details |
|||||
|---|---|---|---|---|---|
| Diagnosis | IQa | Languageb | Adaptive Behaviorc | Brain Imaging | |
| Family 1d | |||||
| III-5 (male) | ASDe: Asperger disorder (ADI-R and ADOS-4) and anxiety | Leiter-R: brief NVIQ = 83 (13th % ile; LA) | OWLS: TL = 68 (2nd % ile; delay) | VABS-I: ABC = 52 (<1st % ile), COM = 43 (<1st % ile), DLS = 63 (1st % ile), and SOC = 65 (1st % ile); he currently takes olanzapine and paroxetine for the anxiety disorder | – |
| I-1 (male) | BAP: shy, reserved, and reluctant to approach people; amassed a large stamp collection; deceased | – | – | – | – |
| IV-1 (male) | ASD: Asperger disorder (ADI-R; SRS: 68T/mild to moderate) | WASI: VIQ = 114 (82nd % ile; HA) and PIQ = 86 (18th % ile; LA) | OWLS: TL = 93, RL = 82 (12th % ile), and EL = 107 (68th % ile) | VABS-II: ABC = 85 (16th % ile), COM = 92 (30th % ile), DLS = 85 (16th % ile), and SOC = 85 (16th % ile) | – |
| PPVT: RV = 97 (42nd % ile) | |||||
| IV-3 (male) | ASD: Asperger disorder (ADI-R and ADOS-3) | WPPSI: FSIQ = 89 (23rd % ile; LA), VIQ = 89 (23rd % ile), and PIQ = 91 (27th % ile) | OWLS: TL = 80 (9th % ile), RL = 78 (7th % ile), and EL = 86 (18th % ile) | VABS-II: ABC = 86 (18th % ile), COM = 91 (27th % ile), DLS = 89 (23rd % ile), SOC = 85 (16th % ile), and MOT = 91 (27th % ile) | – |
| PPVT: RV = 91 (27th % ile) | |||||
| II-4 (female) | non-ASD and non-BAP; anxiety and shyness | – | – | – | – |
| III-2 (female) | non-ASD and non-BAP; social anxiety disorder and generalized anxiety disorder; shy as a child | – | PPVT: RV = 111 (77th % ile) | – | – |
| Family 2f | |||||
| II-1 (male) | ASD with higher functioning (ADI-R; CARS: mild autism) | WISC: FSIQ = 115 (84th % ile; HA), VIQ = 120 (93rd % ile), and PIQ = 100 (50th % ile) (VIQ > PIQ) | – | – | PET: mild hyperfusion temporal left |
Refer to pedigrees in Figure 1 (family 1) and Figure S1 (family 2). The following abbreviations are used: IQ, intelligence quotient; ASD, autism spectrum disorder; % ile, percentile; and PET, positron emission tomography.
IQ was measured with an age-appropriate Weschler scale (WPPSI, Wechsler Preschool and Primary Scale of Intelligence; WISC, Wechsler Intelligence Scale for Children; or WASI, Wechsler Abbreviated Scale of Intelligence). Standard scores and percentiles are presented for full-scale IQ (FSIQ), verbal IQ (VIQ), and/or performance IQ (PIQ). FSIQ is not a valid estimate of IQ when significant discrepancy exists between VIQ and PIQ. Leiter International Performance Scale-Revised (Leiter-R) is a measure of nonverbal IQ (NVIQ) only. Percentile classifications are the following: very superior (VS; >98th % ile), superior (S; 91st–97th % ile), high average (HA; 75th–90th % ile), average (A; 25th–74th % ile), low average (LA; 9th–24th % ile), borderline (B; 2nd–8th % ile), and extremely low (EL; <2nd % ile).
Language was measured with the Oral and Written Language Scales (OWLS). Standard scores and percentiles are presented for total language (TL), receptive language (RL), and/or expressive language (EL). Language was rated as nonverbal, average, or delayed (≤16th % ile). The Peabody Picture Vocabulary Test (PPVT-4th edition) measured receptive vocabulary (RV).
Adaptive Behavior was measured with the Vineland Adaptive Behavior Scales (VABS). Standard score and percentiles are presented for adaptive behavior composite (ABC), communication (COM), daily living skills (DLS), socialization (SOC), and motor (MOT; only for children 7 years old or younger).
Language details for IV-2 (female) in family 1 are the following: (OWLS) RL = 87 (19th % ile) and EL = 108 (70th % ile) and (PPVT) RV = 103 (58th % ile).
The autism-spectrum diagnosis is based on the Autism Diagnostic Interview-Revised (ADI-R) and the Autism Diagnostic Observation Schedule (ADOS; one of four possible modules is administered on the basis of age and language level). In some cases, the Social Responsiveness Scale (SRS) was administered, and reported T-scores represent average skills (≤59T), mild to moderate concerns (60T–75T), or a severe range (76T or higher). Also, the diagnosis for II-1 in family 2 was based on the Childhood Autism Rating Scale (CARS).
II-3 (female) in family 2 was diagnosed with ASD (ADI-R; CARS: mild autism). Her WIPPSI IQ details are the following: PIQ = 99 (50th % ile) and VIQ = nonfunctional language.
Figure 2.
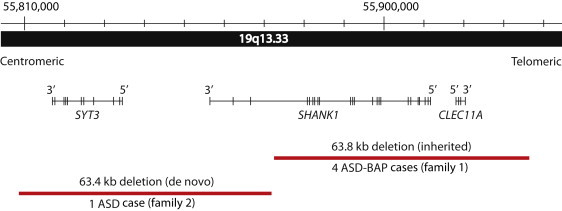
Rare Deletions at SHANK1 Locus in the Two ASD-Affected Families
Chromosomal position of rare deletions of SHANK1 and adjacent genes in ASD. The accurate coordinates for family 1 were mapped by sequencing across the breakpoints and are Chr 19: 55,872,189–55,935,995 (hg18). The de novo deletion of family 2 was detected by microarray and has coordinates Chr 19: 55,808,307–55,871,709 (hg18).
At 16 years of age, proband III-5 was first assessed by a child psychiatrist and was initially given a clinical diagnosis of PDD-NOS. Starting at an early age, there was evidence of impairment in social communication, but there were not enough repetitive stereotyped behaviors for a diagnosis of autism or Asperger disorder. The ADI-R and the ADOS were completed when the proband was 25 years old. The ADI-R indicated that the parents first became concerned when their son was 12–24 months old, a period during which III-5 engaged in repetitive play and speech. He spoke in single words at 24 months of age and spoke in phrases by 36 months of age. He has never lost language or other skills. He has no history of echolalia, pronoun reversal, or neologisms. His eye contact has always been poor, and he has persistently lacked social smiling, facial affect, joint attention, and empathy. His interests during childhood and adolescence included video games, movies, and sports cards. He graduated from high school, and now, at age 32, he lives independently and works in a sheltered workshop. When III-5 was 25 years old, the ADI-R and ADOS diagnosed him with autism according to the cut-offs, but his current best-estimate diagnoses are that of Asperger disorder (in view of the normal language development) and a separate anxiety disorder; anxiety disorders are common comorbid conditions but are not considered part of the ASD clinical spectrum.
An extensive battery of questionnaires and tests were administered to III-5's parents, and both scored in the typical range. His mother (II-4) has exhibited anxiety and shyness for most of her life but would not be considered to have ASD or BAP. His 40-year-old sister (III-2) is married with one son (IV-1) with Asperger disorder, a neurotypical daughter (IV-2), and a son (IV-3) with ASD. III-2 completed university and worked as a school teacher for years. She has a diagnosis of social anxiety and a generalized anxiety disorder for which she has taken antianxiety medication. Assessment by interview and questionnaire indicated that she was typical for all measures and did not show evidence of ASD or the BAP.
III-5's maternal grandfather (I-1) passed away when he was 95 years old. Little is known about his childhood other than that he had difficulty in school. Throughout his adult life, he had been quiet and withdrawn. He did not develop close relationships. He was a truck driver and spent most of his time away from home. There was no history of psychiatric illness. He was an avid stamp collector but did not use this interest to engage in social interactions. His children were interviewed so that a diagnosis of ASD could be determined; based on this interview, our clinical impression is that he most likely had, at the very least, the BAP or, possibly, high-functioning ASD, which could not be determined under the circumstances.
IV-1 was clinically diagnosed with Asperger disorder when he was 8 years old. He was born by cesarean section 10 days late. Early developmental milestones were within normal limits. When he was 3 years old, his parents detected developmental differences by noting that he was not interested in other children and was preoccupied with objects. He had an encyclopedic knowledge of cars. He would approach other children but tended to play beside them and became upset with changes in routine. He exhibited difficulties with eye contact and understanding social cues and rules.
Additional assessments were conducted when he was 10 years old. He met all the cut-offs for an autistic disorder on the ADI-R except for the nonverbal total. The ADOS scores were below the cut-off for a diagnosis of ASD because of his strengths in the communication domain. Descriptive gestures were present but were vague and infrequent, accounting for his communication score of 1 (the cut-off is 2). Impairments in reciprocal social interaction continued to be evident. His score on the performance intelligence quotient (IQ) component (score = 86) of psychometric testing was significantly lower than his score on the verbal IQ subtests (score = 114). IV-1 qualified for a diagnosis of Asperger disorder.
Individual IV-3 was first evaluated when he was 3 years old. When he was 18 months old, his parents became concerned because he was not talking. He developed single words at 24 months. He communicated by leading his parents by the hand and exhibited repetitive behaviors. He did not offer comfort or empathy and did not initiate social interaction, although he would play with his parents. Certain noises such as the washing machine or the toilet flushing would bother him; he became upset if his mother had her hair down or a jacket unzipped.
Assessment when he was 5 years and 8 months old indicated that he was positive on the ADI-R for autism and for ASD on the ADOS. He had made good progress in social interaction and language. His expressive language consisted of short sentences and phrases and some echoed speech and mild articulation difficulties. His IQ and expressive and receptive language scores were in the low-average range, leading to a best-estimate diagnosis of Asperger disorder.
In separate microarray experiments examining 456 ASD-affected individuals from Europe, we identified a 63.4 kb hemizygous CNV in individual F2-II-1 from family 2; this CNV deleted the last three exons of SHANK1 and the entire centromeric synaptotagmin-3 (SYT3 [MIM 600327]) gene, which plays a role in Ca2+-dependent exocytosis of secretory vesicles (Table 1, Figure 2). Haplotype analysis revealed that the deletion resided on the chromosome originating from the mother (who was shown to carry two copies of SHANK1), so it was presumed to be a de novo event (Figure S1). The deletion was not in F2-II-3.
Male individual F2-II-1 (Table 1 and Figure S1) was the first child born to a 20-year-old mother. He has a younger maternal half-sister (F2-II-3) with autism and mild ID. F2-II-1 was born two months before term. Developmental abnormalities were identified during his first year. He did not babble, made no eye contact, and refused to be touched. He started to walk at two years of age, but his motor coordination was poor. He started to talk at 2.5 years of age, which astonished the parents because until then, he had been extremely quiet. He developed a formal, pedantic style of speech with abnormal prosody. He was uninterested in other children. He repeated routines and rituals and accumulated facts on certain subjects such as astronomy. When upset, he flapped his hands or moved his body in a stereotypic fashion. Lately, he has had periods of depression. His IQ is in the normal range and shows that he has good verbal ability. The best estimate diagnosis for F2-II-1 is autism with higher functioning.
No deletion equivalent to those described in families 1 or 2 was observed in 15,122 control individuals. These controls included 2,026 healthy individuals from the Children's Hospital of Philadelphia,27 2,493 controls genotyped at the University of Washington,28 and 10,603 population-based controls8,9,25 whose microarray data were analyzed by our group. This latter dataset included 1,123 controls from northern Germany,29 1,234 Canadian controls from Ottawa,30 1,120 population controls from Ontario,31 1,056 HapMap samples,32 4,783 controls from the Wellcome Trust Case Control Consortium,33 and 1,287 controls recruited by the Study of Addiction: Genetics and Environment consortium.34 Control individuals were predominantly of European ancestry, which was comparable to the ancestry of the ASD subjects. We also examined the Database of Genomic Variants35,36 for previously reported CNVs at the SHANK1 locus. One study37 reported an unvalidated 109.1 kb deletion encompassing the entire SHANK1 gene in a Japanese female HapMap individual (NA18942). We obtained this DNA sample and performed qPCR by using two independent primer pairs at the SHANK1 locus; we were unable to confirm the deletion, indicating that it was a false-positive call (Figure S3).
The frequency of deletions at the SHANK1 locus is significantly higher in ASD cases than in controls (2/1,614 cases versus 0/15,122 controls; Fisher's Exact test two-tailed p = 0.009). No other obvious potentially etiologic CNV was observed in any of the individuals with ASD in families 1 or 2 (Tables S2 and S3). Therefore, at this resolution of analysis, the rare deletions of common segments of SHANK1 were the only common events observed between the two unrelated ASD families.
To test for potential damaging sequence-level mutations in SHANK1, we used Sanger sequencing to examine all 23 exons and splice sites in 509 unrelated ASD (384 male and 125 female) and 340 ID (191 males and 149 females) individuals. The rationale for SHANK1 mutation screening of ASD and ID cases arises from our previous observation of rare truncating nonsense and/or frameshift mutations in SHANK216 and SHANK318,19,38 in both of these disorders. We used Primer3 software v.0.4.0 to design PCR primers. We performed PCRs by using standard conditions, and we purified and sequenced products directly by using the BigDye Terminator sequencing (Applied Biosystems, Foster City, CA, USA). Variant detection was performed with SeqScape software from Applied Biosystems. We validated variants detected in the cases but not previously reported in the Single Nucleotide Polymorphism Database (dbSNP) build 130 by resequencing samples from the proband, both parents, and the siblings when available. All primers used for Sanger sequencing of SHANK1 and PCDHGA11 are listed in Table S4.
We detected 26 rare missense variants in 23 ASD and seven ID cases, which were not found in dbSNP build 130 or in 285 control individuals from the Ontario general population (Table S5 and Figures S4 and S5). However, only two of these missense variants (c.877 G>A [p.Asp293Asn] in families 5 and 6 and c.2207 G>A [p.Arg736Gln] in family 9) are predicted to be damaging on the basis of their alteration of highly conserved residues within the ANK and PDZ domains, respectively. Although they occur in males with ASD, both variants are also found in non-ASD (or BAP) fathers. No significant mutation was found on the nondeleted allele of proband III-5.
We also conducted whole-exome sequencing in individuals III-5 and IV-3 from family 1 to search for potential mutations in other genes (Table S6). We performed target enrichment by utilizing the Agilent SureSelect 50Mb Human All Exon kit (Agilent Technologies, Santa Clara, CA, USA), and we conducted paired-end sequencing on a Life Technologies SOLiD5500XL (Life Technologies, Foster City, CA, USA) platform. Protocols for sequencing and target capture followed specifications from the manufacturers.
A nonsense mutation (Tyr313∗) predicted to introduce a stop codon in the PCDHGA11 prodocadherin gene in chromosomal region 5q31.3 was identified. PCDHGA11 is a member of the protocadherin gamma gene cluster thought to have an important role in establishing connections in the brain.39 The nonsense variant in PCDHGA11 (MIM 606298) was validated by Sanger sequencing. The mutation was found to segregate precisely with the SHANK1 deletion. Because SHANK1 and PCDHGA11 reside on different autosomes, we tested for translocation or transposition and ruled out such linkage (Figure S6). It is possible that the Tyr313∗ mutation in PCDHGA11 works in concert with the SHANK1 deletion to modify (positively or negatively) the extent of the phenotype or that they are just randomly cosegregating (a 1/262,144 chance). We have not detected CNVs or sequence-level mutations in PCDHGA11 in family 2 or in any other ASD subject examined. We have also tentatively ruled out the role of the X chromosome in family 1 given that different X chromosomes were observed in ASD males (by comparing SNP genotypes), and no pathogenic CNV, mutation, or genetic linkage was observed at the X chromosome (Figure S7). The PCDHGA11 and SHANK1 loci at 5q31.3 and 19q13.33, respectively, represented two of five chromosomal regions in which maximal linkage was detected (Figure S7). No other obvious damaging sequence mutations or CNVs were found in genes in the other three putative linkage regions.
We provide here a description of hemizygous deletions of SHANK1 in ASD. Such findings have, perhaps, been anticipated given that mutations leading to haploinsufficiency of SHANK2 and SHANK3 have been previously described in ASD. The striking segregation of ASD in only male SHANK1-deletion carriers in family 1 represents, to our knowledge, the first example of autosomal sex-limited expression in ASD.40 Our finding of an unrelated ASD-affected male carrying an independent de novo deletion of SHANK1 supports our interpretation that the SHANK1 CNV segregating in family 1 is indeed the primary etiologic event leading to ASD. Additional case reports of other multigenerational families will be required for substantiating our findings of gender-influenced autosomal penetrance differences at the SHANK1 locus in ASD.
Our data indicate that SHANK1 deletions are associated with ASD with higher functioning in males. Insofar as all affected males have an IQ in the typical range and have good verbal ability (with a lack of clinically significant language delay), they would also qualify for a diagnosis of Asperger disorder. The one other male (individual I-1) had a diagnosis of the BAP (although this diagnosis was based only on retrospective reports, which preclude the potential for an ASD diagnosis). The female carriers do not show evidence of ASD or the BAP but have suffered from anxiety, which is considered to be an unrelated phenotype to ASD (Table 1). It is surprising that, compared to other studies of inherited CNVs,3 this pedigree shows little variability in clinical features among affected individuals.
We note that the neuronal genes PCDHGA11 and SYT3 could also contribute to aspects of the ASD phenotype in families 1 and 2, respectively. Moreover, subtler mutations,41,42 or combinations of mutations,17 might also be involved. The latter possibility might be particularly relevant given early findings that suggest potential multigenic effects including SHANK2 in ASD risk.17 Paternal silencing or imprinting of SHANK1, as well as potential transmission distortion with respect to cosegregation of SHANK1 and PCDHGA11, also needs to be considered, but at least for the former, there is insufficient evidence for this.43
Consistent with our findings in humans, Shank1-null murines have deficits in several elements of social communication and developmental milestones.44 They also exhibit increased anxiety-related behavior and impaired memory.45 Shank2 and Shank3 offer immature excitatory synapses with unique properties that facilitate synaptogenesis. Shank1 has the important, but perhaps less critical, structural role of “consolidating” novel synaptic contacts via capping Shank2 and Shank3.22,46 Mutations in SHANK1 might be predicted generally to have less of an impact on synapse morphology and stability and, therefore, a less severe phenotype than do similar alterations in SHANK2 or SHANK3. Given that SHANK2 and SHANK3 mutations are observed in ID16,38 and schizophrenia (MIM 181500),47 it is possible that SHANK1 mutations will also be similarly found in other brain disorders.
There are currently three prevailing theories explaining the gender bias in autism. The “extreme brain” theory proposes that autistic behaviors are an exaggeration of typical male personality traits.48 A second theory suggests that given the established genetic basis of autism, it is possible that the sex bias lies in the sex chromosomes. Rare mutations in some X-linked genes such as NLGN3 and NLGN413 and the PTCHD1 locus (MIM 300828)49 have been shown to confer risk to idiopathic ASD in male carriers. A third theory50 posits a sex-specific multiple-threshold model in which females need to carry more genetic liability than males in order to develop ASD. Some protective factor must exist to account for that higher threshold. Our discovery of rare autosomal SHANK1 deletions associated with ASD or the BAP that is limited to males provides further evidence of the existence of this protective factor and has significant implications for elucidating the molecular basis of sex bias in ASD. These results will also have immediate relevance for clinical genetic testing in ASD.
Acknowledgments
This work was supported by grants from the University of Toronto McLaughlin Centre, NeuroDevNet, Genome Canada and the Ontario Genomics Institute, the Canadian Institutes for Health Research (CIHR), the Canadian Institute for Advanced Research, the Canada Foundation for Innovation, the government of Ontario, Autism Speaks, and The Hospital for Sick Children Foundation. S.W.S. holds the GlaxoSmithKline-CIHR Chair in Genome Sciences at the University of Toronto and The Hospital for Sick Children. G.A.R. was supported by Deutsche Forschungsgemeinschaft and the BMBF/NGFNplus (German Mental-Retardation Network). T.B. was supported by Agence Nationale de la Recherche (ANR-08-MNPS-037-01—SynGen), Neuron-ERANET (EUHF-AUTISM), and Fondation Orange. We are indebted to the individuals and their families for participating in this study and to The Centre for Applied Genomics at The Hospital for Sick Children. We thank A. Fiebig, A. Franke, and S. Schreiber at POPGEN (University of Kiel, Kiel, Germany), A. Stewart, R. McPherson, and R. Roberts of the University of Ottawa Heart Institute (University of Ottawa, Ottawa, Canada), the Wellcome Trust Case Control Consortium, and the Study of Addiction: Genetics and Environment consortium for providing control data. A provisional patent has been filed for The Hospital for Sick Children in S.W. Scherer's name.
Contributor Information
Peter Szatmari, Email: szatmar@mcmaster.ca.
Stephen W. Scherer, Email: stephen.scherer@sickkids.ca.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Database of Genomic Variants, http://projects.tcag.ca/variation/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
References
- 1.Losh M., Childress D., Lam K., Piven J. Defining key features of the broad autism phenotype: A comparison across parents of multiple- and single-incidence autism families. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147B:424–433. doi: 10.1002/ajmg.b.30612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillberg C., Cederlund M., Lamberg K., Zeijlon L. Brief report: “The autism epidemic”. The registered prevalence of autism in a Swedish urban area. J. Autism Dev. Disord. 2006;36:429–435. doi: 10.1007/s10803-006-0081-6. [DOI] [PubMed] [Google Scholar]
- 3.Scherer S.W., Dawson G. Risk factors for autism: Translating genomic discoveries into diagnostics. Hum. Genet. 2011;130:123–148. doi: 10.1007/s00439-011-1037-2. [DOI] [PubMed] [Google Scholar]
- 4.Devlin B., Scherer S.W. Genetic Architecture in Autism Spectrum Disorder. Curr. Opin. Genet. Dev. 2012 doi: 10.1016/j.gde.2012.03.002. in press. [DOI] [PubMed] [Google Scholar]
- 5.Cook E.H., Jr., Scherer S.W. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 6.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy D., Ronemus M., Yamrom B., Lee Y.H., Leotta A., Kendall J., Marks S., Lakshmi B., Pai D., Ye K. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toro R., Konyukh M., Delorme R., Leblond C., Chaste P., Fauchereau F., Coleman M., Leboyer M., Gillberg C., Bourgeron T. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010;26:363–372. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Bourgeron T. A synaptic trek to autism. Curr. Opin. Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Jamain S., Quach H., Betancur C., Råstam M., Colineaux C., Gillberg I.C., Soderstrom H., Giros B., Leboyer M., Gillberg C., Bourgeron T., Paris Autism Research International Sibpair Study Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szatmari P., Paterson A.D., Zwaigenbaum L., Roberts W., Brian J., Liu X.Q., Vincent J.B., Skaug J.L., Thompson A.P., Senman L., Autism Genome Project Consortium Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaags A.K., Lionel A.C., Sato D., Goodenberger M., Stein Q.P., Curran S., Ogilvie C., Ahn J.W., Drmic I., Senman L. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am. J. Hum. Genet. 2012;90:133–141. doi: 10.1016/j.ajhg.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berkel S., Marshall C.R., Weiss B., Howe J., Roeth R., Moog U., Endris V., Roberts W., Szatmari P., Pinto D. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat. Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- 17.Leblond C.S., Heinrich J., Delorme R., Proepper C., Betancur C., Huguet G., Konyukh M., Chaste P., Ey E., Rastam M. Genetic and Functional Analyses of SHANK2 Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders. PLoS Genet. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand C.M., Betancur C., Boeckers T.M., Bockmann J., Chaste P., Fauchereau F., Nygren G., Rastam M., Gillberg I.C., Anckarsäter H. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moessner R., Marshall C.R., Sutcliffe J.S., Skaug J., Pinto D., Vincent J., Zwaigenbaum L., Fernandez B., Roberts W., Szatmari P., Scherer S.W. Contribution of SHANK3 mutations to autism spectrum disorder. Am. J. Hum. Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauthier J., Spiegelman D., Piton A., Lafrenière R.G., Laurent S., St-Onge J., Lapointe L., Hamdan F.F., Cossette P., Mottron L. Novel de novo SHANK3 mutation in autistic patients. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 21.Sheng M., Kim E. The Shank family of scaffold proteins. J. Cell Sci. 2000;113:1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- 22.Grabrucker A.M., Schmeisser M.J., Schoen M., Boeckers T.M. Postsynaptic ProSAP/Shank scaffolds in the cross-hair of synaptopathies. Trends Cell Biol. 2011;21:594–603. doi: 10.1016/j.tcb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Risi S., Lord C., Gotham K., Corsello C., Chrysler C., Szatmari P., Cook E.H., Jr., Leventhal B.L., Pickles A. Combining information from multiple sources in the diagnosis of autism spectrum disorders. J. Am. Acad. Child Adolesc. Psychiatry. 2006;45:1094–1103. doi: 10.1097/01.chi.0000227880.42780.0e. [DOI] [PubMed] [Google Scholar]
- 24.Hamdan F.F., Daoud H., Piton A., Gauthier J., Dobrzeniecka S., Krebs M.O., Joober R., Lacaille J.C., Nadeau A., Milunsky J.M. De novo SYNGAP1 mutations in nonsyndromic intellectual disability and autism. Biol. Psychiatry. 2011;69:898–901. doi: 10.1016/j.biopsych.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Lionel A.C., Crosbie J., Barbosa N., Goodale T., Thiruvahindrapuram B., Rickaby J., Gazzellone M., Carson A.R., Howe J.L., Wang Z. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med. 2011;3:95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- 26.Pinto D., Darvishi K., Shi X., Rajan D., Rigler D., Fitzgerald T., Lionel A.C., Thiruvahindrapuram B., Macdonald J.R., Mills R. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat. Biotechnol. 2011;29:512–520. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaikh T.H., Gai X., Perin J.C., Glessner J.T., Xie H., Murphy K., O'Hara R., Casalunovo T., Conlin L.K., D'Arcy M. High-resolution mapping and analysis of copy number variations in the human genome: A data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itsara A., Cooper G.M., Baker C., Girirajan S., Li J., Absher D., Krauss R.M., Myers R.M., Ridker P.M., Chasman D.I. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krawczak M., Nikolaus S., von Eberstein H., Croucher P.J., El Mokhtari N.E., Schreiber S. PopGen: Population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 30.Stewart A.F., Dandona S., Chen L., Assogba O., Belanger M., Ewart G., LaRose R., Doelle H., Williams K., Wells G.A. Kinesin family member 6 variant Trp719Arg does not associate with angiographically defined coronary artery disease in the Ottawa Heart Genomics Study. J. Am. Coll. Cardiol. 2009;53:1471–1472. doi: 10.1016/j.jacc.2008.12.051. [DOI] [PubMed] [Google Scholar]
- 31.Zogopoulos G., Ha K.C., Naqib F., Moore S., Kim H., Montpetit A., Robidoux F., Laflamme P., Cotterchio M., Greenwood C. Germ-line DNA copy number variation frequencies in a large North American population. Hum. Genet. 2007;122:345–353. doi: 10.1007/s00439-007-0404-5. [DOI] [PubMed] [Google Scholar]
- 32.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craddock N., Hurles M.E., Cardin N., Pearson R.D., Plagnol V., Robson S., Vukcevic D., Barnes C., Conrad D.F., Giannoulatou E., Wellcome Trust Case Control Consortium Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bierut L.J., Agrawal A., Bucholz K.K., Doheny K.F., Laurie C., Pugh E., Fisher S., Fox L., Howells W., Bertelsen S., Gene, Environment Association Studies Consortium A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J., Feuk L., Duggan G.E., Khaja R., Scherer S.W. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet. Genome Res. 2006;115:205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 37.Park H., Kim J.I., Ju Y.S., Gokcumen O., Mills R.E., Kim S., Lee S., Suh D., Hong D., Kang H.P. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat. Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamdan F.F., Gauthier J., Araki Y., Lin D.T., Yoshizawa Y., Higashi K., Park A.R., Spiegelman D., Dobrzeniecka S., Piton A., S2D Group Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am. J. Hum. Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips G.R., Tanaka H., Frank M., Elste A., Fidler L., Benson D.L., Colman D.R. Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J. Neurosci. 2003;23:5096–5104. doi: 10.1523/JNEUROSCI.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X., Leotta A., Kustanovich V., Lajonchere C., Geschwind D.H., Law K., Law P., Qiu S., Lord C., Sebat J. A unified genetic theory for sporadic and inherited autism. Proc. Natl. Acad. Sci. USA. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durand C.M., Perroy J., Loll F., Perrais D., Fagni L., Bourgeron T., Montcouquiol M., Sans N. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol. Psychiatry. 2012;17:71–84. doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berkel S., Tang W., Treviño M., Vogt M., Obenhaus H.A., Gass P., Scherer S.W., Sprengel R., Schratt G., Rappold G.A. Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Hum. Mol. Genet. 2012;21:344–357. doi: 10.1093/hmg/ddr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregg C., Zhang J., Weissbourd B., Luo S., Schroth G.P., Haig D., Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wöhr M., Roullet F.I., Hung A.Y., Sheng M., Crawley J.N. Communication impairments in mice lacking Shank1: Reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS ONE. 2011;6:e20631. doi: 10.1371/journal.pone.0020631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung A.Y., Futai K., Sala C., Valtschanoff J.G., Ryu J., Woodworth M.A., Kidd F.L., Sung C.C., Miyakawa T., Bear M.F. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J. Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grabrucker A.M., Knight M.J., Proepper C., Bockmann J., Joubert M., Rowan M., Nienhaus G.U., Garner C.C., Bowie J.U., Kreutz M.R. Concerted action of zinc and ProSAP/Shank in synaptogenesis and synapse maturation. EMBO J. 2011;30:569–581. doi: 10.1038/emboj.2010.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gauthier J., Champagne N., Lafrenière R.G., Xiong L., Spiegelman D., Brustein E., Lapointe M., Peng H., Côté M., Noreau A., S2D Team De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc. Natl. Acad. Sci. USA. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baron-Cohen S., Lombardo M.V., Auyeung B., Ashwin E., Chakrabarti B., Knickmeyer R. Why are autism spectrum conditions more prevalent in males? PLoS Biol. 2011;9:e1001081. doi: 10.1371/journal.pbio.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noor A., Whibley A., Marshall C.R., Gianakopoulos P.J., Piton A., Carson A.R., Orlic-Milacic M., Lionel A.C., Sato D., Pinto D. ). Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci. Transl. Med. 2010;2:49ra68. doi: 10.1126/scitranslmed.3001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szatmari P., Liu X.Q., Goldberg J., Zwaigenbaum L., Paterson A.D., Woodbury-Smith M., Georgiades S., Duku E., Thompson A. Sex differences in repetitive stereotyped behaviors in autism: Implications for genetic liability. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2012;159B:5–12. doi: 10.1002/ajmg.b.31238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


