Abstract
Mitochondria maintain a genome and translation-machinery to synthesize a small subset of subunits of the oxidative phosphorylation system. These organellar gene products must assemble with imported subunits that are encoded in the nucleus to build up functional enzymes. New findings on the early steps in cytochrome oxidase assembly reveal how the mitochondrial translation of its core component Cox1 is directly coupled to the assembly of this respiratory complex.
Introduction
The four respiratory chain complexes in the mitochondrial inner membrane utilize electrons from NADH or FADH2 as energy to generate a proton gradient across the membrane that drives ATP production by the F1Fo-ATP synthase (complex V). Electrons are passed through the redox systems of these complexes and are finally transferred to molecular oxygen by cytochrome oxidase (complex IV). Thus, the energy of the oxyhydrogen reaction is utilized in an indirect way by the mitochondrial respiratory chain to produce ATP.
Three of the mitochondrial respiratory complexes (I, III, IV), and the F1Fo-ATPase, comprise subunits of dual genetic origin. The majority of respiratory chain subunits are encoded by nuclear genes, translated on cytosolic ribosomes, and imported into mitochondria by the translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) import machineries1,2 (FIG. 1). In contrast, the mitochondrial genome encodes only a small number of core complex subunits, most of which are exceedingly hydrophobic. Mitochondrial DNA (mtDNA) encodes seven subunits of complexes III, IV, and V in Saccharomyces cerevisiae, and thirteen subunits of complexes I, III, IV, and V in humans.
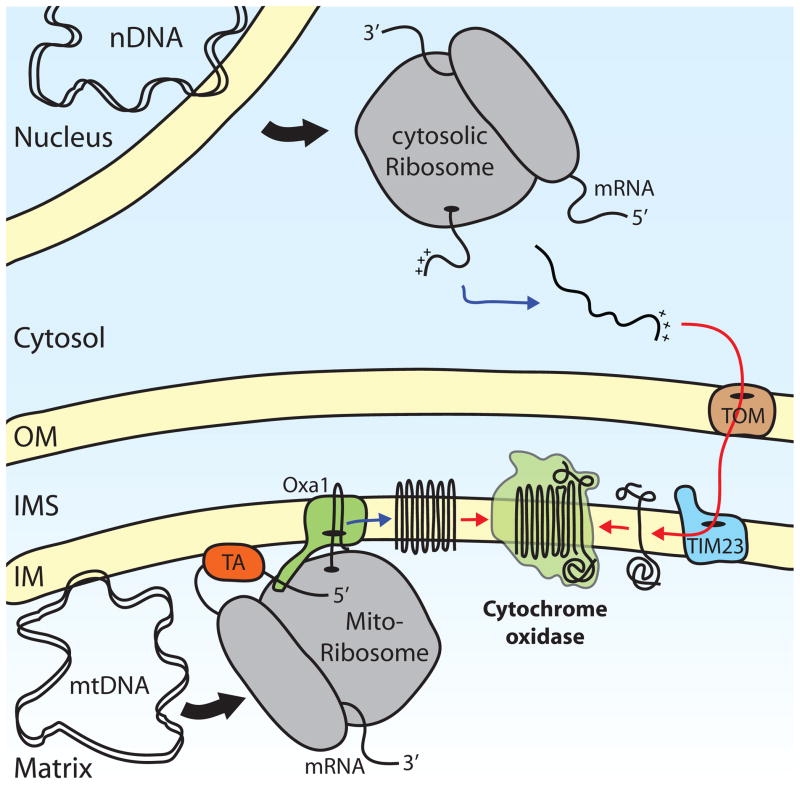
Figure 1. Dual origin of cytochrome oxidase subunits.
Respiratory chain subunits are encoded by both nuclear DNA (nDNA), such as Cox5 and mitochondrial DNA (mtDNA), such as Cox1. Nuclear-encoded proteins are translated on cytosolic ribosomes as precursor proteins that contain information that targets them for mitochondria (+++). Precursors are recognized and transported across the mitochondrial membranes by the translocase of the outer membrane (TOM) complex and the translocase of the inner membrane (TIM23) complex, which cooperate in this process. mtDNA is associated with the inner mitochondrial membrane (IM) and its transcription, as well as translation, occurs in close proximity. Mitochondrial ribosomes translate mRNAs that are bound to the membrane by specific translational activators (TA), such as Pet309 in the case of the mitochondria-encoded Cox1. The translocase Oxa1 inserts nascent polypeptides in a co-translational manner into the membrane and helps these proteins to attain their correct topology. After the insertion of both mitochondria- and nuclear-encoded proteins into the membrane, these subunits can assemble into a functional complex. (OM, outer mitochondrial membrane; IMS, intermembrane space)
To express their small organellar genomes, mitochondria have maintained a transcription–translation apparatus during evolution, the components of which are mainly encoded by the nucleus and represent almost one third of the mitochondrial proteome3. Transcription and translation apparently occur at the matrix side of the mitochondrial inner membrane. Oxa1, a member of a widely conserved family of translocases, is a main constituent of the mitochondrial protein export machinery, which facilitates the insertion of hydrophobic domains into the bilayer and the export of hydrophilic protein domains to the mitochondrial intermembrane space4. (FIG. 1). Its C-terminal domain binds to mitochondrial ribosomes5,6 near their exit-tunnel7,8, consistent with a co-translational mechanism for the insertion of mitochondrially synthesized proteins into the inner membrane. Oxa1 also assists the post-translational insertion of some cytoplasmically synthesized proteins into the inner mitochondrial membrane after their import into the matrix9,10. This is essential for membrane protein domains that do not sort laterally via the TIM23 complex.
It is unclear whether the mitochondrial import of cytoplasmically synthesized respiratory chain subunits is spatially coordinated with the export and insertion into the membrane of those synthesized inside mitochondria. It is clear, however, that respiratory complex subunits do not simply self-assemble in the inner membrane. The assembly of cytochrome oxidase is the best-studied system, where many factors that assist at different steps of the assembly process have been genetically identified in yeast11, and distinct assembly intermediates have been detected in yeast and human organelles (BOX 1).
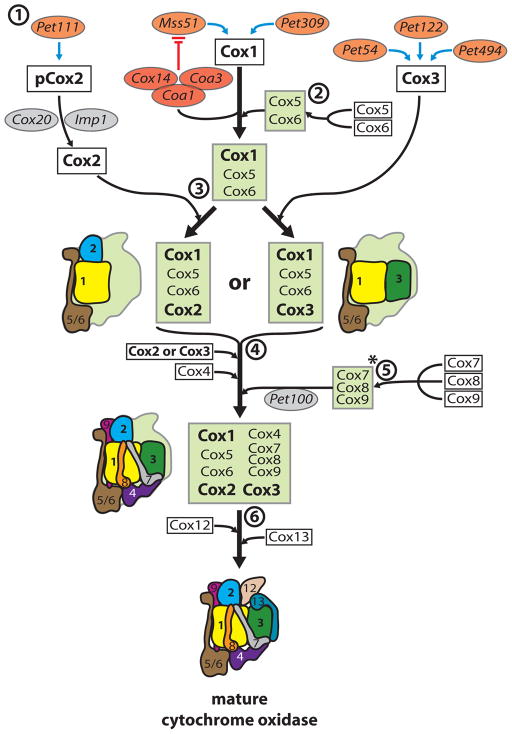
Box 1. The cytochrome oxidase assembly line.
The mRNAs coding for the mitochondria-encoded respiratory chain subunits Cox1, Cox2 and Cox3 have specific translational regulators that have been defined in yeast (orange spheres) and assembly factors (grey and red spheres) that positively or negatively regulate translation (see the figure, part 1). The remaining cytochrome oxidase (Cox) subunits are encoded by the nucleus.
Mss51 inactivation by the Cox14–Coa1–Coa3 complex (red spheres) shuts off Cox1 translation, preventing the assembly of cytochrome oxidase (see main text). In yeast, Cox2 is translated as a precursor (pCox2) with an amino-terminal 15 amino acid extension. Both termini are transported across the membrane4 and Cox2 is kept in an assembly-competent state by Cox2063.
Next, Cox1 assembles with the first nuclear-encoded subunits, Cox6 and Cox547–49(part 2). Whether this association occurs prior to or after the insertion of co-factors into Cox1 is unclear. Mutant analyses revealed that the Cox1–Cox5–Cox6 complex can subsequently form different assembly intermediates in the absence of either Cox2 or Cox3 (part 3). A proposed Cox1–5–6–2 intermediate was identified in patients lacking Cox364, whereas a Cox1–5–6–3 intermediate has been characterized in yeast cox2 mutants48. Copper insertion into Cox2 is a prerequisite for the association with the Cox1–Cox5–Cox6 complex65, whereas incomplete cytochrome oxidase lacking Cox2 was found to associate with complex III of the respiratory chain18.
Little is known about the sequence of events that follow the association of Cox2 and Cox3 with the Cox1–Cox5–Cox6 complex, as no stable assembly intermediates have been found (part 4). However, Cox7, Cox8 and Cox9 form a complex prior to their incorporation, which is mediated by Pet10066 (part 5), although the arrangement of this subcomplex (*) is difficult to reconcile with the structure of mature cytochrome oxidase.
The incorporation of Cox12 and Cox13 concludes the formation of the complex (part 6). However, neither is essential for the enzymatic activity of the complex, which remains stable in their absence67,68. (White boxes, structural Cox subunits; green boxes, reported assembly intermediate complexes; schematic drawings represent assembly intermediates in a structural context, color-code as in Supplementary Figure 1)
The goal of this Progress article is to discuss new insights into the molecular mechanism of cytochrome oxidase biogenesis and to highlight recent findings that link this assembly process to the regulation of the translation of its central subunit Cox1.
The cytochrome oxidase complex
Cytochrome oxidase shuttles electrons from cytochrome c to molecular oxygen to capture energy in the membrane potential by asymmetric proton uptake and proton pumping. The complex is assembled from 13 subunits in humans and 11 subunits in S. cerevisiae. These subunits are termed Cox for cytochrome oxidase, with a non-unified nomenclature (Supplementary Figure 1). The three core subunits, Cox1, Cox2 and Cox3, which are encoded inside mitochondria, are highly conserved among all respiring organisms. The remaining subunits are encoded in the nucleus and imported into mitochondria from the cytosol. The overall dimeric structure of the bovine enzyme reveals that the redox co-factors heme and copper are inserted into the core proteins Cox1 and Cox212 (Supplementary Figure 1). Cox1 carries two heme molecules and a copper ion in the CuB site, while the CuA site is formed by two copper ions in Cox2. The electrons from cytochrome c enter the enzyme from the CuA site in Cox2 and end at the CuB site that together with a heme molecule forms the active center that is deeply embedded in the membrane. Little is known about the enzymatic function of Cox3. As the cytochrome oxidase subunits encoded by the nucleus are less conserved than those encoded by mitochondria, the nomenclature differs depending on the organism (Supplementary Figure 1). We will use the yeast nomenclature in this article.
Given the complexity of this multi-protein, multi-cofactor enzyme, and the fact that its subunits enter the assembly process from two sides of the membrane, it is not surprising that the assembly of cytochrome oxidase requires a large number of assembly factors. While the functions of many assembly factors remain largely enigmatic, some have been assigned to distinct processes such as translational regulation, heme a synthesis and copper or heme insertion (see Supplementary Table 1).
One of the best-characterized and highly conserved assembly factors is Shy1 (SURF1 homolog in yeast). In yeast, loss of Shy1 leads to severe reduction of cytochrome oxidase and a growth defect on non-fermentable carbon sources13. Recent analyses support the idea that Shy1/SURF1 plays a role in the insertion step for heme a3. However, it remains unclear whether it acts as a protein chaperone or has a more direct role in heme insertion, as suggested by heme-binding activity of a bacterial homolog14,15.
Genetic and biochemical studies in yeast, taking Shy1 as a starting point, have led to the identification of several new factors involved in cytochrome oxidase assembly (COA)16–20, including Coa1–Coa4. While Coa2 and Coa4 appear to cooperate with Shy1 for heme insertion16,19,21, Coa1 and Coa3 link Shy1 functionally to early assembly intermediates of Cox1.
In the following sections we will focus on the factors and processes that link the translation of the central subunit Cox1 to these early assembly intermediates.
From mitochondrial mRNA to protein
Translation of mitochondria-encoded mRNAs occurs on mitochondrial ribosomes bound to the inner membrane, where they interact with several membrane proteins5,6,22–25. The intimate link between mitochondrial translation and the inner membrane is underlined by the protein synthesis defects observed in mitochondria deficient in anionic phospholipids26.
In S. cerevisiae, translation of mitochondria-encoded mRNAs specifying subunits of respiratory complexes requires dedicated translational activator proteins, which recognize the 5′-UTRs of their cognate mRNAs27 (BOX 1). In the case of cytochrome oxidase subunits, Pet309 and Mss51 act as translational activators for COX1 mRNA (FIG. 2), Pet111 is required for COX2 translation, and Pet54, Pet122, and Pet494 together promote the translation of COX3 mRNA. While these functions can be separated genetically in S. cerevisiae mutants, in wild-type the activators are actually associated with each other and are believed to co-localize the translation of these COX mRNAs28,29. Indeed, the 5′-UTRs of the COX2 and COX3 mRNAs contain topogenic information presumably recognized by their activator proteins30, which direct the mRNAs to the membrane where subsequent translation occurs.
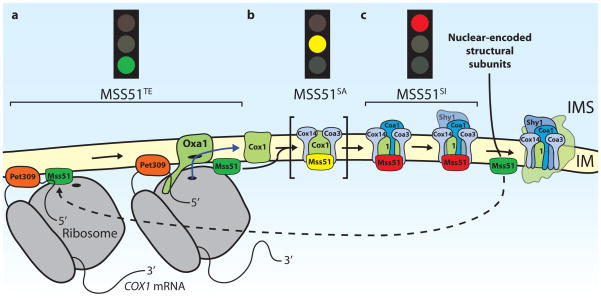
Figure 2. Mechanistic model for the translational regulation of Cox1.
Mss51 regulates the translation of Cox1 by interacting with Cox1 mRNA, and also Cox1 protein that has not yet been assembled into the mature cytochrome oxidase complex. a| Cox1 is synthesized by mitochondrial ribosomes upon activation by the translational activators Pet309 and Mss51. Here Mss51 is in a translation-effective state (MSS51TE). Cox1 is inserted into the inner mitochondrial membrane (IM) co-translationally by the Oxa1 translocase. b| Mss51 is also required for Cox1 translation independently of the 5′UTR. It interacts with newly synthesized Cox1 and the transmembrane proteins Cox14 and Coa3 (cytochrome oxidase assembly), which promote the Cox1–Mss51 interaction. In this sequestered active state (MSS51SA), which has been observed in cells lacking the assembly factor Coa1, Mss51 may be able to initiate further rounds of Cox1 synthesis on mitochondrial ribosomes (not shown) or it might be loosely associated with this complex so that some Mss51 can be released to activate translation. c| The association of Coa1 with this complex in wild type cells converts Mss51 to a sequestered inactive state (MSS51SI), which prevents it from activating translation. Moreover, Coa1 association promotes the binding of Shy1 to Cox1, which might positively regulate the insertion of the heme cofactor into Cox1. (It is currently debated whether Shy1 and Mss51 exist in the same complex with Cox1, since different experimental approaches lead to opposite results.) Subsequent addition of further, nuclear-encoded subunits of cytochrome oxidase, such as Cox6, leads to the release of Mss51 from the assembly intermediates, which allows further rounds of Cox1 synthesis. The mitochondrial Hsp70 Ssc1, which has been suggested to associate with Mss51 in the ‘TE’ and in the ‘SI’ states is not shown.
While little is known about the biochemical mechanism of mitochondrial translational activation, the translational activators of Cox1 have been found to be involved in a regulatory feedback cycle. The COX1 mRNA activator Pet309 is a member of the pentatricopeptide repeat protein family (PPR) that is often associated with RNA metabolism31. Whereas complete loss of Pet309 prevents the translation and stabilization of COX1 mRNA, mutations in the PPR-motifs selectively affect translation31,32. The second COX1-specific activator, Mss51, is a multifunctional pioneer protein whose sequence does not contain any known functional domains or motifs. One target of action, shared with Pet309, is the 5′-UTR of the COX1 mRNA33,34. However, Mss51 also has a second target that maps genetically to the COX1 open reading frame (ORF) allowing it to promote Cox1 synthesis and/or assembly35. Moreover, Mss51 interacts with newly synthesized but unassembled Cox1. Thus, Mss51 appears to interact both with COX1 mRNA and with Cox1 protein that has not been assembled into the cytochrome oxidase complex. These dual activities of Mss51 allow it to couple synthesis of Cox1 to assembly of cytochrome oxidase.
Complex assembly regulates COX1 translation
The synthesis of Cox1 by budding yeast mitochondrial ribosomes is reduced by mutations in most other genes that disrupt the assembly of cytochrome oxidase, e.g. mutants affecting Cox2 maturation. This suggests the existence of a regulatory feedback mechanism coupling Cox1 translation to cytochrome oxidase assembly36. Such a mechanism would ensure that Cox1, the core platform for cytochrome oxidase assembly, is synthesized at an appropriate rate. This is crucial given that certain unassembled Cox1 species are deleterious to cells since they act as pro-oxidants37. The dual activities of Mss51 appear to place it at the nexus of an assembly feedback regulatory system in which the levels of Mss51 available to activate Cox1 synthesis are limited by the amount of Mss51 sequestered by Cox1 protein in cytochrome oxidase assembly intermediate complexes17,18,33,35,36.
Sequestration of Mss51 into Cox1 complexes
It is unclear whether Mss51 molecules that interact with the COX1 mRNA 5′-UTR are transferred directly (in cis) to nascent Cox1 chains emerging from the associated ribosomes. However, when the COX2 mRNA 5′-UTR is placed upstream of the COX1 ORF, Mss51 can still interact with Cox1 in trans33, showing an Mss51 function independent of the 5′-UTR of COX1 mRNA. In any event, newly synthesized Cox1 also rapidly associates with Cox14 and Coa3 (Cox25), two small inner membrane proteins each with domains that are exposed to both the matrix and the intermembrane space20,36,38,39. Newly synthesized Cox1 promotes the dynamic association of Mss51, Cox14, and Coa3 to form a complex with Cox1, which keeps Mss51 in an apparent sequestered active state (MSS51SA; FIG. 2). Based on this, it is tempting to speculate that Mss51 in this complex can associate with ribosomes. In the absence of Cox1 synthesis, interactions between Mss51 and Cox14 or Coa3 are not detected, while the interaction between Cox14 and Coa3 is severely reduced, indicating that these factors only associate upon insertion of Cox1 into the membrane20,33. Furthermore, Cox14 and Coa3 are both required to ensure a stable interaction between Mss51 and newly synthesized Cox120,33. This association is apparently required to prevent Mss51 from activating Cox1 synthesis, since cox14Δ and coa3Δ mutants both exhibit hyperactive Cox1 synthesis that is uncoupled from cytochrome oxidase assembly20,36. However, Cox1 synthesized in the absence of Cox14 or Coa3 is rapidly degraded before it reaches a state, in which it can act as a pro-oxidant, indicating that these assembly factors stabilize Cox1. The degradation process is likely to involve AAA proteases of the inner membrane40,41 or the metalloendopeptidase Oma116,21, as it has been shown for Coa2-deficient cells, where the pro-oxidant Cox1 is already formed. The necessity for the cell to maintain a pool of unassembled Cox1 for complex assembly on the one hand and to degrade a pro-oxidant form on the other, leads to the attractive idea that different activities for Cox1 degradation may exist that are regulated by its assembly state.
Interestingly, the mitochondrial Hsp70 chaperone, Ssc1, is also associated with Mss51 in both a binary complex comprising Mss51 and Ssc1 as well as in larger complexes that also contain Cox1, Coa3 and Cox1439,42. Synthesis of all three mitochondria-encoded cytochrome oxidase subunits was reduced in ssc1 mutants43. However, Ssc1 does not associate with newly synthesized Cox1, but instead with Atp9 and Var143. While it is difficult to imagine Ssc1 playing a role in the folding of the twelve Cox1 transmembrane domains, it could affect the structure of the C-terminal domain of Cox1, which is exposed to the mitochondrial matrix.
A role of Cox1 in its own regulation was directly shown by generating mutations that truncated the Cox1 C-terminus by 11 or 15 residues44. These mutations did not prevent assembly of active cytochrome oxidase. However, they did eliminate assembly feedback regulation of Cox1 synthesis, and weakened the interaction between Mss51 and Cox14 (interaction with Coa3 was not tested). Thus, the C-terminal domain of Cox1 appears to participate in Mss51 sequestration upon its emergence from the ribosome.
Mss51 inactivation by assembly intermediates
The current models for regulation of Cox1 synthesis propose that physical sequestration of Mss51 into assembly intermediate complexes containing newly synthesized Cox1, Cox14, Coa1 and Coa3 prevents it from activating COX1 mRNA translation. Steps in this important pathway have been inferred by the examination of assembly intermediate complexes in solubilized extracts of wild-type and mutant mitochondria from S. cerevisiae using pull-down experiments, blue-native gel electrophoresis and sedimentation14,17,18,20,42,44. These data have generated a hypothetical framework for early events, but not a consensus on the pathway (FIG. 2). Furthermore, it is conceivable that in addition to sequestration, Mss51 translational activity may be altered through structural changes.
In the absence of Coa3 or Cox14, Mss51 does not associate with Cox1 or any other factors except Ssc1, and may be fully in a translation-effective form (MSS51TE). Coa1, an additional player in the regulatory cycle, associates with Cox1, Coa3, Cox14 and Mss5117,18. MSS51SA accumulates in the absence of Coa1. In coa1 mutants, most of the detectable Mss51 is in this complex14,20, but Cox1 synthesis is neither reduced nor subject to normal assembly feedback inhibition17,18. This suggests that in a complex lacking Coa1, Mss51 may not be fully inactivated and that binding of Coa1 is required to generate sequestered, translationally inactive Mss51 (MSS51SI). Interestingly, Coa1 is not absolutely required for cytochrome oxidase assembly, since the coa1Δ phenotype can be suppressed in an unknown mechanism by adding copper to the growth medium17, and minute amounts of mature cytochrome oxidase can be formed in the mutant18.
In the absence of a kinetic analysis of Cox1 biogenesis, the formation of MSS51SA and MSS51SI as distinct pathway intermediates in wild-type S. cerevisiae remains hypothetical. However, the currently available data are consistent with such a stage model: at steady state Mss51 complexes that could correspond to MSS51TE and MSS51SI are detectable14,16,20,42. Thus, under normal conditions, Mss51 likely exists in different states, the equilibrium of which can be shifted according to the requirements for newly synthesized Cox1.
Connecting Mss51 release to complex assembly
The mechanism that promotes Mss51 release from assembly intermediate complexes remains enigmatic, although data suggest that Shy1 might contribute to this process. The presence of Coa1 in the MSS51SI complex promotes the recruitment of Shy1 to Cox114,17,18. Moreover, a lack of Shy1 traps Mss51 together with Coa1 in the sequestered-inactive state17. Shy1, and its homologs in other species, are likely to play a role in the maturation of functional Cox1 through incorporation of heme a314,15,45, either directly15 or by a chaperone-like activity that maintains Cox1 in an assembly competent state14,18. Mutants lacking Shy1 display reduced Cox1 synthesis. This phenotype is partially rescued by the missense mutations mss51-T167R, and mss51-F199I, or by Mss51 overexpression36,46. Moreover, recent studies have shown that overexpression of certain nuclear-encoded Cox-subunits also restores growth of shy1Δ mutants on nonfermentable medium47. Thus, the absence of Shy1 appears to produce a bottleneck in complex assembly that sequesters Mss51, but Shy1 itself may not be necessary for Mss51 release. Interestingly, deletion of the nuclear COX6 gene caused the greatest reduction in Cox1 synthesis among a set of cytochrome oxidase mutants, as judged by pulse-labeling and by the expression of a fused reporter gene44. This suggests that the addition of Cox6, which along with Cox5 is thought to be the first subunit to assemble with newly synthesized yeast and human Cox148,49, may be a key step in the ejection of Mss51 from assembling complexes.
COX1 in humans and disease
The malfunction of cytochrome oxidase has severe implications for cellular energy metabolism and causes increased production of reactive oxygen species with a variety of deleterious consequences in humans50. A number of severe disorders collectively referred to as mitochondrial encephalomyopathies are caused by cytochrome oxidase deficiency due to mutations in mitochondrial genes coding for structural subunits, or impaired assembly of the complex due to nuclear mutations affecting assembly factors. The high demand of certain tissues such as neurons and muscles for oxidative phosphorylation is apparently related to the tissue specific pathology of the patients.
Several assembly factors functioning in yeast have human orthologs (Supplementary Table 1)11. For example, defects in the human copper chaperones Sco1 and Sco2, first identified in yeast, cause a severe cytochrome oxidase deficiency and death shortly after birth51,52. Other cytochrome oxidase deficiencies cause Leigh Syndrome, a neurological disorder with bilaterally symmetrical lesions in subcortical brain regions. The most frequent causes of this disorder are mutations affecting the SURF1 protein53,54 that is associated with early steps of Cox1 biogenesis. However, factors clearly implicated in the translational regulation of Cox1 had not been identified in mammals until very recently.
Since mammalian mitochondrial mRNAs lack significant 5′-UTRs, mechanisms regulating human Cox1 synthesis must differ from those employed in budding yeast. However an activity similar to that of yeast Mss51, whose binding to newly synthesized Cox1 allows continued synthesis and/or assembly35, could be present in mammals and couple cytochrome oxidase assembly to Cox1 synthesis. While no clear Mss51 ortholog is encoded in mammalian genomes, the mammalian protein ZMYND17, of unknown function, is significantly similar.
In any event, post-transcriptional regulation appears to be a feature of mammalian mitochondrial gene expression, based on studies of human patients with Leigh Syndrome and cytochrome oxidase deficiency. The French Canadian variant of this disease is due to mutations affecting the nuclear gene for LRPPRC/LRP130 (leucine-rich pentatricopeptide repeat cassette), a mitochondrial protein distantly related to Pet30955,56. LRPPRC is an RNA-binding protein57 that is associated in a mitochondrial ribonucleoprotein with the stem-loop RNA-binding protein SLIRP and, at least, the mitochondria-encoded COX1 and COX2 mRNAs58. Reduced LRPPRC levels cause lower steady-state levels of most mitochondrially encoded mRNAs and a selective reduction in cytochrome oxidase levels58. More recently, analyses of an unrelated Leigh Syndrome patient revealed a specific reduction in Cox1 synthesis due to mutation in a gene named TACO1, whose protein product appears to be a COX1-specific translational activator in human59. The identification of TACO1 highlights the importance of human genetics in identifying missing assembly factors, and shows that similar mechanisms of translation control may regulate the synthesis of respiratory complex subunits in phylogenetically diverse eukaryotes.
Perspectives
Coupling of organellar gene expression at the level of translation to the assembly of organellar gene products into energy-transducing complexes may be a wide-spread phenomenon. In yeast mitochondria, translation of the bicistronic mRNA encoding the hydrophobic Atp6 and Atp8 subunits of the F0 sector of ATP synthase was recently shown to depend upon the presence of soluble F1 sector subunits α or β, or a partially assembled complex containing them60. While the mechanism of this regulation is unclear, it may prevent the formation of membrane subassemblies containing Atp6 and Atp8, which could dissipate the membrane potential if they are not assembled into a complex with F1. In chloroplasts of Chlamydomonas reinhardtii, translation of organellar encoded mRNAs specifying certain key subunits of photosystem I, photosystem II, and the cytochrome b6f complex is also coupled to the assembly of those complexes61. In parallel to the mechanism proposed for Mss51 function, assembly feedback regulation of cytochrome f synthesis depends upon C-terminal residues of cytochrome f itself, which could interact with a protein that also regulates translation62.
More detailed understanding of assembly feedback control in the mitochondria of model organisms and humans may emerge from further study of the sequences in which assembly intermediates form, their precise compositions and structure, and the interaction dynamics of their constituents. Clearly, a more biochemical understanding will require the development of a true mRNA-dependent in vitro translation system derived from mitochondria (as opposed to translation in isolated intact organelles).
An important health-related goal is to elucidate control mechanisms that operate in human mitochondria. The identification of as-yet-undiscovered nuclear-encoded components may emerge from large-scale gene inactivation studies in cultured cells. The study of their mitochondria-encoded regulatory targets would clearly be advanced by the development of methods to genetically transform and alter animal mitochondrial genomes in a directed fashion.
Supplementary Material
References
- 1.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 2.Vögtle FN, et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 3.Sickmann A, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnefoy N, Fiumera H, Dujardin G, Fox T. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim Biophys Acta. 2009;1793:60–70. doi: 10.1016/j.bbamcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia L, et al. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann J. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia L, Kaur J, Stuart RA. Mapping of the Saccharomyces cerevisiae Oxa1-mitochondrial ribosome interface and identification of MrpL40, a ribosomal protein in close proximity to Oxa1 and critical for oxidative phosphorylation complex assembly. Eukaryotic Cell. 2009;8:1792–1802. doi: 10.1128/EC.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohler R, et al. YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol Cell. 2009;34:344–353. doi: 10.1016/j.molcel.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Hell K, Neupert W, Stuart R. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohnert M, et al. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr Biol. 2010;20:1227–1232. doi: 10.1016/j.cub.2010.05.058. [DOI] [PubMed] [Google Scholar]
- 11.Barrientos A, et al. Cytochrome oxidase in health and disease. Gene. 2002;286:53–63. doi: 10.1016/s0378-1119(01)00803-4. [DOI] [PubMed] [Google Scholar]
- 12.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 13.Mashkevich G, Repetto B, Glerum D, Jin C, Tzagoloff A. SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J Biol Chem. 1997;272:14356–14364. doi: 10.1074/jbc.272.22.14356. [DOI] [PubMed] [Google Scholar]
- 14.Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol Cell Biol. 2010;30:1004–1017. doi: 10.1128/MCB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bundschuh F, Hannappel A, Anderka O, Ludwig B. Surf1, associated with Leigh syndrome in humans, is a heme-binding protein in bacterial oxidase biogenesis. J Biol Chem. 2009;284:25735–25741. doi: 10.1074/jbc.M109.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierrel F, Khalimonchuk O, Cobine PA, Bestwick M, Winge DR. Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis that facilitates the maturation of Cox1. Mol Cell Biol. 2008;28:4927–4939. doi: 10.1128/MCB.00057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierrel F, et al. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mick D, et al. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 2007;26:4347–4358. doi: 10.1038/sj.emboj.7601862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bestwick M, Jeong MY, Khalimonchuk O, Kim H, Winge DR. Analysis of Leigh Syndrome Mutations in the Yeast SURF1 Homolog Reveals a New Member of the Cytochrome Oxidase Assembly Factor Family. Mol Cell Biol. 2010;30:4480–4491. doi: 10.1128/MCB.00228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mick DU, et al. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J Cell Biol. 2010;191:142–154. doi: 10.1083/jcb.201007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bestwick M, Khalimonchuk O, Pierrel F, Winge DR. The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol Cell Biol. 2010;30:172–185. doi: 10.1128/MCB.00869-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frazier A, et al. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J Cell Biol. 2006;172:553–564. doi: 10.1083/jcb.200505060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalimonchuk O, Ostermann K, Rödel G. Evidence for the association of yeast mitochondrial ribosomes with Cox11p, a protein required for the Cu(B) site formation of cytochrome c oxidase. Curr Genet. 2005;47:223–233. doi: 10.1007/s00294-005-0569-1. [DOI] [PubMed] [Google Scholar]
- 24.Ott M, et al. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 2006;25:1603–1610. doi: 10.1038/sj.emboj.7601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauerschmitt H, et al. Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Molecular biology of the cell. 2010;21:1937–1944. doi: 10.1091/mbc.E10-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostrander DB, Zhang M, Mileykovskaya E, Rho M, Dowhan W. Lack of mitochondrial anionic phospholipids causes an inhibition of translation of protein components of the electron transport chain. A yeast genetic model system for the study of anionic phospholipid function in mitochondria. J Biol Chem. 2001;276:25262–25272. doi: 10.1074/jbc.M103689200. [DOI] [PubMed] [Google Scholar]
- 27.Towpik J. Regulation of mitochondrial translation in yeast. Cell Mol Biol Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- 28.Krause K, Lopes de Souza R, Roberts DGW, Dieckmann CL. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Molecular biology of the cell. 2004;15:2674–2683. doi: 10.1091/mbc.E04-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naithani S, Saracco SA, Butler CA, Fox TD. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Molecular biology of the cell. 2003;14:324–333. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchirico ME, Fox TD, Mason TL. Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J. 1998;17:5796–5804. doi: 10.1093/emboj/17.19.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tavares-Carreón F, et al. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J Biol Chem. 2008;283:1472–1479. doi: 10.1074/jbc.M708437200. [DOI] [PubMed] [Google Scholar]
- 32.Manthey G, McEwen J. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Martinez X, Butler C, Shingu-Vazquez M, Fox T. Dual Functions of Mss51 Couple Synthesis of Cox1 to Assembly of Cytochrome c Oxidase in Saccharomyces cerevisiae Mitochondria. Mol Biol Cell. 2009;20:4371–4380. doi: 10.1091/mbc.E09-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zambrano A, et al. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:523–535. doi: 10.1091/mbc.E06-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Martinez X, Broadley S, Fox T. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalimonchuk O, Bird A, Winge DR. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J Biol Chem. 2007;282:17442–17449. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- 38.Glerum D, Koerner T, Tzagoloff A. Cloning and characterization of COX14, whose product is required for assembly of yeast cytochrome oxidase. J Biol Chem. 1995;270:15585–15590. doi: 10.1074/jbc.270.26.15585. [DOI] [PubMed] [Google Scholar]
- 39.Fontanesi F, Clemente P, Barrientos A. Cox25 teams up with Mss51, Ssc1 and Cox14 to regulate mitochondrial cytochrome C oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J Biol Chem. 2010 doi: 10.1074/jbc.M110.188805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arlt H, et al. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 1998;17:4837–4847. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guzelin E, Rep M, Grivell L. Afg3p, a mitochondrial ATP-dependent metalloprotease, is involved in degradation of mitochondrially-encoded Cox1, Cox3, Cob, Su6, Su8 and Su9 subunits of the inner membrane complexes III, IV and V. FEBS Lett. 1996;381:42–46. doi: 10.1016/0014-5793(96)00074-9. [DOI] [PubMed] [Google Scholar]
- 42.Fontanesi F, Soto IC, Horn D, Barrientos A. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol Cell Biol. 2010;30:245–259. doi: 10.1128/MCB.00983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrmann JM, Stuart RA, Craig EA, Neupert W. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol. 1994;127:893–902. doi: 10.1083/jcb.127.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shingu-Vazquez M, et al. The carboxyl-terminal end of Cox1 is required for feedback-assembly regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J Biol Chem. 2010;285:34382–34389. doi: 10.1074/jbc.M110.161976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith D, Gray J, Mitchell L, Antholine W, Hosler J. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J Biol Chem. 2005;280:17652–17656. doi: 10.1074/jbc.C500061200. [DOI] [PubMed] [Google Scholar]
- 46.Barrientos A, Korr D, Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh’s syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fontanesi F, Jin C, Tzagoloff A, Barrientos A. Transcriptional activators HAP/NF-Y rescue a cytochrome c oxidase defect in yeast and human cells. Hum Mol Genet. 2008;17:775–788. doi: 10.1093/hmg/ddm349. [DOI] [PubMed] [Google Scholar]
- 48.Horan S, Bourges I, Taanman J, Meunier B. Analysis of COX2 mutants reveals cytochrome oxidase subassemblies in yeast. Biochem J. 2005;390:703–708. doi: 10.1042/BJ20050598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stiburek L, et al. Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem J. 2005;392:625–632. doi: 10.1042/BJ20050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leary SC, et al. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell Metab. 2007;5:9–20. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Leary SC, Sasarman F, Nishimura T, Shoubridge EA. Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum Mol Genet. 2009;18:2230–2240. doi: 10.1093/hmg/ddp158. [DOI] [PubMed] [Google Scholar]
- 53.Zhu Z, et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- 54.Tiranti V, et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am J Hum Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mootha V, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci U S A. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sterky FH, Ruzzenente B, Gustafsson CM, Samuelsson T, Larsson NG. LRPPRC is a mitochondrial matrix protein that is conserved in metazoans. Biochem Biophys Res Commun. 2010;398:759–764. doi: 10.1016/j.bbrc.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 57.Mili S, Piñol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol Cell Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasarman F, et al. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Molecular biology of the cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weraarpachai W, et al. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat Genet. 2009;41:833–837. doi: 10.1038/ng.390. [DOI] [PubMed] [Google Scholar]
- 60.Rak M, Tzagoloff A. F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc Natl Acad Sci USA. 2009;106:18509–18514. doi: 10.1073/pnas.0910351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choquet Y, Wollman FA. Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 2002;529:39–42. doi: 10.1016/s0014-5793(02)03260-x. [DOI] [PubMed] [Google Scholar]
- 62.Choquet Y, Zito F, Wostrikoff K, Wollman FA. Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell. 2003;15:1443–1454. doi: 10.1105/tpc.011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hell K, Tzagoloff A, Neupert W, Stuart RA. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J Biol Chem. 2000;275:4571–4578. doi: 10.1074/jbc.275.7.4571. [DOI] [PubMed] [Google Scholar]
- 64.Tiranti V, et al. A novel frameshift mutation of the mtDNA COIII gene leads to impaired assembly of cytochrome c oxidase in a patient affected by Leigh-like syndrome. Hum Mol Genet. 2000;9:2733–2742. doi: 10.1093/hmg/9.18.2733. [DOI] [PubMed] [Google Scholar]
- 65.Williams S, Valnot I, Rustin P, Taanman J. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J Biol Chem. 2004;279:7462–7469. doi: 10.1074/jbc.M309232200. [DOI] [PubMed] [Google Scholar]
- 66.Church C, Goehring B, Forsha D, Wazny P, Poyton RO. A role for Pet100p in the assembly of yeast cytochrome c oxidase: interaction with a subassembly that accumulates in a pet100 mutant. J Biol Chem. 2005;280:1854–1863. doi: 10.1074/jbc.M410726200. [DOI] [PubMed] [Google Scholar]
- 67.Massa V, et al. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am J Hum Genet. 2008;82:1281–1289. doi: 10.1016/j.ajhg.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taanman J, Capaldi R. Subunit VIa of yeast cytochrome c oxidase is not necessary for assembly of the enzyme complex but modulates the enzyme activity. Isolation and characterization of the nuclear-coded gene. J Biol Chem. 1993;268:18754–187761. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.