In solving a genetic puzzle posed by George Rizet in 1952 (1), Coustou, Deleu, Saupe, and Begueret report (2) evidence for the first prion (infectious protein) that carries out a normal function. It was studies of scrapie that gave rise to the prion concept, namely, that a normal cellular protein could change to an abnormal form (the prion form) that may be unable to carry out its normal function, but has acquired the ability to convert its normal form into this same abnormal (prion) form. This altered protein, by catalyzing, not its own synthesis, but its own alteration, becomes an infectious agent (a prion) if it can get from cell to cell or from individual to individual (reviewed in refs. 3–6).
Until now, all prions have seemed to cause diseases. Scrapie, Creutzfeldt–Jakob disease, Mad Cow disease, etc. in mammals are invariably lethal neurological diseases involving an altered form of PrP (prion protein), a nonessential cell surface protein (3–6). The non-Mendelian genetic element of Saccharomyces cerevisiae, [URE3] (for ureidosuccinate), is due to a prion change of Ure2p, a regulator of nitrogen metabolism, and results in slow-growing cells (7). [PSI], also a yeast prion, is due to an aggregating form of Sup35p, one of the translation termination proteins whose misfunction in cells carrying [PSI] results in abnormal read-through of translation termination codons (reviewed in ref. 7). This is clearly hazardous to one’s health, but [PSI] strains seem healthy if no suppressor tRNA is around to read the translation termination codon as an amino acid. The [Het-s] prion of the filamentous fungus Podospora anserina causes cell death, but it is a purposeful cell death designed to limit the spread of fungal viruses by preventing cytoplasmic exchange between two colonies.
Podospora Heterokaryon Incompatibility
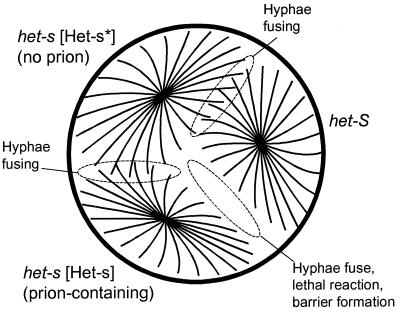
When two fungal colonies grow together the advancing cell processes (hyphae) of the two colonies may fuse (anastomose) to form cells (heterokaryons) with nuclei and cytoplasm from both parent colonies. The two colonies have, in effect, fused to form one interconnecting mat. This hyphal anastomosis or heterokaryon formation is genetically controlled in a different way from the sexual mating that the same two strains may be able to undergo. Whereas sexual mating requires different genotypes at a mating type locus, hyphal anastomosis requires identity at certain other loci, often several loci. In Podospora anserina, these genes are called het (for heterokaryon formation). One such locus, het-s, has alleles het-s and het-S (Table 1). Strains with the same allele can undergo hyphal anastomosis to form heterokaryons. But when het-s and het-S strains grow together, heterokaryon incompatibility is observed. In this case, the peripheral hyphae of the colonies fuse, but the fused hyphae die, the surrounding hyphae are unpigmented, and the line of dead cells between the two colonies acts as a barrier to the colonies growing together (Fig. 1, refs. 1, 8, and 9). The het-S/s locus encodes a protein of 289 amino acid residues, with het-s and het-S alleles differing at 14 residues (10, 11). Remarkably, a single amino acid difference between het-s and het-S is sufficient to produce incompatibility (12).
Table 1.
How many het-s’s are there?
| Name | Meaning |
|---|---|
| het-s | The name of the locus and one of the alleles at that locus |
| het-S | The other allele at the het-s locus |
| [Het-s] | Genotype of cells with the het-s protein in the prion state |
| [Het-s*] | The absence of [Het-s], i.e., the genotype of cells with het-s protein not in the prion state |
| pHET-s | The prion form of the het-s protein |
| pHET-s* | The non-prion form of the het-s protein |
Figure 1.
Diagram of vegetative incompatibility in Podospora. Three strains were inoculated on a plate of growth media. Chromosomal alleles het-s and het-S and the presence ([Het-s]) or absence ([Het-s*]) of the prion form of the het-s protein are shown. After several days, the het-s [Het-s] strain and het-S strain show the incompatibility reaction, marked by death of fused hyphae and lack of pigmentation near the barrier.
Sexual mating is probably a mechanism to shuffle the genetic cards, to generate variability. Therefore, mating with an identical strain makes no sense. Heterokaryon incompatibility is believed to be a mechanism to limit the spread of fungal viruses that spread from one colony to another by hyphal anastomosis. Only strains with identical het genes (which presumably already have the same viruses) can form heterokaryons. In filamentous fungi the spread of viruses is limited in sexual mating because germ cell formation often largely excludes the cytoplasm where the viruses are located (9).
Prions of Yeast Identified by Genetic Properties
[PSI] and [URE3] of yeast were proposed to be prions (13) based on three genetic properties that they share: (i) reversible curability—from strains cured of the genetic element could be isolated rare clones that had again acquired it spontaneously; (ii) overexpression of Sup35p or Ure2p, respectively, increased the frequency with which [PSI] and [URE3] arose; and (iii) Sup35p and Ure2p were necessary for propagation of [PSI] and [URE3], respectively, and yet sup35 and ure2 mutants had the same phenotypes as the presence of [PSI] and [URE3]. These are all properties expected of prions, but not of nucleic acid replicons like viruses or plasmids. This genetic evidence then was supported by finding that Ure2p is protease-resistant in [URE3] strains (14) and Sup35p is aggregated in [PSI] strains (15). [PSI] is eliminated by overexpression of the chaperone Hsp104, a finding that both supports the prion model for [PSI] and introduces a possible route for treatment of the lethal human disease (16). Recently, a [PSI] in vitro system reproducing the main in vivo features of [PSI] also has been reported (17).
[Het-s] Has the Properties of a Prion Form of the het-s Protein
The het-s mystery began in 1952 when Rizet reported that cells with genotype het-s could have either of two phenotypes. One, referred to now as [Het-s], shows the usual heterokaryon incompatibility with het-S colonies. The other, called [Het-s*], shows a neutral phenotype in that it can form heterokaryons with either het-s or het-S cells (Table 1). [Het-s] behaves genetically as a non-Mendelian (cytoplasmic) genetic element and [Het-s*] as its absence. Thus, heterokaryons formed between [Het-s] and [Het-s*] strains eventually become all [Het-s].
Mating het-s [Het-s] cells with het-S strains produced only het-S and het-s [Het-s*] meiotic segregants (a sort of curing; ref. 1). But these [Het-s*] segregants, when grown, gave rise to some [Het-s] segregants (reversible curing) (8). This is one of the genetic criteria for a prion. Coustou et al. (2) now have found that overproduction of the het-s protein increases the frequency with which [Het-s*] strains become [Het-s] (acquire the putative prion). This is a second property expected of a prion. Propagation of the [Het-s] trait requires the het-s protein: het-so strains have the neutral phenotype and cannot propagate [Het-s] (2). Because the [Het-s] phenotype is opposite that of het-so, this result does not point to [Het-s] being a prion. If [Het-s] were a function of a plasmid whose propagation required the het-s protein, the same results would be found. But this result is not inconsistent with the prion model, if the prion form of the het-s protein does something positive to give a phenotype (not just by eliminating the normal form). Finally, the het-s protein is protease resistant in [Het-s] strains when compared with that from [Het-s*] strains (2). Although protease resistance is neither a necessary nor sufficient criterion of a prion change, it certainly argues that the het-s protein is altered in [Het-s] strains. Thus Coustou et al. (2) have made a strong case that [Het-s] represents a prion form of the het-s protein, fulfilling similar genetic and biochemical criteria to those supporting the view of the yeast [URE3] and [PSI] elements as prions.
Early Studies of [Het-s] in View of the Prion Interpretation
In light of the prion explanation of [Het-s], it is of interest to revisit the early studies of het-s/S heterokaryon incompatibility. In meiotic crosses of male het-s [Het-s] and female het-s [Het-s*], the segregants are all [Het-s*] (lacking the prion), but the progeny all carry the prion if the sexes are reversed, i.e., female het-s [Het-s] X male het-s [Het-s*] (1). This shows that the prion can pass through meiosis, but that it is restricted to the cytoplasm, almost none of which is included in the tiny male gametes (microconidia).
Surprisingly, crossing male or female het-s [Het-s] (carrying the prion) with het-S results in all genetically het-s segregants being [Het-s*] (lacking the prion). This means that the het-S protein cures the prion. Could the incorporation of the slightly different het-S protein into a het-s [Het-s] “crystal” poison crystal growth?
Why, in the incompatibility reaction, does the combination of het-s protein in the prion form and het-S protein lead to death of the fused hyphae? Why doesn’t the het-S protein just poison crystal growth here? We can expect many interesting answers to these questions that may tell us important things about the way cells handle prions and incipient prions.
Comparison of [Het-s] and Other Putative Prions (Table 2)
Table 2.
Comparison of putative prions
| Prion | Species | Effect of prion form | Function of normal protein | Structural features |
|---|---|---|---|---|
| Scrapie, CJD... | Mammals | Ataxia, dementia, death | Unknown | Normal form: α helix |
| Scrapie form: β sheet | ||||
| [URE3] | S. cerevisiae | Loss of nitrogen regulation | Nitrogen regulation | Asparagine-rich prion domain |
| [PSI] | S. cerevisiae | Translation read-through | Translation termination | Asparagine-glutamine rich prion domain, octapeptide repeats |
| [Het-s] | Podospora anserina | Heterokaryon incompatibility | Unknown | ?? |
CHD, Creutzfeldt–Jakob disease.
[Het-s] is like scrapie, and unlike [URE3] and [PSI], in that the prion form produces a phenotype by doing mischief, not by simply causing the absence of the active normal form of the het-s protein. The normal form of the protein is dispensable for growth, mating, and heterokaryon formation (10, 11).
Unlike all the other putative prions, [Het-s], the prion form of the het-s protein, is carrying out a normal fungal cell function. Heterokaryon incompatibility systems are widespread among filamentous fungi and usually are controlled by genetic loci showing none of the characteristics suggestive of prions. Is there an advantage to Podospora in using a prion to signal heterokaryon incompatibility? Because this is a purposeful cell death, and many viruses produce apoptosis in their host cells, could this heterokaryon incompatibility reaction be a form of fungal apoptosis?
The het-s protein has no evident similarity to other putative prion proteins. The prion domains of Ure2p and Sup35p are rich in asparagine and glutamine residues, but this is not true of either PrP or the het-s protein. Sup35p and PrP have similar octapeptide repeats, but these appear to be outside the prion domain of PrP and are not found in Ure2p or the het-s protein. Whether structural similarities will be found among the normal or prion forms of these proteins remains to be determined.
Conclusions
Have any of the putative prions been proven to be prions? There continues to be disagreement (e.g., refs. 3, 18, and 19), particularly in the case of scrapie, in part because of the practical difficulties of the animal systems. However, even for the yeast systems, where little rancor exists, the evidence is not conclusive. The exciting new results on the Podospora system widen the scope of application of the prion idea and again demonstrate the value of studying a wide variety of systems. The powerful genetic evidence for the yeast and fungal prions has, at least psychologically, complemented the biochemical evidence in the mammalian systems (which are not without some of their own genetic evidence; refs. 20 and 21), to bring wide acceptance to the prion concept.
Because of the hyphal anastomosis phenomenon, fungi should be quite susceptible to prions. When two fungal colonies grow together, if one is infected, the other will become so. In fact, the vegetative incompatibility systems may, in part, have evolved to block the spread of prions, as well as of fungal viruses and deleterious mitochondrial plasmids.
There are probably many prions in nature. The four cases described thus far were all described as phenomenon 25 to 250 years ago. With knowledge rapidly accumulating about prions, we can expect many new prions to be found in a more directed way. We also can thank the brilliant geneticists, George Rizet ([Het-s]), Brian Cox ([PSI]), and Francois Lacroute ([URE3]) for their pioneering work that has made possible the recent outbreak of prions.
References
- 1.Rizet G. Rev Cytol Biol Veg. 1952;13:51–92. [Google Scholar]
- 2.Coustou V, Deleu C, Saupe S, Begueret J. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prusiner S B. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Raven; 1996. pp. 2901–2950. [Google Scholar]
- 4.Weissmann C, Fischer M, Raeber A, Bueler H, Sailer A, Shmerling D, Rulicke T, Brandner S, Aguzzi A. Cold Spring Harbor Symp Quant Biol. 1996;61:511–522. [PubMed] [Google Scholar]
- 5.Caughey B, Chesebro B. Trends Cell Biol. 1997;7:56–62. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 6.Wickner R B. Prion Diseases of Mammals and Yeast: Molecular Mechanisms and Genetic Features. Austin, TX: Landes; 1997. [Google Scholar]
- 7.Wickner R B. Annu Rev Genet. 1996;30:109–135. doi: 10.1146/annurev.genet.30.1.109. [DOI] [PubMed] [Google Scholar]
- 8.Beisson-Schecroun J. Ann Genet. 1962;4:3–50. [PubMed] [Google Scholar]
- 9.Begueret J, Turcq B, Clave C. Trends Genet. 1994;10:441–446. doi: 10.1016/0168-9525(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 10.Turcq B, Denayrolles M, Begueret J. Curr Genet. 1990;17:297–303. [Google Scholar]
- 11.Turcq B, Deleu C, Denayrolles M, Begueret J. Mol Gen Genet. 1991;288:265–269. doi: 10.1007/BF00282475. [DOI] [PubMed] [Google Scholar]
- 12.Deleu C, Clave C, Begueret J. Genetics. 1993;135:45–52. doi: 10.1093/genetics/135.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickner R B. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 14.Masison D C, Wickner R B. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 15.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 16.Chernoff Y O, Lindquist S L, Ono B-I, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 17.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 18.Chesebro B. Nat Med. 1997;3:491–492. doi: 10.1038/nm0597-491. [DOI] [PubMed] [Google Scholar]
- 19.Lasmezas C I, Deslys J-P, Robain O, Jaegly A, Beringue V, Peyrin J-M, Fournier J-G, Hauw J J, Rossier J, Dormont D. Science. 1997;275:402–405. doi: 10.1126/science.275.5298.402. [DOI] [PubMed] [Google Scholar]
- 20.Prusiner S B, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 21.Bueler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]