Abstract
Bacteria of the genus Bradyrhizobium are able to establish a symbiotic relationship with peanut (Arachis hypogaea) root cells and to fix atmospheric nitrogen by converting it to nitrogenous compounds. Quorum sensing (QS) is a cell-cell communication mechanism employed by a variety of bacterial species to coordinate behavior at a community level through regulation of gene expression. The QS process depends on bacterial production of various signaling molecules, among which the N-acylhomoserine lactones (AHLs) are most commonly used by Gram-negative bacteria. Some previous reports have shown the production of QS signaling molecules by various rhizobia, but little is known regarding mechanisms of communication among peanut-nodulating strains. The aims of this study were to identify and characterize QS signals produced by peanut-nodulating bradyrhizobial strains and to evaluate their effects on processes related to cell interaction. Detection of AHLs in 53 rhizobial strains was performed using the biosensor strains Agrobacterium tumefaciens NTL4 (pZLR4) and Chromobacterium violaceum CV026 for AHLs with long and short acyl chains, respectively. None of the strains screened were found to produce AHLs with short acyl chains, but 14 strains produced AHLs with long acyl chains. These 14 AHL-producing strains were further studied by quantification of β-galactosidase activity levels (AHL-like inducer activity) in NTL4 (pZLR4). Strains displaying moderate to high levels of AHL-like inducer activity were subjected to chemical identification of signaling molecules by high-performance liquid chromatography coupled to mass spectrometry (LC-MS/MS). For each AHL-producing strain, we found at least four different AHLs, corresponding to N-hexanoyl-dl-homoserine lactone (C6), N-(3-oxodecanoyl)-l-homoserine lactone (3OC10), N-(3-oxododecanoyl)-l-homoserine lactone (3OC12), and N-(3-oxotetradecanoyl)-l-homoserine lactone (3OC14). Biological roles of 3OC10, 3OC12, and 3OC14 AHLs were evaluated in both AHL-producing and -non-producing peanut-nodulating strains. Bacterial processes related to survival and nodulation, including motility, biofilm formation, and cell aggregation, were affected or modified by the exogenous addition of increasing concentrations of synthetic AHLs. Our results clearly demonstrate the existence of cell communication mechanisms among bradyrhizobial strains symbiotic of peanut. AHLs with long acyl chains appear to be signaling molecules regulating important QS physiological processes in these bacteria.
Keywords: Bradyrhizobium, AHLs characterization, biofilm, motility, cell aggregation
1. Introduction
Bacteria of the genus Bradyrhizobium are a diverse group of soil microorganisms that have the ability to establish an association with legume (e.g., soybean, peanut) and non-legume plants (e.g., Parasponia) [1]. Peanut (Arachis hypogaea L.) is an economically important legume crop cultivated in tropical, subtropical, and temperate areas worldwide. In Argentina, production of peanut is localized in the central region of Córdoba province. Since strains capable of interacting with this legume are genetically highly diverse, the species identity has not been defined for these rhizobia, and the main peanut-nodulating strains are therefore grouped as Bradyrhizobium sp. [2,3].
The nitrogen-fixing symbiosis is the result of a complex interaction whereby a plant and a type of bacteria (rhizobia) both obtain nutritional benefit: the bacteria supply the plant with reduced nitrogen from atmospheric sources that are not directly available to the plant, while the bacteria (which would starve in the external soil environment) obtain carbon compounds from the plant within the protected root nodule [4,5]. The shift from free-living soil bacteria to endosymbiont bacteria is a dramatic change that involves physiological, metabolic, and ecological alterations. To undergo this change, rhizobia presumably need to use a chemical communication mechanism to coordinate their activities.
Quorum sensing (QS) is a complex environmental sensing system employed by bacteria to communicate among themselves and thereby regulate their population activities in response to various stimuli. The QS mechanism depends on the synthesis and release of chemical signals into the environment and on the detection of these signals as a function of cell population density. Such group behavior results in altered gene expression that drives the activities of the bacteria in a coordinated manner [6,7].
Bacteria synthesize chemical signals that include a wide variety of small molecules [8]. Of these, the N-acylhomoserine lactones (AHLs) are the most commonly used by Gram-negative bacteria for bacterial communication. The AHL molecule consists of a homoserine lactone ring with an amide-linked acylated side-chain having either a keto or hydroxy substituent at the C3 position [9,10]. The biosynthesis and effects of AHLs depend primarily on the activity of the LuxI and LuxR protein families, respectively. After AHLs are produced by LuxI enzymes (AHL synthases), they diffuse across bacterial membranes and accumulate externally until reaching high local concentrations. At a given threshold intracellular concentration, the AHL binds to a LuxR response regulator forming a complex that regulates gene expression [9,11]. AHL-based QS has been shown to be crucial for many plant-associated bacteria, including rhizobia [12–14]. Quorum communication via AHLs in rhizobia affects many metabolic and physiological process, including motility, exopolysaccharide synthesis, biofilm formation, plasmid transfer, root nodulation efficiency, and nitrogen fixing efficiency [15–17].
Most published studies on QS in Bradyrhizobium sp. are controversial and restricted to strains symbiotic with soybean. Studies on soybean-nodulating strains have revealed the use of AHL-like signals [18–21], but not in a widespread manner. Loh et al. [22] described a mechanism in Bradyrhizobium japonicum that depends on cell density and is mediated by a novel signaling molecule named bradyoxetin. Recent studies have demonstrated the production of two new signaling molecules by bacteria of the Bradyrhizobium genus: cinnamoyl-homoserine lactone (an aryl-HSL) in photosynthetic stem-nodulating bradyrhizobia [23] and isovaleryl-homoserine lactone (a branched-chain fatty HSL) in the soybean symbiont Bradyrhizobium japonicum USDA 110 [24].
A few recent studies have explored QS in Bradyrhizobium sp., but none have focused on peanut-nodulating strains. The aims of the present study were to identify and characterize QS signals produced by peanut-nodulating bradyrhizobial strains and to evaluate their effects on bacterial motility and on processes involving cell-cell interaction, e.g., aggregation and biofilm formation.
2. Experimental
2.1. Bacterial Strains and Culture Conditions
The rhizobial strains used in this study are listed in Table A1. Peanut-nodulating strains were routinely grown on TY medium [25] at 28 °C with rotary shaking (Model SI4-2 Shel Lab, 12 mm orbit, Sheldon Manufacturing Inc., Cornelius, OR, USA) at 150 rpm. Chromobacterium violaceum CV026 [26] and Agrobacterium tumefaciens NTL4, and their transconjugants A. tumefaciens NTL4 (pZLR4) and A. tumefaciens NTL4 (pTiC58ΔaccR) [27], were grown on LB medium [28] and agrobacterium medium (AB) [29], respectively, at 28 °C with rotary shaking at 150 rpm. When necessary, cycloheximide (50 μg mL−1), vancomycin (4 μg mL−1), and/or gentamicin (30 μg mL−1) were added.
2.2. Identification and Quantification of AHLs
2.2.1. Bioassays
For determination of AHL-like molecules with short acyl chains, the biosensor C. violaceum CV026 was utilized. This strain is a mini Tn5 double mutant defective in the synthesis of violacein pigment. The production of this pigment in vitro is activated by AHLs with short acyl chains. These autoinducers in peanut-nodulating strains were detected by the method of McClean et al. [26].
A. tumefaciens NTL4 (pZLR4) was utilized to detect AHLs with long acyl chains. This strain carries the plasmid pZLR4, which contains a traG::lacZ fusion and traR. In the presence of AHLs with long acyl chains the TraR protein is activated, transcription of the traG::lacZ fusion is turned on, and LacZ (β-galactosidase) activity can be used as a reporter of traG transcription [30]. AHL-like molecules with long acyl chains in peanut-nodulating strains were detected by the method of Farrand et al. [31].
2.2.2. Preparation of AHL Extracts
Extraction of AHLs was performed as described by Shaw et al. [32]. Peanut-nodulating bacteria were grown to the early stationary phase, and cells were removed from 25 mL growth medium by centrifugation at 12,000 g for 15 min at 4 °C. AHLs were then extracted from culture supernatants with three equal volumes of ethyl acetate, and the extracts were dried and resuspended in 500 μL ethyl actetate.
2.2.3. β-Galactosidase Assay
Aliquots (100 μL) of AHL extracts as above were added to 10 mL cultures of A. tumefaciens NTL4 (pZLR4) grown to OD600 0.5. The cultures were incubated for 6–8 h until reaching OD600 1.0. β-galactosidase activity was determined in Miller units as described by Miller [33]. For each strain extract, the values presented are means of four repeated experiments.
2.2.4. Thin-Layer Chromatography
Aliquots (10 μL) of AHL extracts were analyzed by reverse-phase C18-thin layer chromatography (RP-C18 TLC) using methanol/water (60:40 v/v) as the mobile phase. The plates were air-dried, overlaid with AB medium, 0.7% agar containing X-Gal 40 μg mL−1, and the biosensor A. tumefaciens NTL4 (pZLR4), and incubated overnight at 30 °C [32].
2.2.5. High-Performance Liquid Chromatography and Mass Spectrometry (LC-MS/MS)
AHL extracts were resuspended in 100 μL methanol (100%) for identification and quantification in LC-MS/MS, using a Restek C18 (Restek, USA) column (2.1 × 100 mm, 5 μm) with injection volume 10 μL. AHLs were separated at 28 °C using a gradient solvent system with increasing methanol concentration, constant glacial acetic acid concentration 0.2% (v/v) in water, and initial flow rate 0.2 mL min−1. The gradient was increased linearly from 40% (v/v) methanol/ 60% (v/v) water–acetic acid to 80% (v/v) methanol/ 20% (v/v) water–acetic acid over 25 min.
Mass spectrometry was performed on a quadrupole tandem mass spectrometer (MS–MS, Quattro Ultima™ PT; Micromass, Manchester, UK) equipped with an electrospray ion source (ESI).
AHLs were identified by comparison of retention times and m/z transitions with those of the following standards: N-hexanoyl-dl-homoserine lactone (C6), N-(3-oxodecanoyl)-l-homoserine lactone (3OC10), N-(3-oxododecanoyl)-l-homoserine lactone (3OC12), and N-(3-oxotetradecanoyl)-l-homoserine lactone (3OC14).
2.3. Effect of AHLs on Biological Processes in Peanut-Nodulating Strains
2.3.1. Motility (Swimming) Assay
Bacterial motility was determined in plates containing TY medium and reduced 1/10 TY medium with a final concentration of 0.3% agar. The 3OC10, 3OC12, and 3OC14 AHLs were added at various concentrations. Strains were inoculated by puncture in the center of the plate and incubated for 8 days at 28 °C. Halo diameters as indicators of motility were measured in cm.
2.3.2. Biofilm Formation Assay
Biofilm formation capacity was determined macroscopically by the method of O'Toole and Kolter [34], with some modifications. Glass tubes were inoculated with 800 μL bacterial culture (OD600 0.5) and incubated with agitation for 72 h at 30 °C. Planktonic cells were removed, and each tube was washed three times with saline solution, emptied, stained with crystal violet 0.1% for 15 min, and rinsed three times with distilled water to remove excess crystal violet. Biofilms formed were quantified by adding 1 mL 95% ethanol to the stained tube. The absorbance of solubilized crystal violet was determined by spectrophotometry at 570 nm.
2.3.3. Autoggregation Assay
Each peanut-nodulating strain was grown for 5 days at 28 °C in 25 mL TY medium supplemented with the appropriate AHLs. The bacterial suspension (5 mL) was transferred to a glass tube (10 × 70 mm) and left to settle for 24 h at 4 °C. An aliquot (0.2 mL) of the upper portion of the suspension was carefully transferred to a microtiter plate, and OD600 was measured (ODfinal). A control tube was vortexed for 30 s and OD600 was determined (ODinitial). The autoaggregation percentage was calculated as 100[1 – (ODfinal/ODinitial)] [35].
2.4. Statistical Analysis
Experiments on the effects of AHLs on biological processes were conducted using a completely randomized design using at least fifteen (15) replicates for each treatment. The values presented are means of four repeated experiments. The data were subjected to one-way ANOVA followed by comparison of multiple treatment levels with the control using post hoc Fisher’s Least Significant Difference (LSD) test. To evaluate the overall effect of AHLs on biological functions, we performed a multivariate study with principal components analysis (PCA). Statistical analyses were performed using Infostat software version 2.0 (Group Infostat, Universidad Nacional de Córdoba, Argentina).
3. Results and Discussion
3.1. Detection and Characterization of AHL-Like Molecules in Peanut-Nodulating Strains
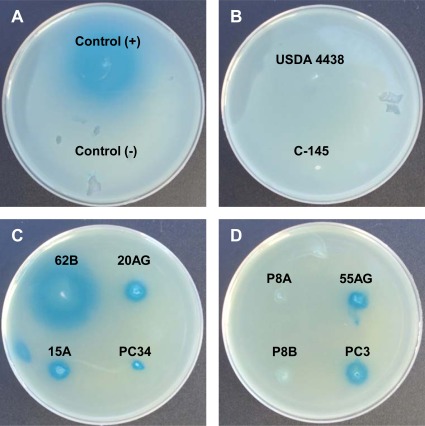
AHL-like molecules in peanut-nodulating strains were detected using the biosensor strains A. tumefaciens NTL4 (pZLR4) and C. violaceum CV026. None of the tested strains showed positive results for the C. violaceum CV026 bioassay, indicating that the strains were unable to synthesize AHL-like molecules having short acyl chains. On the other hand, results of the A. tumefaciens NTL4 (pZLR4) bioassay revealed that some strains were able to produce AHL-like molecules with long acyl chains. A positive result consisted of the presence of a blue halo around a colony resulting from hydrolysis of X-Gal, as shown in Figure 1. Of the 53 strains analyzed, 14 (26%) were able to synthesize AHL-like molecules with long acyl chains (positive result), two (4%, indicated as +/−) showed an undefined result, and the remaining 37 (70%) showed a negative result for the production of AHL-like molecules by A. tumefaciens NTL4 (pZLR4) (Table A1).
Figure 1.
Bioassay for detection of AHL-like molecules with long acyl chains using A. tumefaciens NTL4 (pZLR4) as biosensor strain.
The plates show bioassay results obtained for control + (A. tumefaciens NTL4 pTiC58ΔaccR), control – (A. tumefaciens NTL4) (A), and peanut-nodulating strains (B, C, D). Strains USDA 4438 and C-145 were negative (B); 62B, 20AG, 15A, PC34 (C), 55AG, and PC3 were positive (D); P8A and P8B showed undefined results (D) for AHL production.
The production of AHLs was quantified by measuring β-galactosidase activity using A. tumefaciens NTL4 (pZLR4) in rhizobial strains that showed positive or undefined results in the bioassay (Table A1). The response of the strains was quite diverse: five strains showed low levels of AHL production, three strains showed high production, and the remaining strains showed moderate production compared with the control strain (Table A2).
Considering the origin of the strains (Table A1) and their genetic diversity [36,37], our results are in good agreement with those of Pongslip et al. [18], who reported the production of long-chain AHLs by ∼22% of a geographically and genetically diverse group of soybean-nodulating bacteria. Similarly, Westenberg [21] reported that three out of 12 tested strains of B. japonicum produced AHL-like autoinducers. However, the number of strains evaluated in this study was small, and the biological roles of the autoinducers, if any, were not demonstrated.
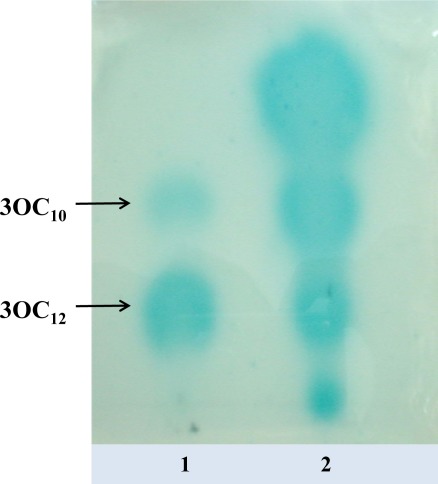
Based on the results of the β-galactosidase assay, we selected strains having moderate (62B) and high (P8A) activity for further characterization of the autoinducers by TLC and LC-MS/MS analysis. Using bacterial extracts of strains 62B and P8A, we detected the presence of four different signaling molecules corresponding to standards of C6 AHL and 3OC10, 3OC12, and 3OC14 AHLs. Figure 2 shows the TLC patterns of AHLs produced by strain 62B detected by the biosensor A. tumefaciens NTL4 (pZLR4). Extracts of strains 62B and P8A (not shown) typically produced four spots detected by NTL4 (pZLR4).
Figure 2.
Detection of AHLs with long acyl chain by thin layer chromatography.
The TLC plate was overlaid with the biosensor A. tumefaciens NTL4 (pZLR4). Lane 1, long chain 3-oxo-standards. Lane 2, 10 μL ethyl acetate extract from supernatant of liquid culture of strain 62B.
Our results showed clearly that some peanut-nodulating strains had the ability to produce various signaling molecules identified as AHLs with long acyl chains. Similarly, other authors have reported the production of different signaling molecules by rhizobia that nodulate legumes, e.g., Rhizobium leguminosarum bv. viciae synthesizes mainly C6 AHL and 3OC8 AHL [38]; Sinorhizobium meliloti produces at least six different AHLs, including C6 AHL and 3OC14 AHL [39].
The analysis of HPLC-MS/MS data allowed us to calculate the concentrations of the molecules produced. In comparison to strain P8A, strain 62B produced higher amounts of all autoinducers, with concentrations of ∼800 nM for C6 AHL and 20–30 nM for 3-oxo AHLs. Autoinducer concentrations in P8A extracts were ∼15 nM for C6 AHL and 0.1–0.5 nM for the other molecules (Table A3). These findings suggest that the biosensor employed is more sensitive to the AHLs produced by P8A than to those produced by 62B (Table A2). It is possible that the production of different types and concentrations of AHL-like molecules may reflect the physiological regulation of different activities in peanut-nodulating bacterial strains.
3.2. Biological Effect of AHLs in Peanut-Nodulating Strains
The biological roles of 3OC10, 3OC12, and 3OC14 AHLs were evaluated in both AHL-producing (62B, P8A) and AHL-non-producing (USDA 4438, P5) peanut-nodulating strains. In view of the lack of information regarding the physiological role of AHLs in various rhizobial processes, we examined the effect of exogenously added synthetic AHLs at various concentrations on activities related to rhizobial survival and nodulation, including motility, biofilm formation, and cell aggregation. To ensure that addition of the solvent (ethyl acetate) employed to dissolve the AHLs did not affect the parameters quantified, we added the same solvent in the control treatments. No effect of the solvent was observed.
3.2.1. Effect of AHLs on Bacterial (Swimming) Motility
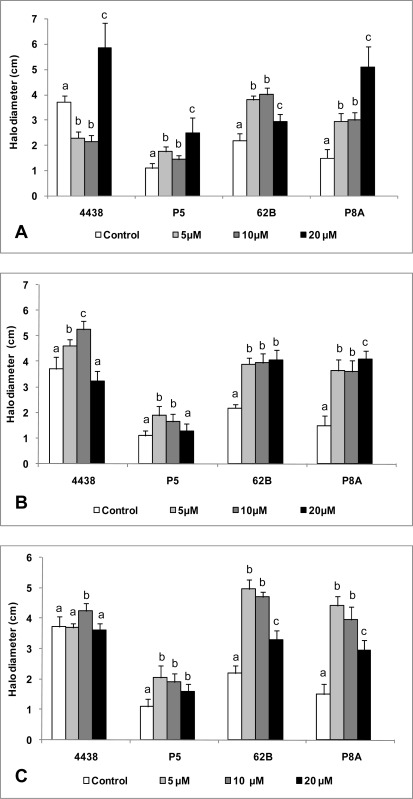
Microorganisms with the ability to move have an ecological advantage in that they are able to find more favorable or less hazardous niches for colonizing and persisting in a given environment. In the case of rhizobia, motility has been shown not to be essential for nodulation, but allows the bacteria to find their specific host legume and establish symbiosis. Bacterial motility is an energetically costly process, and therefore must be finely regulated [40]. Several studies using S. marcescens, V. cholerae, E. coli, and Y. pseudotuberculosis have shown that QS systems are involved in the control of bacterial motility [41–44]. We evaluated for the first time the effect of QS signals on the swimming motility of peanut-nodulating bacterial strains. The optimal experimental condition for evaluating the motility of these strains was found to be limited nutrient availability (reduced 1/10 TY medium), whereby their motility was two-fold higher than in full growth medium (entire TY medium) (data not shown). We therefore added the AHLs to reduced 1/10 TY medium and measured the swimming halo of peanut-nodulating strains.
In general, the motility of 62B, P8A, and P5 was increased as a result of exposure to various types and concentrations of AHLs (Figure 3). In the case of 62B and P8A, a low autoinducer concentration (5 μM) resulted in a significant increase in motility, and this effect was maintained or increased in the presence of higher autoinducer concentrations (10 and 20 μM) (Figure 3(A,B,C)). P5 also responded positively to the presence of autoinducers, but to a lesser degree than 62B or P8A. We concluded that, regardless of the type and concentration of autoinducer, these rhizobial strains show increased motility in the presence of exogenous signaling molecules. Interestingly, USDA 4438, a reference strain that did not induce blue haloes in biosensor strains, displayed a more complex swimming behavior when various exogenous AHLs were added. Low concentrations of 3OC10 AHL (5 or 10 μM) significantly decreased the motility of USDA 4438, whereas the maximal autoinducer concentration tested (20 μM) significantly increased its swimming ability in comparison to controls without addition of exogenous AHLs (Figure 3(A)). The addition to USDA 4438 of 5 or 10 μM 3OC12 AHL caused a slight but significant increase in motility (Figure 3(B)), and the addition of 10 μM 3OC14 AHL also caused a slight increase (Figure 3(C)).
Figure 3.
Effect of AHLs on swimming motility of peanut-nodulating strains.
Swimming motility (expressed as halo diameter; cm) of peanut-nodulating strains in reduced 1/10 TY medium with 0.3% agar, supplemented with various concentrations of 3OC10 AHL (A), 3OC12 AHL (B), and 3OC14 AHL (C). Values indicated by different letters are significantly different from each other according to Fisher’s LSD test (P < 0.05).
Bacterial motility probably occurs as a dispersal strategy in nutrient-limited environments, or to searching for a new host [41–43]. Hoang et al. [45] showed that motility of the symbiotic S. meliloti is negatively regulated by a QS system, ExpR/SinI. When the bacterial population of S. meliloti grows, AHL production increases and motility is repressed. This phenomenon plays an important role in the interaction of S. meliloti with its host (alfalfa); i.e., the accumulation of bacteria around the host root and the scope of a quorum could provide an elegant strategy to suppress motility and simultaneously activate the invasion of the root.
There is no previous evidence for a connection between a QS system and motility control in peanut-nodulating Bradyhizobium sp. strains. We show here for the first time that the motility process in these bacteria is affected by the presence of QS signaling molecules (AHLs). It is clear that a finely regulated mechanism of cell signaling via AHLs coordinates the process of motility in these bacteria. It remains to be determined whether peanut-nodulating strains employ a motility mechanism for dispersion, colonization of new niches, invasion of the legume host, or other activities.
3.2.2. Effect of AHLs on Biofilm Formation
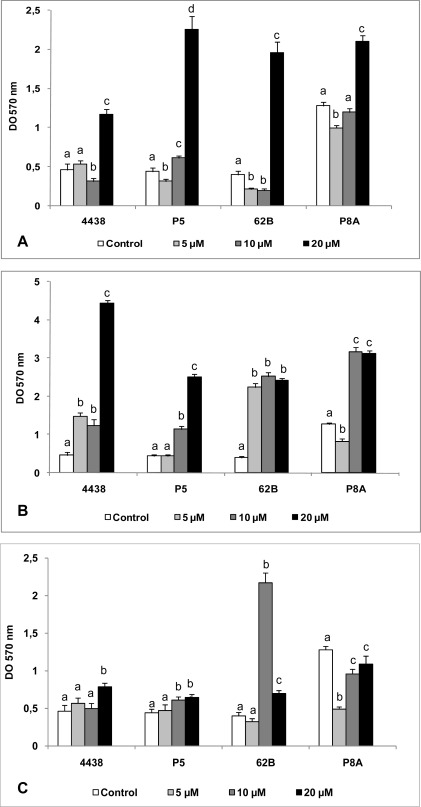
Bacteria are social organisms that live in terrestrial environments and form highly complex and highly organized communities. Such multicellular structures develop on various surfaces and facilitate survival and optimized resource utilization in hostile environments. This structure and associated lifestyle are termed a “biofilm” [6,46,47]. It has been estimated that over 99% of all bacterial activity in natural ecosystems is associated with bacteria organized in biofilms [48]. Thus, understanding the biology of biofilms is crucial in studies of ecosystems, including those in the soil. QS is a key mechanism that regulates several aspects of biofilm development, including adhesion, motility, maturation, and dispersal [49–52]. The present study is the first to examine the relationship between AHL production and biofilm development in peanut-nodulating bacteria.
Regardless of the presence of exogenously-added AHLs, all rhizobial strains tested were able to develop sessile biomass on a glass surface, with OD570 values ranging from 0.4 to ∼5.0 (Figure 4). A high concentration (20 μM) of 3OC10 AHL caused a significant (3–5 fold) increase in the biofilm formation ability of all strains, relative to strains grown without added AHL (Figure 4(A)). With lower concentrations (5, 10 μM) of added 3OC10 AHL, there was either no difference in biofilm formation or a slight inhibitory effect (Figure 4(A)).
Figure 4.
Effect of AHLs on biofilm formation ability of peanut-nodulating strains.
The biofilm formation ability of peanut-nodulating strains was determined after 72 h incubation in TY medium supplemented with various concentrations of 3OC10 AHL (A), 3OC12 AHL (B), and 3OC14 AHL (C). Values indicated by different letters are significantly different from each other according to Fisher’s LSD test (P < 0.05).
In comparison to the results with 3OC10 and 3OC14, all concentrations of 3OC12 AHL tested caused a larger increase in biofilm formation (Figure 4(B)). Similarly to results with 3OC10 AHL, a high concentration (20 μM) of 3OC12 AHL produced the greatest increase in biofilm formation relative to controls. Such an increase was observed even at low concentrations (5, 10 μM) (Figure 4(B)), indicating that, in contrast to results with other signaling molecules, a significant increase in biofilm formation occurred in the presence of 3OC12 AHL regardless of its concentration.
The effect of various concentrations of exogenously added 3OC14 AHL was strain-dependent (Figure 4(C)). This molecule only slightly modified biofilm formation by P5 and USDA 4438. Surprisingly, an intermediate concentration (10 μM) of C14 AHL greatly increased biofilm formation by 62B strain, whereas all C14 AHL concentrations tested reduced biofilm formation by P8A relative to control (Figure 4(C)).
The ability of rhizobia to establish a biofilm can be used as a strategy for survival or for colonization and/or invasion [53,54]. In general, peanut-nodulating strains show increased biofilm formation ability in the presence of added AHLs with long acyl chains, independently of their capacity to synthesize AHLs. However, some strains showed different behaviors depending on the type and concentration of AHL. Since rhizobia are exposed to various and fluctuating environmental conditions, this range of behaviors is presumably employed depending on the prevailing environmental and biological conditions in order to increase rhizosphere fitness and to adapt and survive in a hostile soil ecosystem. The decision to change rhizobial lifestyle from planktonic cell to biofilm cell probably depends on the detection of certain environmental factors that direct the bacteria to activation of cell communication mechanisms that regulate the conduct of the entire population to the new state [6,47,54].
Several recent studies have addressed the regulatory effect of cell signaling mechanisms mediated by AHL on the process of biofilm formation in various Gram-negative bacteria, including Pseudomonas aeruginosa [55,56], Pseudomonas putida [57], Serratia liquefaciens [58], Aeromonas hydrophila [59], and Burkholderia cepacia [60]. Although there is no direct evidence for a mechanistic relationship between QS and biofilm formation in peanut-nodulating strains, AHLs clearly have an effect on biofilm formation. The ability of some AHL-non-producing strains to form biofilms may be due to the presence of signaling molecules (AHLs or other types) that we were not able to detect. However, there was a clear response to added AHLs by both AHL-producing and AHL-non-producing strains in this study. While there is no previous evidence that a sensing system mediated by AHLs is involved in the mechanism of biofilm formation in peanut-nodulating strains, the present results suggest that these signaling molecules play an important role in the regulation of this process. Such QS regulation may be exerted on surface components such as EPS, LPS, flagella, and pili, which are essential for different steps of biofilm development [54].
It is interesting that putative AHL-non-producing strains are able to respond to the exogenous addition of these signaling molecules. This finding is probably attributable to the ability of strains that are incapable of synthesizing AHLs to recognize them. This could be due to the presence of LuxR-like regulator molecules capable of binding the signaling molecules produced by other bacteria within a given population or community. We hypothesize that this mechanism is used by AHL-non-producing strains and confers an adaptive advantage for regulating certain activities, depending on the bacterial signaling molecules produced by other community members. In nature, biofilms are composed of multiple bacterial species and many other types of organisms, resulting in assemblies that are taxonomically and functionally complex and diverse [61]. Perhaps it is a common occurrence that QS signals produced by certain members regulate the activities of the community as a whole, in ways advantageous for bacterial survival [9].
3.2.3. Effect of AHLs on Cell Aggregation
Cell-cell interactions in bacterial cultures (planktonic cells) are manifested as the autoaggregative phenomenon [62]. Cell aggregation allows the establishment of microcolonies as well as the development of mixed microbial communities in natural habitats. Autoaggregation has been proposed as the initial step in biofilm formation; the cell aggregates formed could then attach to surfaces and develop a sessile population, conferring a competitive advantage and promoting bacterial survival in hostile environments [63,64].
Based on this process of physical cell-cell interaction, there may be mechanisms of cellular communication that mediate the autoaggregative phenomenon. No previous studies have addressed this process or its possible relationship with QS systems in Bradyrhizobium sp. We therefore evaluated the effect of AHLs on cell aggregation of peanut-nodulating strains.
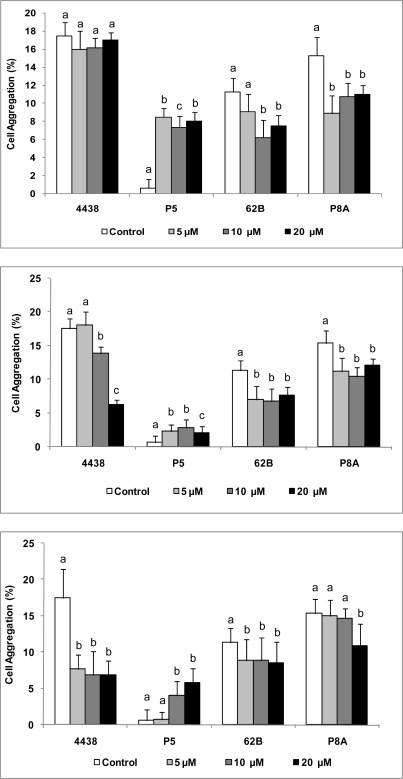
Under our experimental conditions, the peanut-nodulating Bradyrhizobium strains studied showed a low capacity for cell aggregation, with values ranging from ∼5–20%. In comparison to controls, the addition of exogenous AHLs caused decreased autoaggregative ability in all strains except P5, which showed a slight increase (Figure 5(A,B,C)). All concentration of 3OC10 AHL produced the greatest increases in autoaggregation of P5, with values ∼10-fold higher than those of controls (Figure 5(A)). 3OC12 (Figure 5(B)) and 3OC14 AHLs (Figure 5(C)) showed effects that were similar to but smaller than that of 3OC10 AHL. Autoaggregation of USDA 4438 was not affected by addition of 3OC10 AHL, but autoaggregation of 62B and P8A decreased in response to increasing concentrations of this autoinducer (Figure 5(A)). A negative dose-dependent effect by 3OC12 AHL was observed on autoaggregation of USDA 4438, and a negative dose-independent effect by this autoinducer was observed on autoaggregation of 62B and P8A (Figure 5(B)). Exposure to any concentration of 3OC14 AHL significantly reduced autoaggregation of USDA 4438 and 62B, and a high concentration (20 μM) of this autoinducer reduced autoaggregation of P8A (Figure 5(C)).
Figure 5.
Effect of AHLs on cell aggregation of peanut-nodulating strains.
Cell aggregation (%) of peanut-nodulating strains was calculated as described in “Experimental”. For the assay, TY medium was supplemented with various concentrations of 3OC10 AHL (A), 3OC12 AHL (B), and 3OC14 AHL (C). Values indicated by different letters are significantly different from each other according to Fisher’s LSD test (P < 0.05).
Previous studies have shown that QS regulatory systems are involved in control of bacterial autoaggregation, e.g., in Rhodobacter sphaeroides [65], Burkholderia thailandensis [66], Yersinia pseudotuberculosis [67], and Pseudomonas aureofaciens [68]. Our results indicate that the process of autoaggregation of peanut-nodulating strains is reduced or increased by QS signaling molecules. We can therefore speculate regarding the controlling effect of this process on the behavior of bacterial populations in nature. For some bacterial strains, QS signals may induce the dispersal of aggregates, leading individual bacteria to colonize new microniches in the soil. For other strains, QS mechanisms may positively regulate the autoaggregative process, enhancing the survival of the microorganisms in the soil.
3.2.4. Overall Biological Effect of AHLs on Bradyrhizobium Strains
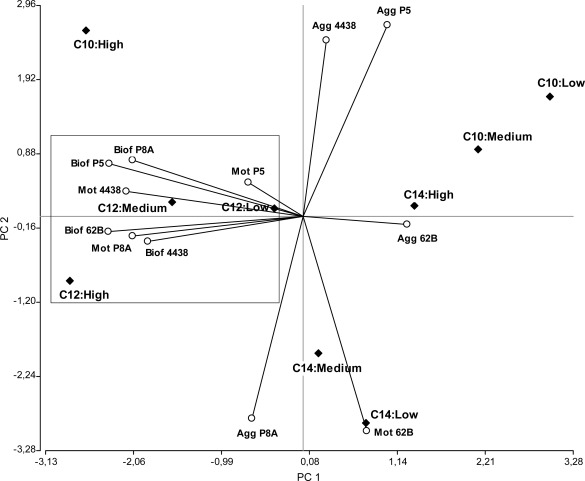
In order to provide a more integrated picture of the impact of AHLs on autoaggregation, biofilm formation, and motility phenotypes of peanut-nodulating strains, we carried out a statistical multivariate PCA. This analysis allowed us to project the data set and variables in a graph in which they could be more easily visualized. The type of AHLs (depending on acyl chain length) associated with the different concentrations tested were the observations (or cases), while the variables were the motility, autoaggregation, and biofilm formation ability of each peanut-nodulating Bradyrhizobium strain.
According to the PCA, the analysis of all data in three dimensions (PC1, PC2, and PC3) explained ∼80% of the total variability in the study. The graph generated according to PC1 and PC2 (which explained ∼60% of the variability) (Figure 6) shows that motility and biofilm formation were processes regulated by the influence of 3OC12 AHL. Similarly (although not conclusively), it appears that 3OC10 and 3OC14 AHLs affect the autoaggregative process.
Figure 6.
Overall effect of AHLs on various biological processes of peanut-nodulating strains.
Graphic obtained from PCA using the software InfoStat version 2.0. Solid diamonds indicate various combinations of AHL type (C10: 3OC10; C12: 3OC12; C14: 3OC14) and concentration (low: 5 μM; medium: 10 μM; high: 20 μM). Open circles indicate the biological variables measured for each strain: motility (Mot), biofilm formation ability (Biof), and cell aggregation (Agg). The angles formed between the straight lines indicate the degree of correlation between variables. PC1: Principal Component 1; PC2: Principal Component 2.
Regarding the correlation between variables, there is a strong positive linkage between the variables of motility and biofilm formation for each strain (acute angle, positive correlation). This is consistent with previous findings that the different mechanisms of bacterial motility play important roles in the colonization of surfaces, which is the first step in the formation of biofilms [69].
Less strongly than for the correlation between motility and biofilm formation, we observed certain linkages between the processes of motility and autoaggregation (acute angles more open). This may reflect the logical necessity of bacterial motility to allow physical contact between the microorganisms prior to cellular aggregation.
In agreement with previous reports [69], motility appears to be a key mechanism for peanut-nodulating bacteria to establish various processes of cell interaction, such as cell aggregation and formation, remodeling, and disassembly of biofilms.
Surprisingly, there were weak or no linkages between biofilm formation and autoaggregation (right or obtuse angles), suggesting that, at least for peanut-nodulating Bradyrhizobium strains, these physiological processes may be regulated differently and probably used differently for the bacteria according to their exposure to environmental signals. Thus, the behavior of the bacteria could be driven to aggregation-disaggregation or biofilm formation-biofilm dispersal depending on the type of signal (physical, chemical, etc.), the presence-absence of a surface (biotic or abiotic), and other factors. The high biofilm formation capacity (Figure 4) compared to the low autoaggregation percentage (Figure 5) determined for these bacteria suggests that the former mechanism is more important in terms of the environmental benefits that these physiological processes confer to these peanut-symbiotic microorganisms.
4. Conclusions
Our results demonstrate for the first time the production of QS signaling molecules by peanut-nodulating bacteria (Bradyrhizobium sp. strains). Such molecules were identified as C6, 3OC10, 3OC12, and 3OC14 AHLs, and were found only in a small percentage of the bacterial population.
The exposure of both AHL-producing and AHL-non-producing strains to exogenous addition of these molecules had a positive effect on physiological processes related to survival and colonization ability, particularly motility and biofilm formation ability.
In view of the finding that various biological effects observed in vitro were affected by different types and concentrations of AHLs, we conclude that the overall behavior of peanut-nodulating soil rhizobial populations is subject to a fine and coordinated regulation of the synthesis of a particular pattern of AHLs, in response to specific environmental factors. We are currently examining the production of AHL-like molecules in the strains that gave negative results in our bioassays, as well as the putative QS mechanism of peanut-nodulating Bradyrhizobium sp. The presence of the legume host is presumably a key factor in the regulation of the behavior of rhizobial populations. These hypotheses will be tested in future studies.
Acknowledgments
This work was supported by grants from the Secretaría de Ciencia y Técnica de la Universidad Nacional de Río Cuarto. WG and PB are Career Members of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), República Argentina. FN and FS have a fellowship from CONICET. We gratefully acknowledge Oscar Masciarelli for his help with the LC-MS/MS analysis, and Stephen Anderson for English editing.
Appendix
Table A1.
Peanut-nodulating rhizobial strains used, and production of AHL-like molecules with long acyl chains.
| Strain | Origin | Source | AHL | Strain | Origin | Source | AHL |
|---|---|---|---|---|---|---|---|
| P1 | Río Cuarto | [70] | − | 35LA | La Aguada | [36] | − |
| P5 | Río Cuarto | [70] | − | 51LA | La Aguada | [36] | − |
| P7 | Río Cuarto | [70] | − | 61LA | La Aguada | [36] | − |
| P8A | Río Cuarto | [70] | +/− | 65LA | La Aguada | [36] | − |
| P8B | Río Cuarto | [70] | +/− | 36A | La Aguada | This study | + |
| PR1 | Río Cuarto | [70] | − | 3A | La Aguada | This study | − |
| 6H | Río Cuarto | [36] | + | 4A | La Aguada | [36] | − |
| PC30 | Río Cuarto | [70] | + | 8A | La Aguada | [36] | − |
| PC27 | Gral. Cabrera | [37] | + | 9A | La Aguada | [36] | − |
| PC28 | Gral. Cabrera | This study | − | 15A | La Aguada | This study | + |
| PC29 | Gral. Cabrera | [37] | + | 40A | La Aguada | [36] | − |
| PC31 | Gral. Cabrera | [37] | + | Ch1 | Chaján | [36] | − |
| PC34 | Gral. Cabrera | [37] | + | Ch4 | Chaján | [36] | − |
| PC3 | Gral. Cabrera | [37] | + | Ch8 | Chaján | [36] | − |
| PC4 | Gral. Cabrera | [37] | − | Ch9 | Chaján | [36] | − |
| VM45 | V. Mackenna | [37] | − | Ch10 | Chaján | [36] | − |
| VM50 | V. Mackenna | [37] | − | Ch12 | Chaján | [36] | − |
| V80 | V. Mackenna | [37] | − | Ch13 | Chaján | [36] | − |
| 62B | V. Mackenna | [37] | + | Ch17 | Chaján | [36] | − |
| 20AG | La Aguada | [37] | + | Ch19 | Chaján | [36] | − |
| 45AG | La Aguada | [37] | − | Ch21 | Chaján | [36] | − |
| 55AG | La Aguada | [37] | + | Ch22 | Chaján | This study | − |
| 62AG | La Aguada | [37] | + | Ch25 | Chaján | [36] | − |
| 62A | La Aguada | [37] | − | C-145 | Reference | NifTAL, USA | − |
| 26LA | La Aguada | [37] | − | USDA 4438 | Reference | USDA, ARS, USA | − |
| 27LA | La Aguada | [37] | + | USDA 3180 | Reference | USDA, ARS, USA | − |
| 33LA | La Aguada | This study | − |
AHL: Acyl homoserine lactone.
(−): Negative result.
(+): Positive result.
(+/−): Undefined result.
Table A2.
Induction of β-galactosidase activity in A. tumefaciens NTL4 (pZLR4) by AHLs obtained from supernatants of peanut-nodulating rhizobial strains.
| Strain | Activity * |
|---|---|
| A. tumefaciens NTL4 (pTiC58ΔaccR) ▪ | 349.6 ± 27.5 |
| A. tumefaciens NTL4 ▴ | 47.0 ± 10.0 |
| A. tumefaciens NTL4 (pZLR4) ♦ | 47.0 ± 5.0 |
| Low AHL-like inducer activity (≤ 100) | |
| PC29 | 68.7 ± 16.0 |
| PC27 | 94.6 ± 6.1 |
| PC30 | 70.2 ± 12.1 |
| PC34 | 94.3 ± 8.7 |
| 6H | 91.9 ± 6.0 |
| Moderate AHL-like inducer activity (≥ 100 < 200) | |
| 15A | 123.5 ± 30.3 |
| 20AG | 129.3 ± 35.8 |
| 62AG | 109.3 ± 22.6 |
| 27LA | 101.8 ± 42.6 |
| 36A | 118.5 ± 17.0 |
| PC3 | 102.2 ± 12.8 |
| 62B | 197.1 ± 10.7 |
| 55AG | 153.9 ± 4.4 |
| High AHL-like inducer activity (≥ 200) | |
| P8A | 369.7 ± 15.5 |
| P8B | 411.8 ± 17.1 |
| PC31 | 678.9 ± 50.4 |
β-Gal activity expressed in Miller units;
Control +;
Control −;
Biosensor strain under control conditions (no AHL added).
Table A3.
AHL concentration (nM) detected by LC-MS/MS for strains 62B and P8A.
|
AHL concentration (nM) |
|||||
|---|---|---|---|---|---|
| Strain | C6 | 3OC10 | 3OC12 | 3OC14 | |
| 62B | 840.1 | 23.2 | 28.0 | 19.6 | |
| P8A | 15.4 | 0.13 | 0.31 | 0.54 | |
References
- 1.Becking J.H. The Rhizobium Symbiosis of the Nonlegume Parasponia. In: Stacey G.S., Evans H.J., editors. Biological Nitrogen Fixation. Routledge; Chapman and Hall, NY, USA: 1992. pp. 497–559. [Google Scholar]
- 2.Urtz B.E., Elkan G.H. Genetic diversity among Bradyrhizobium isolates that efectively nodulate peanut (Arachis hypogaea) Can. J. Microbiol. 1996;188:65–75. doi: 10.1139/m96-144. [DOI] [PubMed] [Google Scholar]
- 3.van Rossum D., Schuurmans F.P., Gillis M., Muyotcha A., van Verselveld H.K. Genetic and phonetic analyses of Bradyrhizobium strains nodulating peanut (Arachis hypogaea L.) roots. Appl. Environ. Microbiol. 1995;61:1599–1609. doi: 10.1128/aem.61.4.1599-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gage D.J. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones K.M., Kobayashi H., Davies B.W., Taga M.E., Walker G.C. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nature Rev. Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuqua W.C., Greenberg E.P. Listening in on bacteria: Acyl-homoserine lactone signaling. Nature Rev. Mol. Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 7.Waters C.M., Bassler B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell. Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 8.Camilli A., Bassler B.L. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology. 2007;153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- 10.Williams P., Winzer K., Chan W., Cámara M. Look who’s talking: Communication and quorum sensing in the bacterial world. Philos. T. Roy. Soc. B. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanzelka B.L., Greenberg E.P. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 1995;177:815–817. doi: 10.1128/jb.177.3.815-817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsek M.R., Greenberg E.P. Acyl-homoserine lactone quorum sensing in Gram negative bacteria: A signaling mechanism involved in associations with higher organisms. Proc. Nat. Acad. Sci. USA. 2000;97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brelles-Marino G., Bedmar E.J. Detection, purification and characterisation of quorum-sensing signal molecules in plant-associated bacteria. J. Biotechnol. 2001;91:197–209. doi: 10.1016/s0168-1656(01)00330-3. [DOI] [PubMed] [Google Scholar]
- 14.Loh J., Pierson E.A., Pierson L.S., Stacey G., Chatterjee A. Quorum sensing in plant-associated bacteria. Curr. Opin. Plant Biol. 2002;5:285–290. doi: 10.1016/s1369-5266(02)00274-1. [DOI] [PubMed] [Google Scholar]
- 15.González J.E., Marketon M.M. Quorum sensing in nitrogen-fixing rhizobia. Microbiol. Mol. Biol. Rev. 2003;67:574–592. doi: 10.1128/MMBR.67.4.574-592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Contreras M., Bauer W.D., Gao M., Robinson J.B., Downie A.J. Quorum-sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Philos. Trans. R Soc. Lond. B Biol. Sci. 2007;362:1149–1163. doi: 10.1098/rstb.2007.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierson L.S., Pierson E.A. Roles of diffusible signals in communication among plant-associated bacteria. Phytopathology. 2007;97:227–232. doi: 10.1094/PHYTO-97-2-0227. [DOI] [PubMed] [Google Scholar]
- 18.Pongsilp N., Triplett E.W., Sadowsky M.J. Detection of homoserine lactone-like quorum sensing molecules in Bradyrhizobium strains. Curr. Microbiol. 2005;51:250–254. doi: 10.1007/s00284-005-4550-5. [DOI] [PubMed] [Google Scholar]
- 19.Loh J., Yuen-Tsai J.P., Stacey M.G., Lohar D., Welborn A., Stacey G. Population density-dependent regulation of the Bradyrhizobium japonicum nodulation genes. Mol. Microbiol. 2001;42:37–46. doi: 10.1046/j.1365-2958.2001.02625.x. [DOI] [PubMed] [Google Scholar]
- 20.Loh J., Lohar D.P., Andersen B., Stacey G. A two-component regulator mediates population-density-dependent expression of the Bradyrhizobium japonicum nodulation genes. J. Bacteriol. 2002;184:1759–1766. doi: 10.1128/JB.184.6.1759-1766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westenberg D.J. Evidence of AHL Autoinducer Production by the Soybean Symbiont Bradyrhizobium japonicum. In: Finan T., O_Brian M., Vessey S., Newton W., editors. Nitrogen Fixation: Global Perspectives; Proceedings of the 13th International Congress on Nitrogen Fixation; Cambridge, UK: CABI Publishing; 2002. p. 409. [Google Scholar]
- 22.Loh J., Carlson R.W., York W.S, Stacey G. Bradyoxetin, a unique chemical signal involved in symbiotic gene regulation. Proc. Natl. Acad. Sci. USA. 2002;99:14446–14451. doi: 10.1073/pnas.222336799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahlgren N.A., Harwood C.S., Schaefer A.L., Giraud E., Greenberg E.P. Aryl-homoserine lactone quorum sensing in stem-nodulating photosynthetic bradyrhizobia. Proc. Natl. Acad. Sci. USA. 2011;108:7183–7188. doi: 10.1073/pnas.1103821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindemann A., Pessi G., Schaefer A.L., Mattmann M.E., Christensen Q.H., Kessler A., Hennecke H., Blackwell H.E., Greenberg E.P., Harwood C.S. Isovaleryl-homoserine lactone, an unusual branched-chain quorum-sensing signal from the soybean symbiont Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA. 2011;108:16765–16770. doi: 10.1073/pnas.1114125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beringer J.E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 26.McClean K.H., Winson M.K., Fish L., Taylor A., Chhabra S.R., Camara M., Daykin M., Lamb J.H., Swift S., Bycroft B.W., Stewart G.S., Williams P. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 27.Luo Z.Q., Clemente T.E., Farrand S.K. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol. Plant Microbe Interact. 2001;14:98–103. doi: 10.1094/MPMI.2001.14.1.98. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J., Fritsch E.F., Maniatis F. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory; New York, NY, USA: 1989. [Google Scholar]
- 29.Chilton N.W., Barbano J.P. Guidelines for reporting clinical trials. J. Periodontal Res. Suppl. 1974;14:207–208. doi: 10.1111/j.1600-0765.1974.tb01786.x. [DOI] [PubMed] [Google Scholar]
- 30.Cha C.E., Gao O, Chen Y.C., Shaw P.D., Farrand S.K. Production of acyl-hosmoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- 31.Farrand S.K., Qin Y., Oger P. Quorum-sensing system of Agrobacterium plasmids: Analysis and utility. Methods Enzymol. 2002;358:452–484. doi: 10.1016/s0076-6879(02)58108-8. [DOI] [PubMed] [Google Scholar]
- 32.Shaw P.D., Ping G., Daly S.L., Cha C., Cronan J.E., Jr., Rinehart K.L. Detecting and characterizing N-acyl-homoserina lactone signal molecules by thin layer chromatography. Proc. Nathl. Acad. Sci. USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; New York, NY, USA: 1972. [Google Scholar]
- 34.O’Toole G.A., Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 35.Sorroche F., Rinaudi L., Zorreguieta A., Giordano W. EPS II-dependent autoaggregation of Sinorhizobium meliloti planktonic cells. Current. Microbiol. 2010;61:465–470. doi: 10.1007/s00284-010-9639-9. [DOI] [PubMed] [Google Scholar]
- 36.Nievas F., Bogino P., Nocelli N., Giordano W. Genotypic analysis of isolated peanut-nodulating rhizobial strains reveals differences among populations obtained from soils with different cropping histories. Appl. Soil. Ecol. 2012;53:74–82. [Google Scholar]
- 37.Bogino P., Banchio E., Giordano W. Molecular diversity of peanut-nodulating rhizobia in soils of Argentina. J. Basic Microbiol. 2010;50:274–279. doi: 10.1002/jobm.200900245. [DOI] [PubMed] [Google Scholar]
- 38.Wisniewski-Dye F., Downie J.A. Quorum-sensing in Rhizobium. A. Van Leeuw. 2002;81:397–407. doi: 10.1023/a:1020501104051. [DOI] [PubMed] [Google Scholar]
- 39.Marketon M.M., Gronquist M.R., Eberhard A., González J.E. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 2002;184:5686–5695. doi: 10.1128/JB.184.20.5686-5695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soutourina O.A., Bertin P.N. Regulation cascade of flagellar expression in gram-negative bacteria. FEMS Microbiol. Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 41.Ang S., Horng T.Y., Shu J.C., Soo P.C., Liu J.H., Yi W.C., Lai H.C., Luh K.T., Ho S.W., Swift S. The role of RsmA in the regulation of swarming motility in Serratia marcescens. J. Biomed. Sci. 2001;8:160–169. doi: 10.1007/BF02256408. [DOI] [PubMed] [Google Scholar]
- 42.Zhu J.M., Miller M.B., Vance R.E., Dziejerman M., Basseler B.L., Mekalanos J.J. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperandio V., Torres A., Kapper J. Quorum sensing Escherichia coli regulators B and C (QseBC): A novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E.coli. Mol. Microbiol. 2002;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 44.Atkinson S., Chang C.Y., Patrick H.L., Buckley C.M., Wang Y., Sockett R.E., Cámara M., Williams P. Functional interplay between the Yersinia pseudotuberculosis YpsRI and YtbRI quorum sensing systems modulates swimming motility by controlling expression of flhDC and fliA. Mol. Microbiol. 2008;69:137–151. doi: 10.1111/j.1365-2958.2008.06268.x. [DOI] [PubMed] [Google Scholar]
- 45.Hoang H.H., Gurich N., González J.E. Regulation of motility by ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J. Bacteriol. 2007;190:861–871. doi: 10.1128/JB.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costerton J.W., Lewandowski Z., Caldwell D.E., Korber D.R., Lappin-Scott H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 47.Parsek M.R., Greenberg E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trend. Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 48.Potera C. Biofilms invade microbiology. Science. 1996;273:1795–1797. doi: 10.1126/science.273.5283.1795. [DOI] [PubMed] [Google Scholar]
- 49.Dong Y., Zhang X., An S., Xu J., Zhang L. A novel two-component system BqsS-BqsR modulates quorum sensing-dependent biofilm decay in Pseudomonas aeruginosa. Commun. Integr. Biol. 2008;1:88–96. doi: 10.4161/cib.1.1.6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasmussen T.B., Bjarnsholt T., Sindersoe M.E., Hentzer M., Kristoffersen P., Kote M., Nielsen J., Eberl L., Givskov M. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 2005;187:1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daniels R., Vanderleyden J., Michiels J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 2004;28:261–289. doi: 10.1016/j.femsre.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 52.Stoodley P., Wilson S., Hall-Stoodley L., Boyle J.D., Lappin-Scott H.M., Costerton J.W. Growth and detachment of cell clusters from mature mixed-species biofilms. App. Environ. Microbiol. 2001;67:5608–5613. doi: 10.1128/AEM.67.12.5608-5613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson L.R. Microcolony and biofilm formation as a survival strategy for bacteria. J. Theor. Biol. 2008;251:24–34. doi: 10.1016/j.jtbi.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 54.Rinaudi L.V., Giordano W. An integrated view of biofilm formation in rhizobia. FEMS Microbiol. Lett. 2010;304:1–11. doi: 10.1111/j.1574-6968.2009.01840.x. [DOI] [PubMed] [Google Scholar]
- 55.Yoon S.S., Hennigan R.F., Hilliard G.M., Ochsner U.A., Parvatiyar K., Kamani M.C., Allen H.L., DeKievit T.R., Gardner P.R., Schwab U., Rowe J.J., Iglewski B.H., McDermott T.R., Mason R.P., Wozniak D.J., Hancock R.E., Parsek M.R., Noah T., Boucher R.C., Hassett D.J. Development and persistence of anaerobic Pseudomonas aeruginosa biofilms: Relevance to pathogenesis and therapy of cystic fibrosis lung disease. Dev. Cell. 2002;3:593–603. doi: 10.1016/s1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 56.Heydorn A., Ersbøll B., Kato J., Hentzer M., Parsek M.R., Tolker-Nielsen T., Givskov M., Molin S. A statistical analysis of Pseudomonas aeruginosa biofilm development: Impact of mutations in genes involved in twitching motility, cell-to-cell signalling and stationary phase gene expression. Appl. Environ. Microbiol. 2002;68:2008–2017. doi: 10.1128/AEM.68.4.2008-2017.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arevalo-Ferro C., Reil G., Görg A., Eberl L., Riedel K. Biofilm formation of Pseudomonas putida IsoF: The role of quorum sensing as assessed by proteomics. Syst. Appl. Microbiol. 2005;28:87–114. doi: 10.1016/j.syapm.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Labatte M., Queck S.Y., Koh K.S, Rice S.A., Givskov M., Kjelleberg S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 2004;186:692–698. doi: 10.1128/JB.186.3.692-698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynch M.J., Swift S., Kirke D.F., Keevil C.W., Dodd C.E.R., Williams P. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 2002;4:18–28. doi: 10.1046/j.1462-2920.2002.00264.x. [DOI] [PubMed] [Google Scholar]
- 60.Huber B., Riedel K., Hentzer M., Heydorn A., Gotschlich A., Givskov M., Molin S., Eberl L. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology. 2001;147:2517–2528. doi: 10.1099/00221287-147-9-2517. [DOI] [PubMed] [Google Scholar]
- 61.Jackson C.R. Changes in community properties. Turing microbial succession. Oikos. 2003;101:444–448. [Google Scholar]
- 62.Schembri M.A., Christianesen G., Klemm P. FimH-mediated autoaggregation of E. coli. Mol. Microbiol. 2001;41:1419–1430. doi: 10.1046/j.1365-2958.2001.02613.x. [DOI] [PubMed] [Google Scholar]
- 63.Nikitina V.E, Ponomareva E.G., Alenkina S.A., Konnova S.A. The role of cell-surface lectins in the aggregation of azospirilla. Microbiology. 2001;70:471–476. [PubMed] [Google Scholar]
- 64.Chandler D.E., Gumbart J., Stack J.D., Chipot C., Schulten K. Membrane curvature induced by aggregates of LH2s and monomeric LH1s. Biophys. J. 2009;97:2978–2984. doi: 10.1016/j.bpj.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Puskas A., Greenberg E.P., Kaplan S., Schaefer A.L. A quorum sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 1997;179:7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chandler J.R., Breck J.R., Duerkop A., Hinz A., Eoin West T., Herman J.P., Churchill M.E.A., Skerrett S.J., Greenberg P.E. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J. Bacteriol. 2009;191:5901–5909. doi: 10.1128/JB.00591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Atkinson S., Throup J.P., Stewart G.S., Williams P. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol. Microbiol. 1999;33:1267–1277. doi: 10.1046/j.1365-2958.1999.01578.x. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Z., Pierson L.S., III A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 2001;67:4305–4315. doi: 10.1128/AEM.67.9.4305-4315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X., Jia J., Popat R., Ortori C.A., Li J., Diggle S.P., Gao K., Cámara M. Characterisation of two quorum sensing systems in the endophytic Serratia plymuthica strain G3: Differential control of motility and biofilm formation according to life-style. BMC Microbiol. 2011;11 doi: 10.1186/1471-2180-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bogino P., Banchio E., Rinaudi L., Cerioni G., Bonfiglio C., Giordano W. Peanut (Arachis hypogaea) response to inoculation with Bradyrhizobium sp. in soils of Argentina. Ann. Appl. Biol. 2006;148:207–212. [Google Scholar]