Abstract
A modular system for detecting protease activity via enzyme-triggered gel formation is described. Protease-specific recognition sequences are utilized to achieve enzyme specificity. Artificial blood clotting is demonstrated by activating endogenous thrombin to trigger gelation in fibrinogen-deficient blood plasma.
The discovery that small peptides can reversibly form gels has sparked intense research into these new biomaterials.1 The gels themselves are being investigated for applications such as tissue engineering2 and lab-on-a-chip platforms.3 The stimuli-responsive nature of the gelation process, on the other hand, is being used in applications including drug delivery4 and regenerative medicine.5 Peptides are advantageous building blocks for all these applications because of their biocompatibility and facile synthesis. Moreover, peptide-based gels can be triggered to form (or breakdown) by simply changing the pH, temperature, or ionic strength.
Because many diseases are correlated with overactive and/or overexpressed enzymes, a simple visual assay based on enzyme-triggered gelation of peptides is appealing for both detection and diagnosis.6 Although several enzyme-triggered gelations have been reported over the last decade,7 few systems are suitable for detecting physiological concentrations of enzymes. For example, Xu and co-workers developed several gelations based on a phosphatase-triggered dephosphorylation reaction, which induces gelation by changing the peptide solubility.8 These systems are not suitable for detection, however, because the reaction is not selective due to the inherent promiscuity of phosphatase enzymes (i.e., most tyrosine phosphatases will trigger gelation) and non-physiological concentrations of enzymes were needed. In a different example, Xu and co-workers were able to use physiologically relevant enzyme concentrations in a β-lactamase-triggered gelation.9 Nevertheless, the system was not demonstrated to be generalizable to other enzymes. The only reported enzyme-triggered gelation that is suitable for sensing involves a protease-triggered gelation using matrix metalloproteinase-7 (MMP-7), a protein that is over-expressed in some cancers.10 Goto and co-workers used a recognition sequence to achieve selectivity and demonstrated that gelation could be triggered under physiologically relevant concentrations.10 The limitation of this system is that the recognition sequence remains on the gelator after the cleavage event. This requirement complicates the application to other proteases because each protease has its own unique recognition sequence and a novel gelator containing the recognition sequence must be identified for every protease of interest. Given that switching a single amino acid residue within a peptide can disrupt gelation,11 this requirement will significantly limit the overall utility of this method.
We hypothesized that an enzyme-triggered cleavage at the N-terminus of a peptide gelator, which separates the recognition sequence from the gelator, would represent a general and modular strategy for detecting enzymatic activity via gelation (Scheme 1). The recognition sequence provides specificity and optimizing the gelator structure should enable detection of physiologically relevant enzyme concentrations. Proteases were selected in this study based on their important role in many biological processes, including blood clotting, apoptosis, and pathogenesis.12
Scheme 1.
Design strategy of a modular sensor for enzymatic activity based on gelation.
The first challenge was to identify a peptide-based gelator with an N-terminal amine. To avoid complications arising from protonation of the amine, p-aminobenzoic acid (PABA), which has an approximate pKa of 5, was selected as the first building block of the peptide-based gelator. At neutral pH, this amine remains uncharged. In addition, peptides containing a related anilide linkage have been successfully cleaved by proteases.13
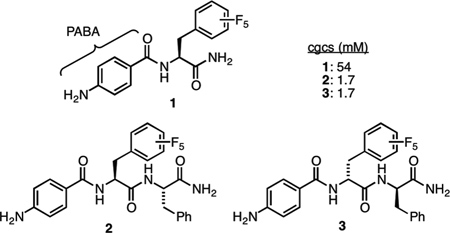
Several different hydrophobic amino acids were then appended to PABA and the resulting compounds were screened for gelation in buffer (100 mM PBS, 10% DMSO, pH = 7.2) using the heat/cool method (ESI). Of these, PABA-F5Phe-NH2 (1) was found to form gels at neutral pH.14 Nevertheless, the critical gel concentration (cgc) was 54 mM, which is insufficient for detecting physiological concentrations of enzyme. To lower the cgc, an additional amino acid residue (Phe) was appended and the resulting compound (2) was found to form gels in buffer (100 mM PBS, 10% DMSO, pH = 7.2) with a cgc of 1.7 mM. A similar dipeptide was then prepared, this time with the unnatural D-amino acids (3). The rationale was that this dipeptide would be more resistant to proteolysis while maintaining an identical ability to gel.15 The resulting gel was comprised of nanometer-sized fibers and exhibited characteristic viscoelastic behavior under external stress and strain, similar to other peptide-based gels (ESI).1
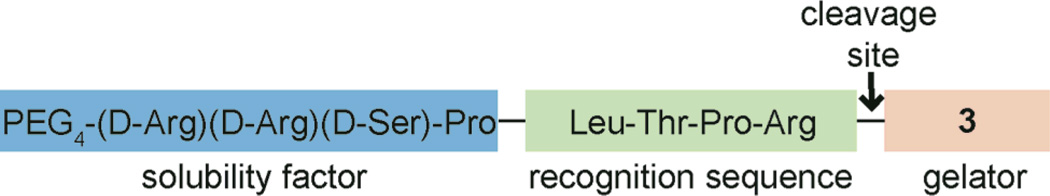
Thrombin, a blood-clotting enzyme, was initially selected because its recognition sequence is well defined, and the enzyme is both specific and highly active.16 To increase solubility and prevent gelation, an oligo(ethylene glycol) unit and three hydrophilic D-amino acids were added to the N-terminal end of the recognition sequence. D-Amino acids were again selected for their resistance to proteolysis.15 The resulting compound (4) did not form a gel under any conditions examined and was soluble in buffer (100 mM PBS, 10% DMSO, pH = 7.2) to concentrations greater than 12 mM (Scheme 2).
Scheme 2.
Thrombin-selective compound 4.
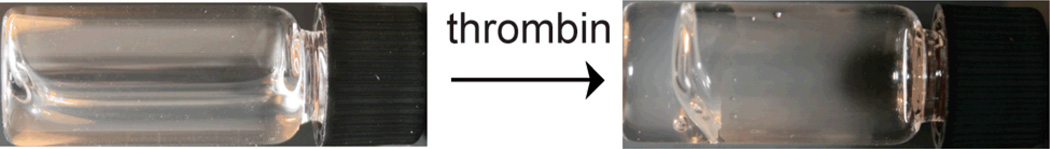
To test the enzyme-triggered gelation, compound 4 (4.4 mM) was treated with thrombin (50 nM) in buffer (400 µL, 100 mM PBS, 10% DMSO, pH = 7.2). Within 10 min, a translucent gel was observed. Aliquots were taken at various times and analyzed by LC-MS to determine conversion. These studies revealed that approximately 40% of compound 4 had been cleaved when gelation occurred, which corresponds to about 1.7 mM of released 3 (ESI). It is important to note that off-target cleavage products were not observed by LC-MS, indicating that the enzyme-triggered gelation is specific to the recognition sequence. The lowest concentration of thrombin that we could detect using this method was 400 pM (ESI). Of note, physiological concentrations of thrombin in the body are low nanomolar (0.1–3 nM), indicating that our system functions at physiological concentrations.17 Adding a thrombin inhibitor (1 mM PMSF) led to the expected decrease in the rate of enzymatic cleavage and gelation was not observed even after 24 h. A second control, wherein no thrombin was added, remained a non-viscous solution, indicating that compound 4 is stable under these conditions. Combined, these controls demonstrate that gelation is dependent upon specific thrombin activity.
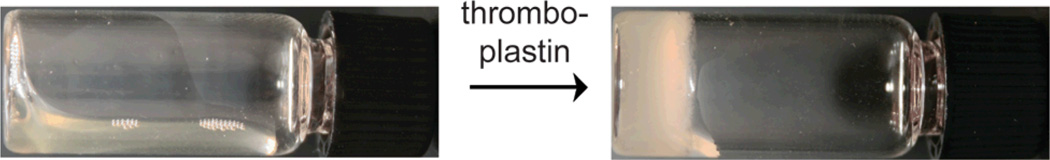
Because thrombin is involved in the blood-clotting cascade,18 we examined whether an artificial clot could be formed via this method. The physiological function of thrombin is to cleave fibrinogen into fibrin, which triggers blood clot formation.18 While rare, deficiencies in fibrinogen can lead to fatal hemorrhages.19 We anticipated that our gelator could be used to cause artificial clotting in patients with low fibrinogen levels. Indeed, an opaque gel was observed within 30 min of adding compound 4 and thrombin (59 nM) to fibrinogen-depleted human blood plasma, indicating that gelation can occur in this complex medium. Control samples missing either compound 4 or thrombin failed to form a gel after 24 h (ESI). The fibrinogen-deficient plasma contains endogenous thrombin, in the form of unactivated prothrombin (coagulation factor II). This endogenous thrombin can be activated by adding either thromboplastin or calcium.18,19 Excitingly, gelation occurred within 2 h of adding thromboplastin and within 20 h of adding calcium to blood plasma containing compound 4 (Figure 2 and ESI). Thus, we were able to create artificial clotting in human blood plasma deficient with fibrinogen, a key component of the blood-clotting pathway.
Figure 2.
An opaque gel is observed within 2 h of adding thromboplastin (170 µL of a 1 mg/mL solution) to compound 4 (12 mM) in fibrinogen-deficient blood plasma (130 µL). This photo was taken after 20 h.
This gelation method was designed to be modular and easily extended to other enzymes. To demonstrate the generality of this approach, two additional proteases with highly distinct recognition sequences were targeted: chymotrypsin (Ala-Ala-Pro-Phe)20 and Glu-C (Asp-Ala-Phe-Glu).21 We reasoned that a highly hydrophobic recognition sequence (as with chymotrypsin) and a dianionic recognition sequence (as found in Glu-C) would test the limits of recognition sequences compatible with our system. Using the same gelator and solubility factor, two new compounds were prepared (ESI). In a series of experiments highlighted in Figure 3, gelation was observed to be specific to each enzyme based solely on the recognition sequence. For example, chymotrypsin was unable to induce gelation of either the thrombin or Glu-C progelator. Moreover, the enzyme concentrations (50 nM) were physiologically relevant. These results demonstrate that the modular system described herein is quite general to other proteases.
Figure 3.
Each vial contains ~ 4 mM of the indicated compound and 50 nM enzyme in buffer (400 µL, 100 mM PBS, 10% DMSO, pH = 7.2) where THR = thrombin and CHY = chymotrypsin. Although all gels were stable to inversion after 2 h, these photos were taken after 20 h.
In summary, a modular system for detecting protease activity using an enzyme-triggered gelation has been developed. Important features include the ability to simply change the recognition sequence to target other proteases and the use of physiologically relevant enzyme concentrations. We further demonstrated that artificial blood clots could be triggered in human blood plasma using gel formation. One could imagine using our thrombin-activated gelation in bandages to promote blood clotting where severe trauma has occurred. Furthermore, we anticipate that this system will lead to facile assays for detection and diagnosis of disease-relevant proteases, and these studies are currently underway.
Supplementary Material
Figure 1.
A clear gel is observed within 10 min of adding thrombin (50 nM) to compound 4 (4.4 mM) in buffer (400 µL, 100 mM PBS, 10% DMSO, pH = 7.2).
Acknowledgments
We thank Ms. Anne Juggernauth for assistance with the AFM measurements, the Office of Naval Research (N00014-09-1-0848 to A.J.M.) and National Institutes of Health (R00RR024366 to M.B.S.) for support of this work.
Footnotes
Electronic Supplementary Information (ESI) available: experimental data; gel rheology and morphology characterization data; control experiments. See DOI: 10.1039/b000000x/
References
- 1.For recent reviews of peptide-based gels, see: Adams DJ. Macromol. Biosciences. 2011;11:160–173. doi: 10.1002/mabi.201000316. Truong WT, Su Y, Meijer JT, Thordarson P, Braet F. Chem. Asian J. 2011;6:30–42. doi: 10.1002/asia.201000592. Jung JP, Gasiorowski JZ, Collier JH. Biopolymers. 2010;94:49–59. doi: 10.1002/bip.21326. Adams DJ, Topham PD. Soft Matter. 2010;6:3707–3721. Cui H, Webber MJ, Stupp SI. Biopolymers. 2010;94:1–18. doi: 10.1002/bip.21328.
- 2.For reviews of tissue engineering applications, see: Ryan DM, Nilsson BL. Polym. Chem. 2012;3:18–33. Collier JH, Rudra JS, Gasiorowski JZ, Jung JP. Chem. Soc. Rev. 2010;39:3413–3424. doi: 10.1039/b914337h.
- 3.For a review of hydrogel-based microarrays, see: Ikeda M, Ochi R, Hamachi I. Lab Chip. 2010;10:3325–3334. doi: 10.1039/c004908e.
- 4.For recent examples of drug-delivery, see: Mao L, Wang H, Tan M, Ou L, Kong D, Yang Z. Chem. Commun. 2012;48:395–397. doi: 10.1039/c1cc16250k. Wang H, Yang C, Wang L, Kong D, Zhang Y, Yang Z. Chem. Commun. 2011;47:4439–4441. doi: 10.1039/c1cc10506j. Li X, Li J, Gao Y, Kuang Y, Shi J, Xu B. J. Am. Chem. Soc. 2010;132:17707–17709. doi: 10.1021/ja109269v. Zhao Y, Tanaka M, Kinoshita T, Higuchi M, Tan T. J. Controlled Release. 2010;147:392–399. doi: 10.1016/j.jconrel.2010.08.002. Zhao F, Ma ML, Xu B. Chem. Soc. Rev. 2009;38:883–891. doi: 10.1039/b806410p.
- 5.For recent reviews on regenerative medicine, see: Matson JB, Stupp SI. Chem. Commun. 2012;48:26–33. doi: 10.1039/c1cc15551b. Matson JB, Zha RH, Stupp SI. Curr. Opin. Solid State Mater. Sci. 2011;15:225–235. doi: 10.1016/j.cossms.2011.08.001.
- 6.For a recent review, see: Razgulin A, Ma N, Rao J. Chem. Soc. Rev. 2011;40:4186–4216. doi: 10.1039/c1cs15035a.
- 7.For recent reviews of enzyme-triggered gelations, see: Zhang Y, Kuang Y, Gao Y, Xu B. Langmuir. 2011;27:529–537. doi: 10.1021/la1020324. Williams RJ, Mart RJ, Ulijn RV. Biopolymers. 2010;94:107–117. doi: 10.1002/bip.21346. Gao Y, Yang Z, Kuang Y, Ma ML, Li J, Zhao F, Xu B. Bipolymers. 2010;94:19–31. doi: 10.1002/bip.21321.
- 8.For recent examples, see: Li X, Gao Y, Kuang Y, Xu B. Chem. Commun. 2010;46:5364–5366. doi: 10.1039/c0cc00163e. Wang Q, Yang Z, Gao Y, Ge W, Wang L, Xu B. Soft Matter. 2008;4:550–553. doi: 10.1039/b715439a. See also: Yang Z, Liang G, Xu B. Acc. Chem. Res. 2008;41:315–326. doi: 10.1021/ar7001914. Zhang Y, Kuang Y, Gao Y, Xu B. Langmuir. 2011;27:529–537. doi: 10.1021/la1020324.
- 9.Yang Z, Ho P-L, Liang G, Chow KH, Wang Q, Cao Y, Guo Z, Xu B. J. Am. Chem. Soc. 2007;129:266–267. doi: 10.1021/ja0675604. [DOI] [PubMed] [Google Scholar]
- 10.Koda D, Maruyama T, Minakuchi N, Nakashima K, Goto M. Chem. Commun. 2010;46:979–981. doi: 10.1039/b920359a. [DOI] [PubMed] [Google Scholar]
- 11.Seemingly minor structural changes can convert peptide-based gelators into nongelators. For recent examples, see: Muro-Small ML, Chen J, McNeil AJ. Langmuir. 2011;27:13248–13253. doi: 10.1021/la202702r. Wang H, Yang C, Tan M, Wang L, Kong D, Yang Z. Soft Matter. 2011;7:3897–3905. Ma M, Kuang Y, Gao Y, Zhang Y, Gao P, Xu B. Am. Chem. Soc. 2010;132:2719–2728. doi: 10.1021/ja9088764. Adams DJ, Mullen LM, Berta M, Chen L, Frith WJ. Soft Matter. 2010;6:1971–1980. Chen L, Revel S, Morris K, Serpell LC, Adams DJ. Langmuir. 2010;26:13466–13471. doi: 10.1021/la102059x.
- 12.For recent reviews of the therapeutic relevance of proteases, see: Drag M, Salvesen GS. Nat. Rev. Drug Discovery. 2010;9:690–701. doi: 10.1038/nrd3053. Turk B. Nat. Rev. Drug Discovery. 2006;5:785–799. doi: 10.1038/nrd2092.
- 13.Carl PL, Chakravarty PK, Katzenellenbogen JA, Weber MJ. Proc. Natl. Acad. Sci. U.S.A. 1980;77:2224–2228. doi: 10.1073/pnas.77.4.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The influence of halogen substituents on Fmoc-Phe was previously reported. For reference, see: Ryan DM, Anderson SB, Nilsson BL. Soft Matter. 2010;6:3220–3231. Ryan DM, Anderson SB, Senguen FT, Youngman RE, Nilsson BL. Soft Matter. 2010;6:475–479.
- 15.Miller SM, Simon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH. Drug Dev. Res. 1995;35:20–32. [Google Scholar]
- 16.Coughlin SR. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 17.Shuman MA, Levine SP. J. Clin. Invest. 1978;61:1102–1106. doi: 10.1172/JCI109010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davie EW, Fujikawa K, Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 19.(a) Rijken DC, Lijnen HR. J. Thromb. Haemostasis. 2009;7:4–13. doi: 10.1111/j.1538-7836.2008.03220.x. [DOI] [PubMed] [Google Scholar]; (b) Falati S, Gross P, Merrill-Skoloff G, Furie BC, Furie B. Nat. Med. 2002;8:1175–1180. doi: 10.1038/nm782. [DOI] [PubMed] [Google Scholar]
- 20.Bergmann M, Fruton JS. J. Biol. Chem. 1937;118:405–415. [Google Scholar]
- 21.Drapeau GR, Boily Y, Houmard J. J. Biol. Chem. 1972;247:6720–6726. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.