Summary
Innate immunity relies entirely upon germ-line encoded receptors, signalling components and effector molecules for the recognition and elimination of invading pathogens. The fruit fly Drosophila melanogaster with its powerful collection of genetic and genomic tools has been the model of choice to develop ideas about innate immunity and host–pathogen interactions. Here, we review current research in the field, encompassing all layers of defence from the role of the microbiota to systemic immune activation, and attempt to speculate on future directions and open questions.
Keywords: innate immunity, Drosophila, host defence
2. Introduction
The study of Drosophila immunity was initiated at Umeå University, Sweden in the laboratory of microbiologist Hans Boman. In their seminal study, Boman et al. [1] clarified very early the humoral nature of the response, its inducibility and lack of specificity. It soon became apparent, however, that before the genetic backdrop of the response could be explored, it would be necessary to purify the factors responsible for this immune response. Because of its size Drosophila was not a good model in which to do this, so for the next 15 years Boman, co-workers and alumni of his research team started to investigate the giant silk moth Hyalophora cecropia opening the molecular era for the field of insect immunity (see [2,3] as examples of their work). Some of the tenants of this inducible immune reaction were found to be secreted antimicrobial peptides (AMPs), several classes of which were subsequently cloned and studied in several other species of Lepidoptera and Diptera (see [4] for review). It was still Drosophila, however, that gave the impetus to study in-depth defence reactions in insects and relate them to mammals. AMP gene promoters contained NF-κB binding sites, crucial for their induction [5,6] and Drosophila Toll controlled AMP gene expression through NF-κB [7]. Following this finding, the hypothetical receptors that Charles Janeway postulated being mediators of innate immunity were found to be homologues of Toll [8,9] a finding that not only re-defined the field of innate immunity as a whole, but also placed its evolution under a new perspective. Below, we attempt a current synthesis of Drosophila immunity highlighting its enormous progress as well as pinpoint some of the challenges that remain ahead.
3. Where does infection come from?
Like all organisms, insects live in a world containing an almost unquantifiable amount of micro-organisms. Some insects, however, are exposed considerably more than the average organism as they feed, lay their eggs and develop on decomposing media. These insects include Drosophila where part of its microbial load is introduced in the gut through the digestive process. Subsequently, a part of the digested microbes reach and may colonize the gastrointestinal epithelial wall. These micro-organisms may then become part of the commensal flora or induce pathogenicity and systemic immunity. In addition, systemic activation may occur through septic injury by nematodes or by wasps depositing their eggs on fruit fly larvae.
4. Epithelial responses and gut flora
Anatomically, the Drosophila gut can be divided into foregut, midgut and hindgut. The upper digestive system is used for food uptake and storage while processing and absorption takes place in the mid and posterior regions of the midgut. In this continuous system typical of higher Diptera, some of the meal is completely processed and defecated before some has even entered the digestive section of the midgut. The availability of gut-specific GAL4 lines combined with the advent of genome-wide RNAi libraries initiated the functional cell biology of the midgut (see below). It soon became apparent that the presence of intestinal stem cells (ISCs) ensures gut homeostasis with the supply of differentiated enterocytes (ECs). A characteristic of ECs is their rapid turnover where apoptotic cells are replaced by the compensatory proliferation of ISCs. ISCs were first described by the Spradling and Perrimon laboratories [10,11]. Similarly to mammals, the Notch, Wingless, platelet-derived growth factor (PDGF), epidermal growth factor (EGF), and insulin receptor pathways have been implicated in the maintenance, proliferation and/or differentiation of ISCs (see [12] for a review). In addition, Hippo signalling is used to restrict stem-cell proliferation in the gut of both Drosophila and mammals [13]. Recently, a controversy in the field was settled by recording the absence of active stem cells but presence of Wingless-expressing cells within the anterior pylorus, the proliferation of which provides homeostasis following serious damage [14].
In parallel to studies of gut physiology, intense investigation has been directed towards the elucidation of the Drosophila microbiota in both laboratory and field populations [15–20]. It was found that Drosophila is harbouring a community of gut bacteria that is much simpler compared with vertebrates and it is now possible to extract and cultivate these bacteria, use them in re-colonization experiments and produce mutants to interrogate host–pathogen interactions (see table 1). Combining functional cell biology and the knowledge of microbiota, several digestive infection models have been developed; these will be summarized below.
Table 1.
Bacterial species associated with life stages of D. melanogaster from laboratory populations and collected from the wild.
| bacterial genera | [15] | [17] | [18] | [20] | [19] | [16] |
|---|---|---|---|---|---|---|
| Acetobacter | +w, l | +l | +w | +l | +w, l | +l |
| Acidovorax | +w | |||||
| Acinetobacter | +w | |||||
| Agrobacterium | +w | |||||
| Alcaligenes | +w | |||||
| Arcobacter | +w | |||||
| Azospirillum | ||||||
| Bacillus | +? | |||||
| Bordetella | +w | |||||
| Bradyrhizobium | +? | |||||
| Chitinophaga | +? | |||||
| Citrobacter | +? | |||||
| Cladosporium | +l | |||||
| Commensalibacter | +w, l | |||||
| Corynebacterium | +w | |||||
| Dysgonomonas | +w, l | |||||
| Enterobacter | +? | +w | +w, l | |||
| Enterococcus | +w, l | +w | +w | |||
| Erwinia | +? | +w | ||||
| Frateuria | +w | |||||
| Gluconacetobacter | +w | +l | ||||
| Gluconobacter | +? | +w | ||||
| Klebsiella | +? | |||||
| Lactobacillus | +? | +l | +w | +l | +w, l | +l |
| Leuconostoc | +? | +w | ||||
| Morganella | +? | |||||
| Pantoea | +? | + w | ||||
| Providencia | +w | +w, l | ||||
| Pseudomonas | +? | +w | ||||
| Serratia | +? | +w, l | ||||
| Shigella | +w, l | |||||
| Spiroplasma | +w | |||||
| Staphylococcus | +? | +l | +w | +l | ||
| Stenotrophomonas | +? | +w | ||||
| Vagococcus | ||||||
| Weissella | +? | |||||
| Wolbachia | +w | +w | +l | +w |
4.1. Commensal bacteria
The first observation of the possible role of flora to the development of Drosophila occurred more than 40 years ago. Bakula observed that axenic cultures of Drosophila larvae showed elongated developmental times [21]. Many years later, Brummel et al. [22] showed that the lifespan of adult flies under axenic conditions was reduced and that reintroducing bacteria during the first week of adult life could restore wild-type longevity. Bacterial flora seems to be necessary for optimal larval development upon nutrient scarcity. Lactobacillus plantarum is sufficient on its own to recapitulate the natural microbiota growth-promoting effect. Lactobacillus plantarum exerts its benefit by acting genetically upstream of the target of rapamycin (TOR)-dependent host nutrient sensing system controlling hormonal growth signalling [23].
Recently, Shin et al. [24] attempted to identify the molecular aspect of the above relationship between the development of the host and the flora. They showed the role of pyrroloquinoline quinone-dependent alcohol dehydrogenase (PQQ-ADH) of the commensal bacterium Acetobacter pomorum interacts with insulin/insulin-like growth factor signalling (IIS) in Drosophila to maintain the gut–microbe mutualism. The modulation of host IIS by the PQQ-ADH defines developmental factors like body size, energy metabolism and ISC activity of the host. Germ-free animals infected by PQQ-ADH-deficient bacteria showed deregulation of developmental and metabolic homeostasis. Both enhancement of the host IIS or enrichment of the diet with acetic acid (the metabolic product of PQQ-ADH) proved capable of reversing the above defects.
Studies above that determined the microbiota also showed the ability of commensal bacteria (like L. plantarum, Lactobacillus brevis, A. pomorum, Enteroccocus faecalis, Gluconobacter sp. and a bacterium in the family Acetobacteraceae, strain A911 of Commensalibacter intestini) to colonize germ-free adults [15–19,24]. In contrast, non-commensal bacteria like Erwinia carotovora carotovora and Escherichia coli did not exhibit the same capacity. Interestingly, the NF-κB homologue Relish (see immune deficiency pathway below) was detected in the nucleus of intestinal cells in the presence of the microbiota [20]. The question was, therefore, how the host manages to maintain low levels of AMP and preserve the structure of its flora. Ryu et al. [20] showed that the intestinally expressed homeobox gene Caudal represses the NF-κB-dependent AMP genes, in this way regulating commensal-gut homeostasis.
4.2. Non-commensal (pathogenic and non-pathogenic) bacteria
In 2000, the first natural bacterial infection of Drosophila larvae revealed the activation of host immune responses by different bacteria of the genus Erwinia [25]. It was the first time that systemic AMP production was recorded using an ingestion model. Importantly, the non-pathogenic strain E. carotovora carotovora-15 (Ecc-15) has proved to be a valuable tool in exploring gut homeostasis. Tzou et al. [26], using the strain Ecc-15, showed that AMP production was following a tissue-specific pattern. For example, diptericin expression in larvae upon infection was observed in the proventriculus and part of the midgut, while no AMP expression was observed in this tissue. Foley & O'Farrell [27] showed the important signalling role of nitric oxide (NO) to innate immunity by using Ecc-15 and E. coli in their feeding experiments. Nitric oxide synthase (NOS) was upregulated upon infection while its inactivation compromised host survival.
In their quest for a bacterium that can naturally infect and kill Drosophila, Bruno Lemaitre's laboratory isolated a previously uncharacterized bacterial species, Pseudomonas entomophila (Pe) that can orally infect and kill Drosophila larvae and adults [28]. The same group sequenced and assembled its genome [29] and interrogated Pe mutants for virulence factors [28,30,31]. From the side of the host, Vodovar et al. [28] showed the importance of an Immune deficiency (Imd)-dependent (see later for Imd signalling) local response against Pe as opposed to systemic immunity underlying the importance of local AMP expression against food-borne pathogens.
Using Serratia marcescens as a pathogenic bacterium Nehme et al. [32] confirmed the induction of both local and systemic immune responses and the importance of the consequent Imd-dependent local AMPs production to fight off infection. The availability of RNAi strains for more than 90 per cent of the Drosophila genome directed Cronin et al. [33] to follow a genome-wide in vivo RNAi screen revealing host genes involved in susceptibility or resistance to intestinal infection with S. marcescens. Applying whole-organism and tissue-specific knock down these authors uncovered that the JAK-STAT signalling pathway participated in intestinal defence by regulating stem cell proliferation. Participation of the JAK-STAT pathway along with Imd in gut immunity was also confirmed by conducting oral infections with Ecc-15 [31]. This study showed that gut homeostasis includes inflection of the stress response, increased ISC proliferation and epithelia renewal in response to bacterial infection. Using Pe, Jiang et al. [34] showed that activation of JAK-STAT in ISCs was due to the production of cytokines (Upd, Upd2 and Upd3) by ECs in the midgut.
In addition, oral challenge by pathogenic bacteria revealed new information about the effects of the physical barrier of the peritopic matrix (PM), which lines the intestinal lumen. PM forms a layer of chitins and glycoproteins protecting the epithelium from rough food particles and microbes. Infection by Ecc-15 showed that a gene for a putative eye-lens protein called drosocrystallin (Dcy) was strongly up-regulated upon infection but its expression was not controlled by the Imd pathway. The role of Dcy in adult PM formation was recently elucidated. Dcy-deficient flies showed an increased susceptibility to oral infections with the entomopathogenic bacteria P. entomophila and S. marcescens [35].
Experiments in parallel with the above established ingestion models led to the identification of the important role of reactive oxygen species (ROS) in the gut immune response of Drosophila. Oral ingestion of bacteria induces the rapid synthesis of ROS in the gut by an NADPH oxidase called duox oxidase (DUOX). In cases of suppressed DUOX expression, an increased mortality rate upon minor infection in adults is recorded [36,37]. A signalling network that controls both positively and negatively the expression and activity of DUOX, important for the host response to commensal and pathogenic bacteria, was thus identified [38].
4.3. Fungi
Ingestion of Cryptococcus neoformans caused the death of the fly in contrast to the injection of Saccharomyces cerevisae or the nonpathogenic Cryptococcus kuetzingii or Cryptococcus laurentii. The Toll pathway did not show any role in Drosophila adult defense upon ingestion of C. neoformans [39]. However, Toll showed important roles to both clearance of C. neoformans cells and survival of adults after systemic infection by the yeast [39]. Recently, our laboratory developed a Drosophila model to study Candida albicans gastrointestinal (GI) infection [40]. Candida albicans GI infection caused extensive JNK-mediated death of gut cells and induced systemic activation of AMP activity in the larval fat body. Both phenomena were partially mediated through fungal proteases. From the side of the host, NO and blood cells influenced systemic AMP responses. The system is now ready for isolating both pathogen and host factors that influence gut pathogenesis and activation of systemic immunity.
The above, as well as parallel studies, have emphasized the integration of gut responses, blood cells and AMP systemic immunity in host defence through both paracrine and autocrine signals recently involving TGF-β signalling and tissue-specific regulation of AMPs by FOXO and Drifter/ventral veinless [41–43].
5. Layers of host defence in systemic immunity
5.1. Haemocytes
Drosophila counters systemic infection through the wide-ranging action of haemocytes, considered as the insect equivalent to vertebrate blood cells. Recent studies along with a classic paper by Hartenstein and colleagues [44] have delineated the ontogeny of these cells from embryonic development (plasmatocytes and crystal cells) to larval stages, where they persist and form circulating and sessile subpopulations, and then through metamorphosis to adults (for review, see [45]). Following the first phase of haematopoiesis in embryos, there is a second phase in larvae directed by a specialized compartmentalized organ situated in the dorsal aorta, namely the lymph gland (for review, see [46]). This organ contains progenitors (pro-haemocytes) for three types of functional haemocytes including the plasmatocytes, which are monocyte-like cells involved in phagocytosis of apoptotic bodies and pathogens, and crystal cells, which are required for melanization (see below). These two haemocyte types are released in the haemolymph upon dispersal of the lymph gland at the onset of the larva to pupa transition. The haematopoietic organ also gives rise to a third type of haemocyte, the lamellocyte, devoted to encapsulation of foreign bodies that are too large to be phagocytosed. Lamellocytes do not differentiate in normal developmental conditions but only in response to specific immune challenges such as wasp parasitism or stress conditions mediated by an increase of ROS. Mutant backgrounds with increased haemocyte proliferation lead to formation of ‘melanotic tumours’ that result from encapsulation of larval tissue by lamellocytes. In this context, large-scale screens to identify melanotic-tumour-suppressor genes have been published uncovering new genes and gene networks controlling haemocyte homeostasis [47–49].
One question that has long remained unanswered in the field was the possible interconnectedness of haemocyte responses to fat-body-directed AMP gene regulation. An early study proposed there was no such connection [50]. These results were based on the use of the domino (dom) mutant, which lacked more than 90 per cent of circulating haemocytes and a similar proportion of the sessile subpopulation [50]. Dom is a member of the SWI2/SNF2 family of DNA-dependent ATP-ases functioning as a global transcriptional regulator of proliferative tissues [51]. Larvae, carrying strong dom mutant alleles died in late larval/early pupal stages in the absence of infection and earlier when infected [51]. However, the experimental set-up precluded use of those early larvae including only those that survived immune challenge for measuring AMP gene expression, which was found to be comparable to wild-type larvae [50]. One additional caveat of the analysis was the general effect the mutation had on cell proliferation in many tissues other than haemocytes. Nevertheless, dom mutants failed to induce diptericin during Gram-negative GI infection [25], suggesting that blood cells could relay a signal emanating from the gut to activate the Imd pathway that controlled diptericin expression in the fat body. This signal may be NO as both bacterial and fungal GI infection need haemocytes to relay the NOS-generated signal to the fat body and induce systemic activation of AMP gene expression [27].
Additional evidence for the contribution of blood cells towards fat body antimicrobial responses came with the description of psidin by Brennan et al. [52]. Identified in a genetic screen for mutants with a reduced AMP response, psidin encodes a lysosomal protein required in haemocytes for degradation of engulfed bacteria as well as expression of the AMP gene defencin in the fat body, establishing thus a connection between pathogen detection by phagocytes and fat body AMP gene induction. This led to the proposition that haemocytes were internalizing and subsequently presenting non-self antigens to fat body cells [52], shifting the debate from whether there was a connection between haemocytes and fat body to whether the connection was antigen presentation or secreted signal(s). A problem with the Brennan paper, however, was that the rescue of the mutant with a wild-type copy of psidin was performed using peroxidasin-GAL4, which is also expressed in the fat body [53]. Therefore, it may be that psidin is needed in both tissues although the authors detected only expression of psidin in haemocytes [52].
Three studies published in 2009 redressed the debate by following a different approach. This was to genetically eliminate plasmatocytes by targeting their apoptosis through forced expression of pro-apoptotic genes [53–55]. It was found that haemocytes were indispensable for embryonic development [54] but surprisingly, their absence did not influence post-embryonic development [53–55]. This was interesting given the belief that haemocytes participated in extensive tissue remodelling during pupariation and reinforced the argument that larval lethality seen in dom and psidin mutants was not linked to blood cells but to other tissues. Haemocyte-ablated larvae were unable to mount a full systemic response following GI infection [53], while larval responses to systemic challenge were also dependent on the presence of haemocytes [53]. Silencing the Toll ligand spz in haemocytes produced the same result, namely, the significant reduction of Toll-dependent AMP responses [53]. Spz expressed by haemocytes could have both a paracrine as well as an autocrine function in AMP induction and is the first signal identified in the crosstalk between haemocytes and fat body in larvae. Evidence from a parallel study gave impetus to the idea of Spz as a pro-inflammatory cytokine in a feedback between haemocytes and fat body [56].
In contrast to larvae, absence of blood cells did not influence AMP gene induction in adults [54,55]. Haemocyte-deficient flies were significantly more susceptible to infection owing to the absence of phagocytosis, confirming early experiments which used latex beads to saturate the phagocytic machinery [54,55]. The fact that recent studies have shown that phagocytosis and AMP induction (through the Toll pathway, for example) had additive effects [57] but did not influence each other, indicated that, in adults, these are two independent systems which nevertheless act together to fight off infection. The idea, however, of the ‘internal milieu’ [58] and how immune homeostasis is indeed a result of metabolism interacting with other processes through secreted signals, has been explored in significant work implicating the effect of insulin signalling in Mycobacterium marinum infection [59]. In addition, recent work has shown that TGF-β signals emanating from specific subsets of adult haemocytes modulate infection-induced melanization and AMP gene expression in time [41]. The relation, therefore, between haemocytes and fat body in both larvae and adults remains an evolving picture.
5.2. Phagocytosis
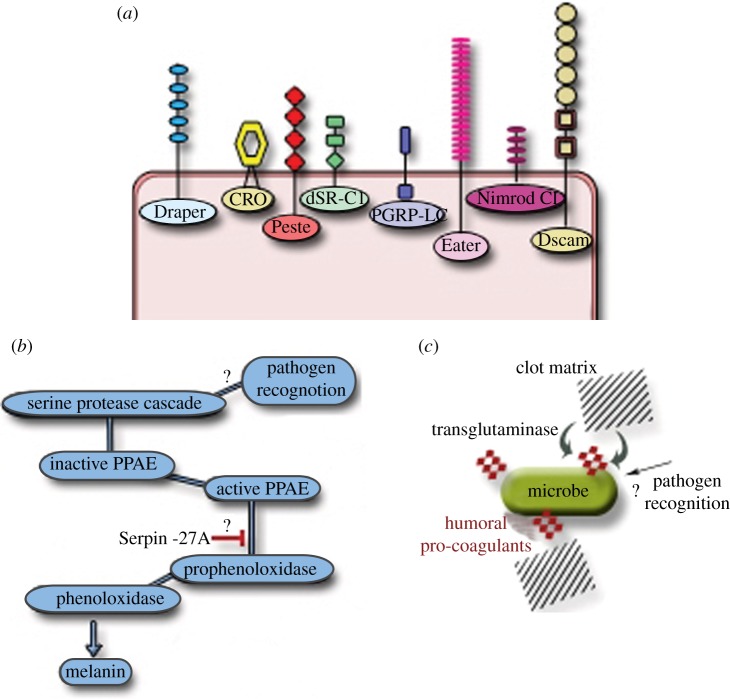
One of the most powerful and immediate ways for fruit flies to eliminate apoptotic bodies, bacterial infection or fungal spores in the haemolymph is by their removal through receptor-mediated recognition and phagocytosis. Drosophila phagocytes have been used as a model for ‘professional’ mammalian phagocytosis (for review see [60]). This is because, during development, dead cells are recognized by evolutionary-conserved receptors such as Croquemort (CRO, the CD36 paralogue) [61] and Draper (the LPS recognition protein (RP) paralogue) [62], although the latter also recognizes lipoteichoic acid from Staphylococcus aureus and mediates uptake of this bacterium [63]. Studies of Drosophila S2 cells, which share many features with mammalian macrophages and are amenable to RNAi, identified phagocytic receptors relevant to host immunity, such as members of the scavenger receptor family Peste and dSR-C1 [64,65], peptidoglycan PGRP-LC [66], members of the Nimrod family of proteins Eater [67] and Nimrod C1 [68] and the IgSF-domain protein Dscam [69]. A summary of these receptors is schematically presented in figure 1a. However, the question of which components of the bacterial cell wall are recognized, and how, by these receptors is still open (for PGRP-LC see below). Nonetheless, significant advances have been made in the elucidation of intracellular signalling and actin regulation [70]. Measurements of time needed to eliminate pathogens by phagocytosis have resulted in describing an impressive capacity: systemically infected larvae with 3000 bacteria can eliminate almost 95 per cent of them in 30 min [53]. It is some hours later that AMP gene expression peaks and therefore a pertinent question was why larvae need AMPs at all. An interesting proposition came not from Drosophila but from Tenebrio molitor where the same time-course was observed in adults [71]. Rolf and co-workers proposed that the timing was crucial in order for AMPs to ‘meet’ a dramatically reduced number of bacteria and thus diminish the possibility for induction of resistance [71]. Moreover, their sustained expression and presence in the haemolymph long after the infection was cleared provided protective immunity.
Figure 1.
Layers of Drosophila immunity: (a) receptors found on the surface of Drosophila macrophages, (b) schematic of the melanization reaction and (c) coagulation. The link to pathogen recognition in both (b) and (c) still remains elusive. PPAE, pro-phenoloxidase activating enzyme.
5.3. Melanization
This is considered to be the earliest and most acute reaction of insects against pathogens breaching the cuticle and invading through septic injury. It is visible by the blackening of the wound site and the surface of the pathogen and is used to encapsulate and sequester pathogens too large to be phagocytosed, as seen with mosquito responses against the malaria parasite [72]. In addition, the intermediates of the reaction are directly toxic to microbes (for review, see [73]). In Drosophila, however, there was literature disputing the importance of melanization in fighting off infection [74,75]. Yet, a significant paper [76] showed elegantly through infection with various Gram-positive and Gram-negative bacteria, which induce strong systemic melanization in fruit flies, that melanization has a considerable impact on host survival following immune challenge. Knock down (or knock out) of one player in the proteolytic cascade leading to melanization (MP2; see below) was sufficient to significantly modulate survival after infection by either increasing susceptibility or augmenting tolerance [76]. Interestingly, even in the cases where there was no change in host survival there was a significant increase in bacterial load suggesting a different balance between resistance and tolerance [76]. An alternative interpretation of course could be that MP2 has roles additional to melanization as has been previously suggested [75].
Mechanistically, melanin synthesis is the final product of this proteolytic cascade involving the sequential activity of serine proteases MP1 and MP2, leading to the cleavage of prophenoloxidase (proPO) to phenoloxidase ([75]; see also figure 1b). The Drosophila genome encodes three proPOs, two expressed in crystal cells (DoxA1 and CG8193) and one in lamelocytes (DoxA3) [77]. Activation of melanization is inhibited by Serpin-27A [78,79]. Although the target of Serpin-27A is thought to be prophenoloxidase activating enzyme as it inhibits the relevant beetle enzyme in vitro [79] the endogenous target of Serpin-27A is not known. An additional open question is the link between pathogen recognition and activation of the cascade. There is very detailed biochemical work in other insects (see [80,81]) but in Drosophila, where in vivo work is possible, these links have not been established.
5.4. Coagulation
An additional layer of innate responses to restrict pathogen dissemination from a wound is the process of haemolymph clotting. In the clot, various proteins form characteristic filaments which cross-link the bacteria and prevent their spread. Experiments following this early reaction in vitro indicated that initial clot formation was independent of melanization since it happened in proPO mutants [82]. In vivo, however, larvae lacking crystal cells had a reduced ability for clot formation and decreased capacity for wound healing [83]. These results showed that proPO may not be crucial for the formation of the clot per se but is important for the hardening of the larval coagulum as well as for healing a septic injury. Proteomic analysis has identified several proteins involved in clotting [84]. These proteins include Hemolectin, a large protein and a major component of the clot, produced by plasmatocytes [85]; the humoral pro-coagulants lipophorin, hexamerin and its receptor (also called fat body protein 1) [84]; Fondue, a haemolymph protein with its production regulated by Toll, which is not involved in initial clot formation but in cross-linking of clot fibers [86]; and Transglutaminase (TG), providing the connection between bacterial surfaces and the clot matrix [87]. TG binding was observed in a variety of bacterial surfaces although TG RNAi affected host survival in a limited number of infections [87]. The presence and role of TG, however, is widely conserved and has been shown to contribute to clot formation in almost every species where clotting has been studied in any detail (see [88] for review), suggesting that there might be qualitative differences in the binding of TG to different bacterial surfaces that ultimately produce differences in host survival. Whether the process of TG binding to microbial surfaces, which in turn aids clot matrix and pro-coagulant assembly to entrap pathogens, is connected to pathogen recognition is not yet clear. Conceptually, both microbial surface components and host–pathogen recognition receptors could serve as substrates for TG (summarized in figure 1c).
5.5. Fat-body-dependent antimicrobial peptide gene induction
Fat-body-dependent AMP gene induction, the hallmark of the systemic response, is the synthesis and secretion in the haemolymph of powerful effector molecules collectively known as AMPs. These are mostly small cationic peptides that directly attack the cell wall of microbes [4]. The cloning and characterization of their promoters paved the way to a series of now classic papers (see below) revealing the signalling pathways that controlled AMP gene expression, starting with the discovery that AMP gene expression was regulated by NF-κB promoter elements (see [5,6] as examples of this work).
6. Signalling in systemic immunity
6.1. The Toll pathway
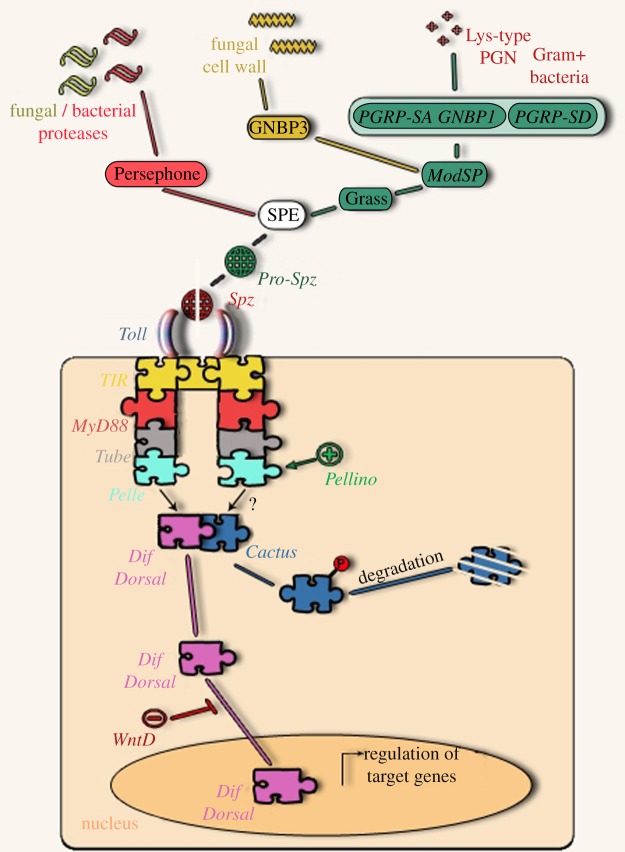
In contrast to its mammalian counterparts, Drosophila Toll is not activated by direct interaction with microbial molecules but through an endogenous ligand, namely the Nerve Growth Factor-related cytokine Spätzle (Spz) [89]. Binding is achieved by two Spz dimers, each interacting with the N terminus of one Toll molecule. This triggers a conformational change in what is now a dimeric Toll receptor, to activate downstream signalling [90]. Spz is in turn activated to bind to Toll via proteolytic cascades, which culminate in processing of its N-terminal pro-domain by the Spz-activating enzyme (SPE) [91]. It is still an open question whether the Spz pro-domain is separated from the hydrophobic C-106 domain when cleaved, as has been suggested in vivo [92], or remains attached through disulphide bonds, as seen in biochemical experiments, to be finally displaced when bound to Toll [93]. SPE is the point where pathogen recognition information is integrated through the activation of three recognition pathways: one triggered by fungal or bacterial proteases that directly activate the host serine protease Persephone [94,95], which in this context acts as a sensor of virulence [95,96]; one induced by recognition of fungal cell wall [96]; and one activated by Lysine (Lys)-type bacterial PG (see below). Both these last two recognition pathways converge to the modular serine protease (ModSP) [97], which in turn activates—not directly—the serine protease Grass [97,98]. Proteases Spirit, Spheroide and Sphinx1/2 were also identified as necessary for a host responding to both fungi and Gram-positive bacteria [98].
The recognition events that initiate the ModSP-Grass-SPE axis are mediated by two PGRPs, namely PGRP-SA and PGRP-SD and the glucan-binding protein GNBP1 [100,101]. These three molecules recognize Lys-type PG, a major component of Gram-positive bacteria [101]. Upon recognition, PGRP-SA and GNBP1 physically interact, forming a complex [101]. We have found that depending on the extent of PG cross-linking GNBP1 acts as an endomuramidase hydrolysing Lys-type PG with low cross-linking thus producing new glycan reducing ends, which are presented to PGRP-SA [102,103]. In contrast, Buchon et al. [97] suggested that full-length GNBP1 had no enzymatic activity. Crucially, however, these authors did not test the functionality of their recombinant GNBP1 in rescuing the relevant mutant, an important element when relating biochemical data to an in vivo hypothesis. Nevertheless, they suggested a (not mutually exclusive) role for GNBP1 as a linker between PGRP-SA and ModSP [97]. PGRP-SD functions as a receptor for Gram-positive bacteria with partial redundancy to the PGRP-SA–GNBP1 complex [98]. A pertinent question nevertheless is how a relatively small number of proteins recognize the vast variability in the cell wall of Gram-positive bacteria and how PG is even accessible as it is ‘buried’ under various cell-wall glycopolymers and bulky modifications. A strategy could be the use of more than one PGRP and/or various layers of different responses (see above) all linked to pathogen recognition.
Our results indicate that when accessibility to PG in the bacterial cell wall is not blocked by glycopolymers such as teichoic acids, then PGRP-SD becomes redundant [104]. It is interesting to note that when teichoic acids are not present in the bacterial cell wall the Toll pathway (but not PGRP-SA itself) becomes redundant as well, indicating that PGRP-SA has Toll-independent functions [104]. The glucan-binding GNBP3 is responsible for yeast recognition [96] and its N-terminal domain has been the only GNBP family of proteins with a crystal structure [105], revealing an immunoglobulin-like fold in which the glucan-binding site is masked by a loop. This loop is displaced during binding representing a novel mechanism for beta-glucan recognition [105].
Following Spz–Toll interaction a receptor–adaptor complex that will transmit the signal from the cell surface to the nucleus is formed. This complex comprises the MyD88 protein, which interacts with Toll through their respective Toll/Interleukin-1 receptor domains [106] and connects with Tube via death domain contacts that will in turn recruit the Drosophila IRAK homologue, the kinase Pelle [107]. The latter will directly or indirectly phosphorylate the IκB homologue Cactus, which is thus targeted for degradation. Upon Cactus degradation, the NF-κB homologues Dorsal or Dif are free to move to the nucleus and regulate hundreds of target genes [108,109]. A positive regulator of the pathway is the RING-domain containing Pellino, acting presumably at the level of Pelle in parallel to mammalian Pellinos that modulate IRAK action [110]. In contrast, a negative regulator is WntD, which reduces Toll activity by preventing translocation of Dorsal to the nucleus [111]. In addition, it has recently been shown that endocytosis is paramount for efficient Toll signalling [112]. A schematic summary of Toll pathway signalling is presented in figure 2.
Figure 2.
Summary of Toll signalling; see text for details.
6.2. The immune deficiency pathway
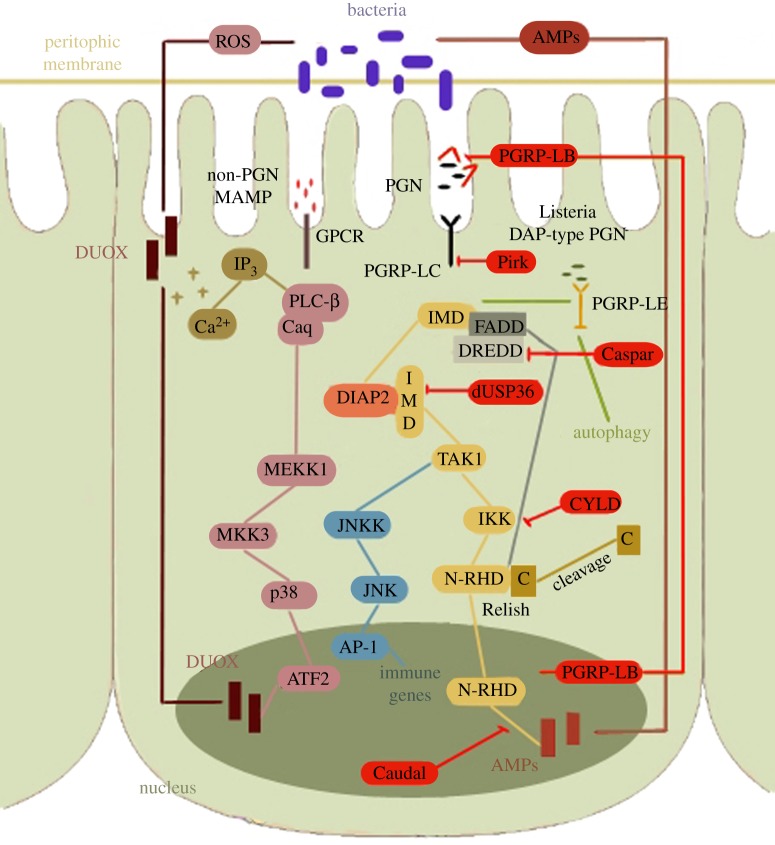
In addition to Toll there is another pathway, which is primarily activated by DAP-type bacterial PG, namely the immune deficiency (IMD) pathway (see figure 3 for summary of both systemic signalling and network of gut defences). DAP-type PG forms the cell wall of Gram-negative bacteria as well as some Gram-positive Bacilli [113]. Pathogen recognition in IMD occurs through the transmembrane PGRP-LC and the intracellular PGRP-LE [114,115]. PGRP-LC is a type-2 transmembrane receptor, with an extracellular PGRP domain that is critical for recognizing extracellular bacteria, while PGRP-LE lacks a transmembrane domain and functions as an intracellular receptor, although an extracellular cleaved form of PGRP-LE made only of the PGRP domain has also been reported in cell culture [114].
Figure 3.
Summary of gut defences and Imd signalling; see text for details.
Flies deficient in both PGRP-LC and -LE are unable to induce AMPs in response to Gram-negative bacteria, being highly susceptible to these infections [113,114]. PGRP-LC encodes three receptors via alternative splicing, namely PGRP-LCx, PGRP-LCy and PGRP-LCa [116]. All three proteins share the same intracellular signalling domain while the extracellular recognition part is unique for each receptor variant [116]. In contrast to PGRP-LCy, whose role remains unclear, it is well established that the other two PGRP-LC splice forms have important functions in activating IMD. On its own, PGRP-LCx is sufficient to respond to E. coli PG whereas both PGRP-LCx and PGRP-LCa form a heterodimer upon recognition of a monomeric disaccharide–tetrapeptide fragment of PG known as tracheal cytotoxin (TCT) [114]. With an as yet unknown mechanism, TCT is able to enter cells and is then sensed by PGRP-LE. This interaction induces the formation of head-to-tail homo-oligomers of PGRP-LE [117]. In addition, PGRP-LE acts as recognition receptor for intracellular bacteria such as Listeria monocytogenes. In this case, PGRP-LE induces autophagy through an IMD-independent pathway [118] in keeping with the ability of Listeria to trigger such responses in mammalian epithelial cells [119].
Subsequent intracellular signalling is transduced through the RHIM-like motif found in PGRP-LC and -LE [114,115]. However, the molecular mechanism by which the RHIM-like domains in PGRP-LC and -LE regulate signalling is unclear. A protein, which binds both LC and LE, is Imd itself, a death-domain-containing protein with homology to mammalian RIP1 (minus the kinase domain) [120]. In turn, Imd associates with the Drosophila FADD (FAS-associated death-domain protein) homologue via a homotypic death-domain interaction [121]. FADD then recruits and interacts with the homologue of mammalian caspase-8, apical caspase death-related Ced-3/Nedd2-like protein (DREDD) [122], via the death-effector domains found in these proteins [123,124]. It is not known whether recruitment of DREDD to the PGRP–IMD–FADD complex is sufficient for its activation.
DREDD cleaves Imd thus unmasking a domain of interaction of the latter with the Drosophila Inhibitor of apoptosis-2 (dIAP-2) [125]. In its turn, dIAP-2, through its RING domain, ubiquitinates and stabilizes Imd, which then acts as a scaffold for the recruitment of downstream components. It is conceivable that the ubiquitin-specific protease 36 (dUSP36) acts to suppress the pathway by reversing this ubiquitination [126]. Components downstream of Imd are TAK1 [127] and its adaptor TAB2 [128]. It is not yet shown whether TAK1 is recruited in an Imd complex but this seems to be the working hypothesis [125]. Once recruited, TAK1 would trigger activation of the IκB-Kinase (IKK) complex, which in turn phosphorylates the NF-κB protein Relish [129]. Relish is a composite protein made of a C-terminal IκB domain and an N-terminal NF-κB part [130]. DREDD is the most probable protein that mediates Relish cleavage resulting in the uncoupling of two Relish domains, thereby allowing the N-terminal to translocate into the nucleus [129,131]. Although Relish phosphorylation is dispensable for its cleavage, it appears to enhance the activity of Relish as a transcription factor in the nucleus [129]. Separately, TAK1 also activates the JNK kinase, which initiates the phosphorylation and nuclear translocation of the transcription factor AP-1 [132].
As mentioned earlier, the Imd pathway is also involved in gut infection. In this context, a number of negative regulators (both intra- as well as extracellular) have been identified. These include the secreted PGRP-LB [133], which has an amidase catalytic activity cleaving DAP-type PG, limiting availability of ligand for PGRP-LC and thus dampening the Imd signal. Inside cells, a protein interacting with PGRP-LC, namely Pirk, has been shown to negatively regulate the Imd pathway not only in the gut but also during systemic activation [134–136]. Flies lacking Pirk exhibited higher levels of AMPs although a resolution of the response was still observed. However, double pirk;PGRP-LB mutants resulted in a further increase showing the synergy of those two factors to control gut defenses [137]. Additionally, the three members of the PGRP-SC locus negatively regulate the pathway in systemic mode [137] and triple pirk;PGRP-SC;PGRP-LB mutants (where the whole PGRP-SC locus has been deleted) showed low viability and a level of AMPs that was 8 times higher at 24 h post infection compared with wild-type flies [137]. The triple mutant also had compromised life span even in unchallenged conditions suggesting that persistent activation of the pathway (presumably mediated by the gut flora or by ingested bacteria) was deleterious [137]. Another negative regulator of the pathway suggested to act at the level of DREDD is a homologue of the Fas-associated factor FAF-1 [138].
Caspar-deficient flies upregulate AMPs in the absence of immune challenge and are more resistant to bacterial infections [138]. An additional intracellular negative regulator of the Imd pathway is Cylindromatosis (CYLD), probably at the level of IKK [139]. It is intriguing that every step of the intracellular part of the pathway has its own negative regulator; until now only TAK1 has been devoid of such a partner, although POSH has been identified as a protein limiting the amount of activated TAK1 and thus restricting the timing of the JNK branch of the TAK1 signal [140].
7. Emerging complexities in Drosophila immunity
The Toll–Imd pathway dichotomy that, as a (very powerful) working hypothesis, has dominated the field for the best part of the 1990s and early 2000s has run its course. There is well-documented evidence of cross-reaction by using elicitors that were traditionally thought as triggers of only one pathway [141–143]. In addition, through genome-wide screening in S2 cells, an array of new genes that influence expression of Toll-dependent or Imd-dependent AMP gene expression have been identified, although their relationship to the core pathways remains to be explored [144–146].
It has also become increasingly obvious that different pathogens elicit different host response strategies, which although dependent on the two pathways and many defences described above, have a connection to physiology and behaviour. Insulin signalling, food uptake and circadian rhythms [59,147,148] have been found to have a significant effect on host survival in parallel to mammalian models. These results have certainly introduced a holistic view of host defence as part of the life history of the organism, while introducing (through the study of microbiota) an ecological perspective that was absent during the intense years of gene discovery. In addition, host responses to viral infection induce RNAi and involve JAK/STAT signalling [149–152]. However, the measure of involvement of the latter pathway has not been tested using all available mutants. Finally, both the Toll and Imd pathways have been implicated in antiviral responses [153,154].
8. Outlook
Far from being a ‘fill-in-the-blanks’ exercise after the positioning of the pathways and systems involved, Drosophila immunity has been used as a model for wide-ranging biology and continues to be so. The directions of study on the interaction of the microflora with the host are endless and tap on any number of physiological/developmental issues [23] and recently even mating [155]. Results are fascinating, especially in parallel to the human microbiome project as Drosophila can be a much simpler organism. At the same time, the host–pathogen interaction aspect at the molecular level is the one that has not been systematically explored. We know a lot about the host reaction but do not know enough about how this reaction is altered when the pathogen changes. So a systematic genome-wide exploration of pathogen mutants and their interaction with fruit fly immunity is important. An additional aspect that has not been explored sufficiently is interaction with natural parasites, despite some early efforts on the subject [156–159].
Finally, the elephant in the room: the hallmark of vertebrate responses is memory, which shapes the almost absolute specificity of the defence. Insects have many of the characteristics of vertebrate immunity (discrimination of self versus non-self, amplification and dissemination of defences throughout the body) but seem to lack the more sophisticated aspects of immunological memory. Or do they? There has been evidence of some form of memory in insects since the beginnings of the field in the classic work of Metalnikow [160]. One much more recent report in Drosophila studied memory following infection by Streptococcus pneumoniae [161] and found that fruit flies better survived lethal doses of the microbe when a previous challenge with the same pathogen ‘primed’ them. However, what it is specifically with S. pneumoniae that provokes a memory response (or whether this is a more general phenomenon) remains to be determined.
Future exploration of Drosophila immunity on the open questions above and beyond them will generate exciting biology revealing new aspects in the evolution and regulation of host defences.
9. Acknowledgements
Work in our laboratory has been supported by the Medical Research Council and is currently funded by the Wellcome Trust and the National Centre for the Replacement Refinement and Reduction of animals in research (NC3Rs). I.K. was supported by a postdoctoral fellowship of the Bodossaki Foundation, Greece.
References
- 1.Boman HG, Nilsson I, Rasmuson B. 1972. Inducible antibacterial defence system in Drosophila. Nature 237, 232–235 10.1038/237232a0 (doi:10.1038/237232a0) [DOI] [PubMed] [Google Scholar]
- 2.Faye I, Pye A, Rasmuson T, Boman HG, Boman IA. 1975. Insect immunity. II. Simultaneous induction of antibacterial activity and selective synthesis of some proteins in diapausing pupae of Hyalophora cecropia and Samia cynthia. Infect. Immun. 12, 1426–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hultmark D, Engström Y, Anderson K, Steiner H, Bennich H, Boman HG. 1983. Insect immunity. Attacins, a family of antibacterial proteins from Hyalophora cecropia. EMBO J. 2, 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulet P, Hetru C, Dimarcq JL, Hoffmann D. 1999. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 23, 329–344 10.1016/S0145-305X(99)00015-4 (doi:10.1016/S0145-305X(99)00015-4) [DOI] [PubMed] [Google Scholar]
- 5.Engström Y, Kadalayil L, Sun SC, Samakovlis C, Hultmark D, Faye I. 1993. Kappa B-like motifs regulate the induction of immune genes in Drosophila. J. Mol. Biol. 232, 327–333 10.1006/jmbi.1993.1392 (doi:10.1006/jmbi.1993.1392) [DOI] [PubMed] [Google Scholar]
- 6.Kappler C, Meister M, Lagueux M, Gateff E, Hoffmann JA, Reichhart JM. 1993. Insect immunity. Two 17 bp repeats nesting a kappa B-related sequence confer inducibility to the diptericin gene and bind a polypeptide in bacteria-challenged Drosophila. EMBO J. 12, 1561–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. 1996. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 10.1016/S0092-8674(00)80172-5 (doi:10.1016/S0092-8674(00)80172-5) [DOI] [PubMed] [Google Scholar]
- 8.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 10.1038/41131 (doi:10.1038/41131) [DOI] [PubMed] [Google Scholar]
- 9.Poltorak A, et al. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 10.1126/science.282.5396.2085 (doi:10.1126/science.282.5396.2085) [DOI] [PubMed] [Google Scholar]
- 10.Micchelli CA, Perrimon N. 2006. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 10.1038/nature04371 (doi:10.1038/nature04371) [DOI] [PubMed] [Google Scholar]
- 11.Ohlstein B, Spradling A. 2006. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 10.1038/nature04333 (doi:10.1038/nature04333) [DOI] [PubMed] [Google Scholar]
- 12.Jiang H, Edgar BA. 2011. Intestinal stem cells in the adult Drosophila midgut. Exp. Cell Res. 317, 2780–2788 10.1016/j.yexcr.2011.07.020 (doi:10.1016/j.yexcr.2011.07.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poernbacher I, Baumgartner R, Marada SK, Edwards K, Stocker H. 2012. Drosophila Pez acts in Hippo signaling to restrict intestinal stem cell proliferation. Curr. Biol. 22, 389–396 10.1016/j.cub.2012.01.019 (doi:10.1016/j.cub.2012.01.019) [DOI] [PubMed] [Google Scholar]
- 14.Fox DT, Spradling AC. 2009. The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell 5, 290–297 10.1016/j.stem.2009.06.003 (doi:10.1016/j.stem.2009.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox CR, Gilmore MS. 1997. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75, 1565–1576 10.1128/IAI.01496-06 (doi:10.1128/IAI.01496-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong CN, Ng P, Douglas AE. 2011. Low diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13, 1889–1900 10.1111/j.1462-2920.2011.02511.x (doi:10.1111/j.1462-2920.2011.02511.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren C., et al. 2007. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 6, 144–152 10.1016/j.cmet.2007.06.006 (doi:10.1016/j.cmet.2007.06.006) [DOI] [PubMed] [Google Scholar]
- 18.Corby-Harris V., et al. 2007. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl. Environ. Microbiol. 73, 3470–3479 10.1128/AEM.02120-06 (doi:10.1128/AEM.02120-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandler JA, et al. 2011. Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 7, e1002272. 10.1371/journal.pgen.1002272 (doi:10.1371/journal.pgen.1002272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu JH, et al. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782 10.1126/science.1149357 (doi:10.1126/science.1149357) [DOI] [PubMed] [Google Scholar]
- 21.Bakula M. 1969. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J. Inv. Path. 14, 365–374 10.1016/0022-2011(69)90163-3 (doi:10.1016/0022-2011(69)90163-3) [DOI] [PubMed] [Google Scholar]
- 22.Brummel T, Ching A, Seroude L, Simon AF, Benzer S. 2004. Drosophila lifespan enhancement by exogenous bacteria. Proc. Natl Acad. Sci. USA 101, 12 974–12 979 10.1073/pnas.0405207101 (doi:10.1073/pnas.0405207101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14, 403–414 10.1016/j.cmet.2011.07.012 (doi:10.1016/j.cmet.2011.07.012) [DOI] [PubMed] [Google Scholar]
- 24.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674 10.1126/science.1212782 (doi:10.1126/science.1212782) [DOI] [PubMed] [Google Scholar]
- 25.Basset A, Khush RS, Braun A, Gardan L, Boccard F, Hoffmann JA, Lemaitre B. 2000. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl Acad. Sci. USA 97, 3376–3381 10.1073/pnas.070357597 (doi:10.1073/pnas.070357597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, Hoffmann JA, Imler JL. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13, 737–748 10.1016/S1074-7613(00)00072-8 (doi:10.1016/S1074-7613(00)00072-8) [DOI] [PubMed] [Google Scholar]
- 27.Foley E, O'Farrell PH. 2003. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 17, 115–125 10.1101/gad.1018503 (doi:10.1101/gad.1018503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, Spellman P, Boccard F, Lemaitre B. 2005. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl Acad. Sci. USA 102, 11 414–11 419 10.1073/pnas.0502240102 (doi:10.1073/pnas.0502240102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vodovar N, et al. 2006. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat. Biotechnol. 24, 673–679 10.1038/nbt1212 (doi:10.1038/nbt1212) [DOI] [PubMed] [Google Scholar]
- 30.Liehl P, Blight M, Vodovar N, Boccard F, Lemaitre B. 2006. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog. 2, e56. 10.1371/journal.ppat.0020056 (doi:10.1371/journal.ppat.0020056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. 2009. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333–2344 10.1101/gad.1827009 (doi:10.1101/gad.1827009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nehme NT, Liégeois S, Kele B, Giammarinaro P, Pradel E, Hoffmann JA, Ewbank JJ, Ferrandon D. 2007. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 3, e173. 10.1371/journal.ppat.0030173 (doi:10.1371/journal.ppat.0030173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cronin SJ, et al. 2009. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325, 340–343 10.1126/science.1173164 (doi:10.1126/science.1173164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. 2009. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 10.1016/j.cell.2009.05.014 (doi:10.1016/j.cell.2009.05.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuraishi T, Binggeli O, Opota O, Buchon N, Lemaitre B. 2011. Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 108, 15 966–15 971 10.1073/pnas.1105994108 (doi:10.1073/pnas.1105994108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ha EM, Lee KA, Park SH, Kim SH, Nam HJ, Lee HY, Kang D, Lee WJ. 2009. Regulation of DUOX by the Galphaq-phospholipase Cbeta-Ca2+ pathway in Drosophila gut immunity. Dev. Cell. 16, 386–397 10.1016/j.devcel.2008.12.015 (doi:10.1016/j.devcel.2008.12.015) [DOI] [PubMed] [Google Scholar]
- 37.Ha EM, Oh CT, Bae YS, Lee WJ. 2005. A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850 10.1126/science.1117311 (doi:10.1126/science.1117311) [DOI] [PubMed] [Google Scholar]
- 38.Ha EM, Lee KA, Seo YY, Kim SH, Lim JH, Oh BH, Kim J, Lee WJ. 2009. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in Drosophila gut. Nat. Immunol. 10, 949–957 10.1038/ni.1765 (doi:10.1038/ni.1765) [DOI] [PubMed] [Google Scholar]
- 39.Apidianakis Y, Rahme LG, Heitman J, Ausubel FM, Calderwood SB, Mylonakis E. 2003. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot. Cell 3, 413–419 10.1128/EC.3.2.413-419.2004 (doi:10.1128/EC.3.2.413-419.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glittenberg MT, Kounatidis I, Christensen D, Kostov M, Kimber S, Roberts I, Ligoxygakis P. 2011. Pathogen and host factors are needed to provoke a systemic host response to gastrointestinal infection of Drosophila larvae by Candida albicans. Dis. Model Mech. 4, 515–525 10.1242/dmm.006627 (doi:10.1242/dmm.006627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark RI, Woodcock KJ, Geissmann F, Trouillet C, Dionne MS. 2011. Multiple TGF-β superfamily signals modulate the adult Drosophila immune response. Curr. Biol. 21, 1672–1677 10.1016/j.cub.2011.08.048 (doi:10.1016/j.cub.2011.08.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becker T, Loch G, Beyer M, Zinke I, Aschenbrenner AC, Carrera P, Inhester T, Schultze JL, Hoch M. 2010. FOXO-dependent regulation of innate immune homeostasis. Nature 463, 369–373 10.1038/nature08698 (doi:10.1038/nature08698) [DOI] [PubMed] [Google Scholar]
- 43.Junell A, Uvell H, Davis MM, Edlundh-Rose E, Antonsson A, Pick L, Engström Y. 2010. The POU transcription factor Drifter/Ventral veinless regulates expression of Drosophila immune defense genes. Mol. Cell Biol. 30, 3672–3684 10.1128/MCB.00223-10 (doi:10.1128/MCB.00223-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tepass U, Fessler LI, Aziz A, Hartenstein V. 1994. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development 120, 1829–1837 [DOI] [PubMed] [Google Scholar]
- 45.Crozatier M, Meister M. 2007. Drosophila haematopoiesis. Cell Microbiol. 9, 1117–1126 10.1111/j.1462-5822.2007.00930.x (doi:10.1111/j.1462-5822.2007.00930.x) [DOI] [PubMed] [Google Scholar]
- 46.Grigorian M, Mandel L, Hartenstein V. 2011. Hematopoiesis at the onset of metamorphosis: terminal differentiation and dissociation of the Drosophila lymph gland. Dev. Genes Evol. 221, 121–131 10.1007/s00427-011-0364-6 (doi:10.1007/s00427-011-0364-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avet-Rochex A, et al. 2010. An in vivo RNA interference screen identifies gene networks controlling Drosophila melanogaster blood cell homeostasis. BMC Dev. Biol. 10, 65–75 10.1186/1471-213X-10-65 (doi:10.1186/1471-213X-10-65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bina S, Wright VM, Fisher KH, Milo M, Zeidler MP. 2010. Transcriptional targets of Drosophila JAK/STAT pathway signalling as effectors of haematopoietic tumour formation. EMBO Rep. 11, 201–207 10.1038/embor.2010.1 (doi:10.1038/embor.2010.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stofanko M, Kwon SY, Badenhorst P. 2008. A misexpression screen to identify regulators of Drosophila larval hemocyte development. Genetics 180, 253–267 10.1534/genetics.108.089094 (doi:10.1534/genetics.108.089094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braun A, Hoffmann JA, Meister M. 1998. Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc. Natl Acad. Sci. USA 95, 14 337–14 342 10.1073/pnas.95.24.14337 (doi:10.1073/pnas.95.24.14337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruhf ML, Braun A, Papoulas O, Tamkun JW, Randsholt N, Meister M. 2001. The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development 128, 1429–1441 [DOI] [PubMed] [Google Scholar]
- 52.Brennan CA, Delaney JR, Schneider DS, Anderson KV. 2007. Psidin is required in Drosophila blood cells for both phagocytic degradation and immune activation of the fat body. Curr. Biol. 17, 67–72 10.1016/j.cub.2006.11.026 (doi:10.1016/j.cub.2006.11.026) [DOI] [PubMed] [Google Scholar]
- 53.Shia AK, Glittenberg M, Thompson G, Weber AN, Reichhart JM, Ligoxygakis P. 2009. Toll-dependent antimicrobial responses in Drosophila larval fat body require Spätzle secreted by haemocytes. J. Cell Sci. 122, 4505–4515 10.1242/jcs.049155 (doi:10.1242/jcs.049155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Defaye A, Evans I, Crozatier M, Wood W, Lemaitre B, Leulier F. 2009. Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J. Innate Immun. 1, 322–334 10.1159/000210264 (doi:10.1159/000210264) [DOI] [PubMed] [Google Scholar]
- 55.Charroux B, Royet J. 2009. Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response. Proc. Natl Acad. Sci. USA 106, 9797–9802 10.1073/pnas.0903971106 (doi:10.1073/pnas.0903971106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paddibhatla I, Lee MJ, Kalamarz ME, Ferrarese R, Govind S. 2010. Role for sumoylation in systemic inflammation and immune homeostasis in Drosophila larvae. PLoS Pathog. 6, e1001234. 10.1371/journal.ppat.1001234 (doi:10.1371/journal.ppat.1001234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nehme NT, Quintin J, Cho JH, Lee J, Lafarge MC, Kocks C, Ferrandon D. 2011. Relative roles of the cellular and humoral responses in the Drosophila host defense against three Gram-positive bacterial infections. PLoS ONE 6, e14743. 10.1371/journal.pone.0014743 (doi:10.1371/journal.pone.0014743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leopold P, Perrimon N. 2007. Drosophila and the genetics of the internal milieu. Nature 450, 186–188 10.1038/nature06286 (doi:10.1038/nature06286) [DOI] [PubMed] [Google Scholar]
- 59.Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS. 2006. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila. Curr. Biol. 16, 1977–1985 10.1016/j.cub.2006.08.052 (doi:10.1016/j.cub.2006.08.052) [DOI] [PubMed] [Google Scholar]
- 60.Stuart LM, Ezekowitz RA. 2008. Phagocytosis and comparative innate immunity: learning on the fly. Nat. Rev. Immunol. 8, 131–141 10.1038/nri2240 (doi:10.1038/nri2240) [DOI] [PubMed] [Google Scholar]
- 61.Franc NC, Dimarcq J-L, Lagueux M, Hoffmann J, Ezekowitz RAB. 1996. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4, 431–443 10.1016/S1074-7613(00)80410-0 (doi:10.1016/S1074-7613(00)80410-0) [DOI] [PubMed] [Google Scholar]
- 62.Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. 2006. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50, 855–867 10.1016/j.neuron.2006.04.027 (doi:10.1016/j.neuron.2006.04.027) [DOI] [PubMed] [Google Scholar]
- 63.Hashimoto Y, Tabuchi Y, Sakurai K, Kutsuna M, Kurokawa K, Awasaki T, Sekimizu K, Nakanishi Y, Shiratsuchi A. 2009. Identification of lipoteichoic acid as a ligand for draper in the phagocytosis of Staphylococcus aureus by Drosophila hemocytes. J. Immunol. 183, 7451–7460 10.4049/jimmunol.0901032 (doi:10.4049/jimmunol.0901032) [DOI] [PubMed] [Google Scholar]
- 64.Philips JA, Rubin EJ, Perrimon N. 2005. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science 309, 1251–1253 10.1126/science.1116006 (doi:10.1126/science.1116006) [DOI] [PubMed] [Google Scholar]
- 65.Rämet M, Pearson A, Manfruelli P, Li X, Koziel H, Göbel V, Chung E, Krieger M, Ezekowitz RAB. 2001. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity 15, 1027–1038 10.1016/S1074-7613(01)00249-7 (doi:10.1016/S1074-7613(01)00249-7) [DOI] [PubMed] [Google Scholar]
- 66.Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli . Nature 416, 644–648 10.1038/nature735 (doi:10.1038/nature735) [DOI] [PubMed] [Google Scholar]
- 67.Kocks C, et al. 2005. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123, 335–346 10.1016/j.cell.2005.08.034 (doi:10.1016/j.cell.2005.08.034) [DOI] [PubMed] [Google Scholar]
- 68.Kurucz E, et al. 2007. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr. Biol. 17, 649–654 10.1016/j.cub.2007.02.041 (doi:10.1016/j.cub.2007.02.041) [DOI] [PubMed] [Google Scholar]
- 69.Watson FL, Püttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D. 2005. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 309, 1874–1878 10.1126/science.1116887 (doi:10.1126/science.1116887) [DOI] [PubMed] [Google Scholar]
- 70.Stuart LM, et al. 2007. A systems biology analysis of the Drosophila phagosome. Nature 445, 95–101 10.1038/nature05380 (doi:10.1038/nature05380) [DOI] [PubMed] [Google Scholar]
- 71.Haine ER, Moret Y, Siva-Jothy MT, Rolff J. 2008. Antimicrobial defense and persistent infection in insects. Science 322, 1257–1259 10.1126/science.1165265 (doi:10.1126/science.1165265) [DOI] [PubMed] [Google Scholar]
- 72.Volz J, Müller HM, Zdanowicz A, Kafatos FC, Osta MA. 2006. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 8, 1392–1405 10.1111/j.1462-5822.2006.00718.x (doi:10.1111/j.1462-5822.2006.00718.x) [DOI] [PubMed] [Google Scholar]
- 73.Cerenius L, Söderhäll K. 2004. The prophenoloxidase-activating system in invertebrates. Immunol. Rev 198, 116–126 10.1111/j.0105-2896.2004.00116.x (doi:10.1111/j.0105-2896.2004.00116.x) [DOI] [PubMed] [Google Scholar]
- 74.Leclerc V, Pelte N, Chamy LE, Martinelli C, et al. 2006. Prophenoloxidase activation is not required for survival to microbial infections in Drosophila. EMBO Rep. 7, 231–235 10.1038/sj.embor.7400592 (doi:10.1038/sj.embor.7400592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang H, Kambris Z, Lemaitre B, Hashimoto C. 2006. Two proteases defining a melanization cascade in the immune system of Drosophila. J. Biol. Chem. 281, 28097–28104 10.1074/jbc.M601642200 (doi:10.1074/jbc.M601642200) [DOI] [PubMed] [Google Scholar]
- 76.Ayres JS, Schneider DS. 2008. A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS Biol. 6, 2764–2773 10.1371/journal.pbio.0060305 (doi:10.1371/journal.pbio.0060305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pentz ES, Black BC, Wright TR. 1990. Mutations affecting phenol oxidase activity in Drosophila: quicksilver and tyrosinase-1. Biochem. Genet. 28, 151–171 10.1007/BF00561334 (doi:10.1007/BF00561334) [DOI] [PubMed] [Google Scholar]
- 78.Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, Jiang H, Hoffmann JA, Reichhart JM. 2002. A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J. 21, 6330–6337 10.1093/emboj/cdf661 (doi:10.1093/emboj/cdf661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.De Gregorio E, et al. 2002. An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev. Cell 3, 581–592 10.1016/S1534-5807(02)00267-8 (doi:10.1016/S1534-5807(02)00267-8) [DOI] [PubMed] [Google Scholar]
- 80.Jiang R, et al. 2009. Three pairs of protease–serpin complexes cooperatively regulate the insect innate immune responses. J. Biol. Chem. 284, 35652–35658 10.1074/jbc.M109.071001 (doi:10.1074/jbc.M109.071001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roh KB, et al. 2009. Proteolytic cascade for the activation of the insect toll pathway induced by the fungal cell wall component. J. Biol. Chem. 284, 19474–19481 10.1074/jbc.M109.007419 (doi:10.1074/jbc.M109.007419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scherfer C, Karlsson C, Loseva O, Bidla G, Goto A, Havemann J, Dushay MS, Theopold U. 2004. Isolation and characterization of hemolymph clotting factors in Drosophila melanogaster by a pullout method. Curr. Biol. 14, 625–629 10.1016/j.cub.2004.03.030 (doi:10.1016/j.cub.2004.03.030) [DOI] [PubMed] [Google Scholar]
- 83.Bidla G, Lindgren M, Theopold U, Dushay MS. 2005. Hemolymph coagulation and phenoloxidase in Drosophila larvae. Dev. Comp. Immunol. 29, 669–679 10.1016/j.dci.2004.11.007 (doi:10.1016/j.dci.2004.11.007) [DOI] [PubMed] [Google Scholar]
- 84.Karlsson C, Korayem AM, Scherfer C, Loseva O, Dushay MS, et al. 2004. Proteomic analysis of the Drosophila larval hemolymph clot. J. Biol. Chem. 279, 52033–52041 10.1074/jbc.M408220200 (doi:10.1074/jbc.M408220200) [DOI] [PubMed] [Google Scholar]
- 85.Lesch C, Goto A, Lindgren M, Bidla G, Dushay MS, et al. 2007. A role for Hemolectin in coagulation and immunity in Drosophila melanogaster. Dev. Comp. Immunol. 31, 1255–1263 10.1016/j.dci.2007.03.012 (doi:10.1016/j.dci.2007.03.012) [DOI] [PubMed] [Google Scholar]
- 86.Scherfer C, Qazi MR, Takahashi K, Ueda R, Dushay MS, Theopold U,, Lemaitre B. 2006. The Toll immune-regulated Drosophila protein Fondue is involved in hemolymph clotting and puparium formation. Dev. Biol. 295, 156–163 10.1016/j.ydbio.2006.03.019 (doi:10.1016/j.ydbio.2006.03.019) [DOI] [PubMed] [Google Scholar]
- 87.Wang Z, et al. 2010. Pathogen entrapment by transglutaminase—a conserved early innate immune mechanism. PLoS Pathog. 6, e1000763. 10.1371/journal.ppat.1000763 (doi:10.1371/journal.ppat.1000763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lorand L, Graham RM. 2003. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140–156 10.1038/nrm1014 (doi:10.1038/nrm1014) [DOI] [PubMed] [Google Scholar]
- 89.Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelièvre E, Gascan H, Ray KP, Morse MA, Imler JL, Gay NJ. 2003. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat. Immunol. 4, 794–800 10.1038/ni955 (doi:10.1038/ni955) [DOI] [PubMed] [Google Scholar]
- 90.Gangloff M, et al. 2008. Structural insight into the mechanism of activation of the Toll receptor by the dimeric ligand Spätzle. J. Biol. Chem. 283, 14629–14635 10.1074/jbc.M800112200 (doi:10.1074/jbc.M800112200) [DOI] [PubMed] [Google Scholar]
- 91.Jang IH, et al. 2006. A Spätzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev. Cell 10, 45–55 10.1016/j.devcel.2005.11.013 (doi:10.1016/j.devcel.2005.11.013) [DOI] [PubMed] [Google Scholar]
- 92.Morisato D. 2001. Spätzle regulates the shape of the Dorsal gradient in the Drosophila embryo. Development 128, 2309–2319 [DOI] [PubMed] [Google Scholar]
- 93.Weber AN, Gangloff M, Moncrieffe MC, Hyvert Y, Imler JL, Gay NJ. 2007. Role of the Spatzle pro-domain in the generation of an active toll receptor ligand. J. Biol. Chem. 282, 13 522–13 531 10.1074/jbc.M700068200 (doi:10.1074/jbc.M700068200) [DOI] [PubMed] [Google Scholar]
- 94.Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. 2002. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 297, 114–116 10.1126/science.1072391 (doi:10.1126/science.1072391) [DOI] [PubMed] [Google Scholar]
- 95.El Chamy L, Leclerc V, Caldelari I, Reichhart JM. 2008. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways 'upstream’ of Toll. Nat. Immunol. 9, 1165–1170 10.1038/ni.1643 (doi:10.1038/ni.1643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, Belvin M, Hoffmann JA, Ferrandon D. 2006. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell 127, 1425–1437 10.1016/j.cell.2006.10.046 (doi:10.1016/j.cell.2006.10.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buchon N, Poidevin M, Kwon HM, Guillou A, Sottas V, Lee BL, Lemaitre B. 2009. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc. Natl Acad. Sci. USA 106, 12 442–12 447 10.1073/pnas.0901924106 (doi:10.1073/pnas.0901924106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. 2006. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr. Biol. 16, 808–813 10.1016/j.cub.2006.03.020 (doi:10.1016/j.cub.2006.03.020) [DOI] [PubMed] [Google Scholar]
- 99.Michel T, Reichhart JM, Hoffmann JA, Royet J. 2001. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414, 756–759 10.1038/414756a (doi:10.1038/414756a) [DOI] [PubMed] [Google Scholar]
- 100.Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, Hoffmann JA, Ferrandon D. 2003. Dual activation of the Drosophila Toll pathway by two pattern recognition receptors. Science 302, 2126–2130 10.1126/science.1085432 (doi:10.1126/science.1085432) [DOI] [PubMed] [Google Scholar]
- 101.Wang L, Gilbert RJ, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. 2008. Peptidoglycan recognition protein-SD provides versatility of receptor formation in Drosophila immunity. Proc. Natl Acad. Sci. USA 105, 11 881–11 886 10.1073/pnas.0710092105 (doi:10.1073/pnas.0710092105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang L, Weber AN, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. 2006. Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J. 25, 5005–5014 10.1038/sj.emboj.7601363 (doi:10.1038/sj.emboj.7601363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Filipe SR, Tomasz A, Ligoxygakis P. 2005. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 6, 327–333 10.1038/sj.embor.7400371 (doi:10.1038/sj.embor.7400371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Atilano ML, Yates J, Glittenberg M, Filipe SR, Ligoxygakis P. 2011. Wall teichoic acids of Staphylococcus aureus limit recognition by the Drosophila peptidoglycan recognition protein-SA to promote pathogenicity. PLoS Pathog. 7, e1002421. 10.1371/journal.ppat.1002421 (doi:10.1371/journal.ppat.1002421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mishima Y, et al. 2009. The N-terminal domain of Drosophila Gram-negative binding protein 3 (GNBP3) defines a novel family of fungal pattern recognition receptors. J. Biol. Chem. 284, 28 687–28 697 10.1074/jbc.M109.034587 (doi:10.1074/jbc.M109.034587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. 2002. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat. Immunol. 3, 91–97 10.1038/ni747 (doi:10.1038/ni747) [DOI] [PubMed] [Google Scholar]
- 107.Towb P, Bergmann A, Wasserman SA. 2001. The protein kinase Pelle mediates feedback regulation in the Drosophila Toll signaling pathway. Development 128, 4729–4736 [DOI] [PubMed] [Google Scholar]
- 108.Rutschmann S, Jung AC, Hetru C, Reichhart JM, Hoffmann JA, Ferrandon D. 2000. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 12, 569–580 10.1016/S1074-7613(00)80208-3 (doi:10.1016/S1074-7613(00)80208-3) [DOI] [PubMed] [Google Scholar]
- 109.Manfruelli P, Reichhart JM, Steward R, Hoffmann JA, Lemaitre B. 1999. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J. 18, 3380–3391 10.1093/emboj/18.12.3380 (doi:10.1093/emboj/18.12.3380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haghayeghi A, Sarac A, Czerniecki S, Grosshans J, Schoöck F. 2010. Pellino enhances innate immunity in Drosophila. Mech. Dev 127, 301–307 10.1016/j.mod.2010.01.004 (doi:10.1016/j.mod.2010.01.004) [DOI] [PubMed] [Google Scholar]
- 111.Gordon MD, Dionne MS, Schneider DS, Nusse R. 2005. WntD is a feedback inhibitor of Dorsal/NF-kappaB in Drosophila development and immunity. Nature 437, 746–749 10.1038/nature04073 (doi:10.1038/nature04073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang HR, Chen ZJ, Kunes S, Chang GD, Maniatis T. 2010. Endocytic pathway is required for Drosophila Toll innate immune signaling. Proc. Natl Acad. Sci. USA 107, 8322–8327 10.1073/pnas.1004031107 (doi:10.1073/pnas.1004031107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kaneko T, et al. 2004. Monomeric and polymeric Gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637–649 10.1016/S1074-7613(04)00104-9 (doi:10.1016/S1074-7613(04)00104-9) [DOI] [PubMed] [Google Scholar]
- 114.Kaneko T, et al. 2006. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 7, 715–723 10.1038/ni1356 (doi:10.1038/ni1356) [DOI] [PubMed] [Google Scholar]
- 115.Choe KM, et al. 2005. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal transducing innate immune receptor. Proc. Natl Acad. Sci. USA 102, 1122–1126 10.1073/pnas.0404952102 (doi:10.1073/pnas.0404952102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. 2000. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl Acad. Sci. USA 97, 13 772–13 777 10.1073/pnas.97.25.13772 (doi:10.1073/pnas.97.25.13772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lim JH, et al. 2006. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 281, 286–295 [DOI] [PubMed] [Google Scholar]
- 118.Yano T, et al. 2008. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 9, 908–916 10.1038/ni.1634 (doi:10.1038/ni.1634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cossart P. 2011. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl Acad. Sci. USA 108, 19 484–19 491 10.1073/pnas.1112371108 (doi:10.1073/pnas.1112371108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Georgel P, et al. 2001. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 1, 503–514 10.1016/S1534-5807(01)00059-4 (doi:10.1016/S1534-5807(01)00059-4) [DOI] [PubMed] [Google Scholar]
- 121.Naitza S, et al. 2002. The Drosophila immune defense against Gram-negative infection requires the death protein dFADD. Immunity 17, 575–581 10.1016/S1074-7613(02)00454-5 (doi:10.1016/S1074-7613(02)00454-5) [DOI] [PubMed] [Google Scholar]
- 122.Leulier F, et al. 2000. Drosophila caspase Dredd is required to resist Gram-negative bacterial infection. EMBO Rep. 1, 353–358 10.1093/embo-reports/kvd073 (doi:10.1093/embo-reports/kvd073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hu S, Yang X. 2000. dFADD, a novel death domain-containing adapter protein for the Drosophila caspase DREDD. J. Biol. Chem. 275, 30 761–30 764 10.1074/jbc.C000341200 (doi:10.1074/jbc.C000341200) [DOI] [PubMed] [Google Scholar]
- 124.Leulier F, et al. 2002. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr. Biol. 12, 996–1000 10.1016/S0960-9822(02)00873-4 (doi:10.1016/S0960-9822(02)00873-4) [DOI] [PubMed] [Google Scholar]
- 125.Paquette N, et al. 2010. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol. Cell 37, 172–182 10.1016/j.molcel.2009.12.036 (doi:10.1016/j.molcel.2009.12.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thevenon D, et al. 2009. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe 6, 309–320 10.1016/j.chom.2009.09.007 (doi:10.1016/j.chom.2009.09.007) [DOI] [PubMed] [Google Scholar]
- 127.Vidal S, et al. 2001. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 15, 1900–1912 10.1101/gad.203301 (doi:10.1101/gad.203301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kleino A., et al. 2005. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 24, 3423–3434 10.1038/sj.emboj.7600807 (doi:10.1038/sj.emboj.7600807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Erturk-Hasdemir D, et al. 2009. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc. Natl Acad. Sci. USA 106, 9779–9784 10.1073/pnas.0812022106 (doi:10.1073/pnas.0812022106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hedengren M, et al. 1999. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4, 827–837 10.1016/S1097-2765(00)80392-5 (doi:10.1016/S1097-2765(00)80392-5) [DOI] [PubMed] [Google Scholar]
- 131.Stoven S, et al. 2003. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc. Natl Acad. Sci. USA 100, 5991–5996 10.1073/pnas.1035902100 (doi:10.1073/pnas.1035902100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Park JM, et al. 2004. Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 18, 584–594 10.1101/gad.1168104 (doi:10.1101/gad.1168104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zaidman-Remy A, et al. 2006. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473 10.1016/j.immuni.2006.02.012 (doi:10.1016/j.immuni.2006.02.012) [DOI] [PubMed] [Google Scholar]
- 134.Kleino A, et al. 2008. Pirk is a negative regulator of the Drosophila Imd pathway. J. Immunol. 180, 5413–5422 [DOI] [PubMed] [Google Scholar]
- 135.Lhocine N, et al. 2008. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe 4, 147–158 10.1016/j.chom.2008.07.004 (doi:10.1016/j.chom.2008.07.004) [DOI] [PubMed] [Google Scholar]
- 136.Aggarwal K, et al. 2008. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 4, e100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Paredes JC, Welchman DP, Poidevin M, Lemaitre B. 2011. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35, 770–779 10.1016/j.immuni.2011.09.018 (doi:10.1016/j.immuni.2011.09.018) [DOI] [PubMed] [Google Scholar]
- 138.Kim M, et al. 2006. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc. Natl Acad. Sci. USA 103, 16 358–16 363 10.1073/pnas.0603238103 (doi:10.1073/pnas.0603238103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tsichritzis T, et al. 2007. A Drosophila ortholog of the human cylindromatosis tumor suppressor gene regulates triglyceride content and antibacterial defense. Development 134, 2605–2614 10.1242/dev.02859 (doi:10.1242/dev.02859) [DOI] [PubMed] [Google Scholar]
- 140.Tsuda M, et al. 2005. The RING-finger scaffold protein Plenty of SH3 s targets TAK1 to control immunity signalling in Drosophila. EMBO Rep. 6, 1082–1087 10.1038/sj.embor.7400537 (doi:10.1038/sj.embor.7400537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lau GW, Goumnerov BC, Walendziewicz CL, Hewitson J, Xiao W, Mahajan-Miklos S, Tompkins RG, Perkins LA, Rahme LG. 2003. The Drosophila Toll pathway participates in resistance to infection by the Gram-negative human pathogen Pseudomonas aeruginosa . Infect. Immun. 71, 4059–4066 10.1128/IAI.71.7.4059-4066.2003 (doi:10.1128/IAI.71.7.4059-4066.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lazzaro BP, Sceurman BK, Clark AG. 2004. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science 303, 1873–1876 10.1126/science.1092447 (doi:10.1126/science.1092447) [DOI] [PubMed] [Google Scholar]
- 143.Mansfield BE, Dionne MS, Schneider DS, Freitag NE. 2003. Exploration of host–pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 5, 901–911 10.1046/j.1462-5822.2003.00329.x (doi:10.1046/j.1462-5822.2003.00329.x) [DOI] [PubMed] [Google Scholar]
- 144.Valanne SH, et al. 2010. Genome-wide RNA interference in Drosophila cells identifies G protein-coupled receptor kinase 2 as a conserved regulator of NF-kappaB signaling. J. Immunol. 184, 6188–6198 10.4049/jimmunol.1000261 (doi:10.4049/jimmunol.1000261) [DOI] [PubMed] [Google Scholar]
- 145.Reed DE, Huang XM, Wohlschlegel JM, Levine MS, Senger K. 2008. DEAF-1 regulates immunity gene expression in Drosophila. Proc. Natl Acad. Sci. USA 105, 8351–8356 10.1073/pnas.0802921105 (doi:10.1073/pnas.0802921105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Goto A, et al. 2008. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in Drosophila and mice. Nat. Immunol. 9, 97–104 10.1038/ni1543 (doi:10.1038/ni1543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ayres JS, Schneider DS. 2009. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 7, e1000150. 10.1371/journal.pbio.1000150 (doi:10.1371/journal.pbio.1000150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stone EF, Fulton BO, Ayres JS, Pham LN, Ziauddin J, Shirasu-Hiza MM. 2012. The circadian clock protein timeless regulates phagocytosis of bacteria in Drosophila. PLoS Pathog. 8, e1002445. 10.1371/journal.ppat.1002445 (doi:10.1371/journal.ppat.1002445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sabatier L, et al. 2003. Pherokine-2 and -3. Eur. J. Biochem. 270, 3398–3407 10.1046/j.1432-1033.2003.03725.x (doi:10.1046/j.1432-1033.2003.03725.x) [DOI] [PubMed] [Google Scholar]
- 150.Zambon RA, et al. 2006. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 8, 880–889 10.1111/j.1462-5822.2006.00688.x (doi:10.1111/j.1462-5822.2006.00688.x) [DOI] [PubMed] [Google Scholar]
- 151.Galiana-Arnoux D, et al. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 7, 590–597 10.1038/ni1335 (doi:10.1038/ni1335) [DOI] [PubMed] [Google Scholar]
- 152.Dostert C, et al. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat. Immunol. 6, 946–953 10.1038/ni1237 (doi:10.1038/ni1237) [DOI] [PubMed] [Google Scholar]
- 153.Costa A, et al. 2009. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS ONE 4, e7436. 10.1371/journal.pone.0007436 (doi:10.1371/journal.pone.0007436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zambon RA, et al. 2005. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl Acad. Sci. USA 102, 7257–7262 10.1073/pnas.0409181102 (doi:10.1073/pnas.0409181102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sharon G, et al. 2011. Commensal bacteria play a role in mating preference of Drosophila. Proc. Natl Acad. Sci. USA 107, 20 051–20 056 10.1073/pnas.1009906107 (doi:10.1073/pnas.1009906107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Boulanger N, Ehret-Sabatier L, Brun R, Zachary D, Bulet P, Imler JL. 2001. Immune response of Drosophila melanogaster to infection with the flagellate parasite Crithidia spp. Insect Biochem. Mol. Biol. 31, 129–137 10.1016/S0965-1748(00)00096-5 (doi:10.1016/S0965-1748(00)00096-5) [DOI] [PubMed] [Google Scholar]
- 157.Rowton ED, McGhee RB. 1978. Population dynamics of Herpetomonas ampelophilae, with a note on the systematic of Herpetomonas spp, in Drosophila. J. Protozool. 25, 232–235 [Google Scholar]
- 158.Rowton ED, McGhee RB. 1983. Transmission of Herpetomonas in laboratory populations of Drosophila melanogaster. J. Protozool. 30, 669–671 [Google Scholar]
- 159.Roxstrom-Lindquist K, Terenius O, Faye I. 2004. Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep. 5, 207–212 10.1038/sj.embor.7400073 (doi:10.1038/sj.embor.7400073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Metalnikow S. 1929. Immunité d'adaptation et immunité de defense SR. Soc. Biol. 101, 34–67 [Google Scholar]
- 161.Pham LN, Dionne MS, Shirasu-Hiza M, Schneider D. 2007. A specific primed response in Drosophila is dependent on hemocytes. PLoS Pathog. 3, e26. 10.1371/journal.ppat.0030026 (doi:10.1371/journal.ppat.0030026) [DOI] [PMC free article] [PubMed] [Google Scholar]