Abstract
We present Bayesian analyses of matrix-variate normal data with conditional independencies induced by graphical model structuring of the characterizing covariance matrix parameters. This framework of matrix normal graphical models includes prior specifications, posterior computation using Markov chain Monte Carlo methods, evaluation of graphical model uncertainty and model structure search. Extensions to matrix-variate time series embed matrix normal graphs in dynamic models. Examples highlight questions of graphical model uncertainty, search and comparison in matrix data contexts. These models may be applied in a number of areas of multivariate analysis, time series and also spatial modelling.
Some key words: Gaussian graphical model, Graphical model search, Hyper-inverse Wishart distribution, Marginal likelihood, Matrix normal model, Matrix-variate dynamic graphical model, Parameter expansion
1. Introduction
We introduce and analyze matrix normal graphical models; that is, matrix normal distributions (Dawid, 1981; Gupta & Nagar, 2000) in which each of the two characterizing covariance matrices reflects conditional independencies consistent with an underlying graphical model (Whittaker, 1990; Lauritzen, 1996). We present fully Bayesian analysis of the matrix normal model as the special case of full graphs, and develop computational methods for marginal likelihood computation on a specified graphical model. This enables graphical model search and comparison for posterior inferences about conditional independence structures. The random sampling framework is extended to matrix-variate time series models that inherit the graphical model structure to represent conditional independencies in matrix time series. We focus on decomposable graphs although the general approach will also apply to nondecomposable models.
Matrix-variate normal distributions have been studied in analysis of two-factor linear models for cross-classified multivariate data (Finn, 1974; Galecki, 1994; Naik & Rao, 2001), in spatio-temporal models (Mardia & Goodall, 1993; Huizenga et al., 2002) and other areas. Some computational and inferential developments, including iterative calculation of maximum likelihood estimates (Dutilleul, 1999; Mitchell et al., 2006) and empirical Bayesian methods for Procrustes analysis with matrix models (Theobald & Wuttke, 2006) have been published. Our work appears to be the first to develop fully Bayesian analysis of the basic matrix normal model alone, though that is only a necessary first step to the broader framework of matrix graphical models.
In time series, graphical modelling of the covariance matrix of multivariate data appears in Carvalho & West (2007a, 2007b). Here we generalize that earlier work to time series of matrix data, providing fully Bayesian inference and graphical model search related to both row and column intra-dependencies in the cross-sectional structure of a matrix-valued time series.
2. Matrix variate normals, graphs and notation
The q × p random matrix Y is matrix normal, Y ∼ N (M, U, V), with mean M (q × p), column and row covariance matrices U = (uij) (q × q) and V = (υij) (p × p), respectively, when
| (1) |
where k(U, V) = (2π)−qp/2|U|−p/2|V|−q/2. The rows yi★ (i = 1, . . ., p), and columns y★j (j = 1, . . ., q), have margins yi★ ∼ N (mi★, uii V) and y★j ∼ N (m★j, υjj U) with precision matrices Λ = V−1 = (λij) and Ω = U−1 = (ωij), respectively. The normal conditional distributions have mean vectors and covariance matrices
for rows i = 1, . . ., q and columns j = 1, . . ., p. Zeros in Λ and Ω define conditional independencies. If (i, j) ≠ (s, t) then yij and yst may, conditional upon y−(ij,st) be dependent through either rows or columns; conditional independence is equivalent to: at least one zero among λtj and ωis when s ≠ i, j ≠ t; ωis = 0 when s ≠ i, j = t; λjt = 0 when s = i, j ≠ t. With no loss of generality, in this section we set M = 0.
Undirected graphical models can be applied to each of Λ and Ω to represent strict conditional independencies. A graph GV on nodes {1, . . ., p} has edges between pairs of column indices (j, t) for which λjt ≠ 0; Λ has off-diagonal zeros corresponding to within-row conditional independencies. Similarly, a graph GU on nodes {1, . . ., q} lacks edges between row indices (i, s) for which ωis = 0. We focus here on decomposable graphs GU and GV. The theory of graphical models can be now overlaid to define conditional factorizations of the matrix normal density over graphs. Over GV, for example, we have
| (2) |
where 𝒫V is the set of complete prime components, or cliques, of GV and 𝒮V is the set of separators. For each subgraph g ∈ {𝒫V, 𝒮V}, Y★g is the q × |g| matrix with variables from the |g| columns of Y defined by the subgraph, and Vg the corresponding submatrix of V. Each term in equation (2) is matrix normal, Y★g ∼ N (0, U, Vg) with having no off-diagonal zeros. We can similarly factorize the joint density over GU.
Now, U and V are not uniquely identified since, for any c > 0, p(Y | U, V) = p(Y | cU, V/c). There are a number of approaches to imposing identification constraints such as tr(V) = p (Theobald & Wuttke, 2006), and possible strategies that use unconstrained parameters; we discuss the latter in § 8. Our use of hyper-Markov priors over each of U and V with underlying graphical models, discussed below, makes it desirable to adopt an explicit constraint and we enforce υ11 = 1 from here on.
3. Matrix graphical modelling
Hyper-inverse Wishart priors are conjugate for covariance matrices in multivariate normal graphical models (Dawid & Lauritzen, 1993). Hyper-inverse Wishart distributions are compatible and consistent across graphs, which is critical when admitting uncertainty about graph structures (Giudici & Green, 1999; Jones et al., 2005). On decomposable graphs, the implied priors on sub-covariance matrices on all components and separators are inverse Wishart. Use of independent hyper-inverse Wishart priors for U, V in the current context is a natural choice, and maintains compatibility and consistency across graphs GU, GV. To incorporate the identification constraint υ11 = 1, we use a parameter expansion approach. Parameter expansion involves expanding the parameter space by adding new nuisance parameters, and has been used purely algorithmically to accelerate Markov chain Monte Carlo samplers (Liu et al., 1998; Liu & Wu, 1999), but can also be used to induce new priors (Gelman, 2004, 2006) as is germane here.
We assume the prior p(U, V) = p(U)p(V) where, using the hyper-inverse Wishart notation of Giudici & Green (1999) and Jones et al. (2005), the margins are defined by
| (3) |
The density function for U is, following Dawid & Lauritzen (1993),
where each component is an inverse Wishart density; p(V*) has a similar form.
The parameter expansion concept relates to as an added parameter that converts column scales in V to those relative to the scale of the first column. As we move across graphs GV, the priors p(V | GV) have the same induced priors over subgraph correlation structures but are no longer in complete agreement for V = V*/ due to the different parameterizations and interpretations. This is natural and appropriate. Suppose GV and G′V are two graphs with a common clique C. Each element in diag(VC) represents the relative scale of variance of that column to the variance of the first column so that, if GV and G′V imply different conditional dependencies between the first column and columns linked to C, then the induced priors over VC should indeed be different.
The prior p(V) is obtained by transformation from V*. On any graph GV, V is determined only by those free elements appearing in the submatrices corresponding to the cliques of the graph, and the nonfree elements of V are deterministic functions of the free elements (Carvalho et al., 2007). Let ν be the number of free elements; then the transformation from V * to (V, ) has Jacobian leading to
Coupled with the prior p(U) on GU, this defines a class of conditionally conjugate priors in the expanded parameter space.
4. Posterior and marginal likelihood computation
4.1. Gibbs sampling on given graphs
Assume an initial random sampling context with q × p data matrices Yi (i = 1, . . ., n), drawn independently from equation (1), and write Y for the full set of data. It is easy to see that, on specified graphs (GU, GV), the posterior p(U, V, |Y) has conditional distributions:
where a = ΣPV|PV|(2|PV| + d) − ΣSV|SV|(2|SV| + d). These distributions form the basis of Gibbs sampling for the target posterior p(U, V, | Y). This involves iterative resampling from the hyper-inverse Wishart, inverse gamma and new conditional hyper-inverse Wishart distributions. Simulation of the former is based on Carvalho et al. (2007), while sampling the latter can be done as follows. From Lemma 2.18 of Lauritzen (1996), we can always find a perfect ordering of the nodes in GV so that node 1 is in the first clique, say C, and then initialize the hyper-inverse Wishart sampler of Carvalho et al. (2007) to begin with a simulation of the implied conditional inverse Wishart distribution for the covariance matrix on that first clique. Sampling VC from an inverse Wishart distribution conditional on the first diagonal element set to unity is straightforward.
4.2. Marginal likelihood
Exploration of uncertainty about graphical model structures involves consideration of the marginal likelihood function over graphs. For any pair (GU, GV), this is
The priors in the integrand depend on the graphs although for clarity we drop that from the notation. In multivariate models, marginal likelihoods can be evaluated in closed form on decomposable graphs (Giudici, 1996; Giudici & Green, 1999; Jones et al., 2005; Carvalho & West, 2007a, 2007b). In our matrix models, the integral cannot be evaluated but we can generate useful approximations via use of the candidate’s formula (Besag, 1989; Chib, 1995). Write Θ = {U, V, } for all parameters, and suppose that we can evaluate p(θ| Y) for some sub-set of parameters θ ∈ Θ; the candidate’s formula gives the marginal likelihood via the identity p(Y) = p(Y | θ)/p(θ| Y). Applying this requires that we estimate components of the numerator or denominator. Choosing θ to maximally exploit analytic integration is key, and different choices that integrate over different subsets of parameters will lead to different, parallel approximations of p(Y) that can be compared. We use two approximations based on marginalization over desirably disjoint parameter subsets, namely, (A): p(Y) = p(Y, , U)/p( , U | Y) at any chosen value of θ = { , U}, and (B): p(Y) = p(Y, V)/p(V | Y) at any value of θ = V. We estimate the components of these equations that have no closed form, then insert chosen values U, V, , such as approximate posterior means, to provide two estimates of p(Y).
For (A), first rewrite as
The numerator terms are each easily computed at any {V, , U}. The second denominator term p(V | , U, Y) has an easily evaluated closed form, as in the Gibbs sampling step. The first denominator term may be approximated by
where the sum is over posterior draws Vj; this is easy to compute as it is a sum of the product of inverse gamma and hyper-inverse Wishart densities.
For (B), the numerator can be analytically evaluated as
where
the H(·, ·, G·) terms are normalizing constants of the corresponding hyper-inverse Wishart distributions (Giudici & Green, 1999; Jones et al., 2005) and
The density function in the denominator is approximated as
where the sum over posterior draws (Uj, ) can be easily performed, with terms given by conditional hyper-inverse Wishart density evaluations.
4.3. Graphical model uncertainty and search
Now admit uncertainty about graphs (GU, GV) using sparsity-encouraging priors in which edge inclusion indicators are independent Bernoulli variates (Dobra et al., 2004; Jones et al., 2005). We now extend Markov chain Monte Carlo simulation for multivariate graphical models (Giudici & Green, 1999; Jones et al., 2005) to learning on (GU, GV) in the above matrix model analysis. Our analysis generates multiple graphs with values of approximate posterior probabilities, using the Markov chain simulation for model search. This relies on the computation of the unnormalized posterior over graphs, p(GU, GV| Y) ∝ p(Y | GU, GV) p(GU, GV) involving the marginal likelihood value for any specified model (GU, GV) at each search step. For the latter, we average the approximate marginal likelihood values from methods (A) and (B). The work in Jones et al. (2005) includes evaluation of the performance of various stochastic search methods in single multivariate graphical models; for modest dimensions, they recommend simple local-move Metropolis–Hastings steps. Here, given a current pair (GU, GV), we can apply local moves in GU space based on the conditional posterior p(GU | Y, GV), and vice-versa. A candidate G′U is sampled from a proposal distribution q(G′U; GU) and accepted with probability
our examples use the simple random add/delete edge move proposal of Jones et al. (2005). We then couple this with a similar step using p(GV | Y, GU) at each iteration. This requires a Markov chain analysis on each graph pair visited in order to evaluate marginal likelihood, so implying a substantial computational burden.
5. Example: a simulated random sample
A sample of size n = 48 was drawn from the (q = 8) × (p = 7) dimensional N (0, U, V) distribution, where, using · to denote zeros to highlight structure, the precision matrices are
First consider analysis on the true graphs under priors with b = d = 3 and B = 5I8, D = 5I7 and simulation sample size 8000 after an initial, discarded burn-in of 2000 iterations. Convergence is rapid and apparently fast-mixing in this as in other simulated examples. The corresponding posterior means of the precision matrices are
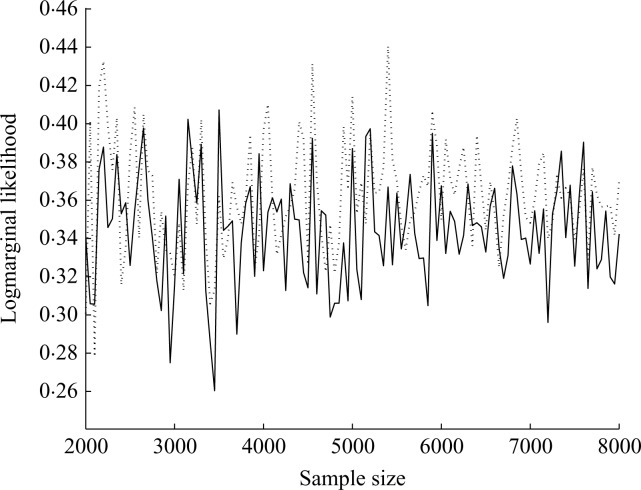
Figure 1 gives an implementation check on the concordance of the two marginal likelihood estimates. These are very close and differ negligibly on the log probability scale even at small Monte Carlo sample sizes.
Fig. 1.
Logmarginal likelihood values in the example of § 5. The two estimates A (full line) and B (dashed line) of § 4.2 were successively re-evaluated and plotted here at differing simulation sample sizes. The vertical scale has been adjusted by addition of 3583 for clarity.
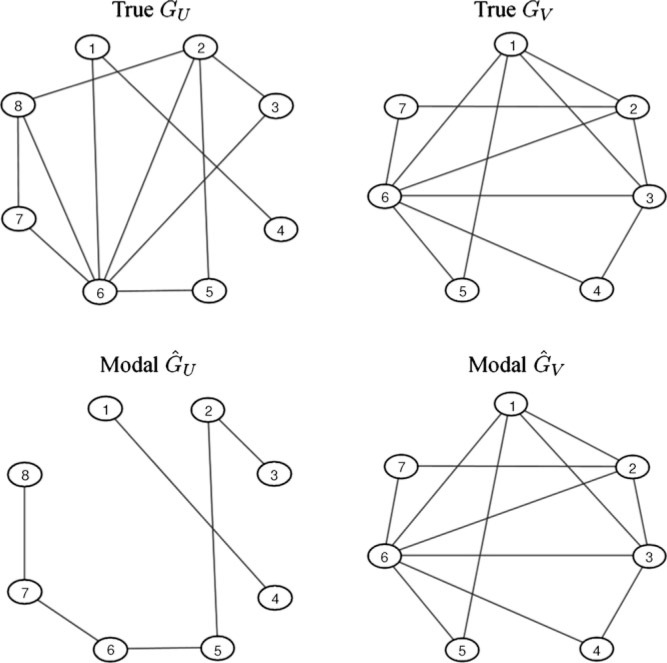
Consider graphical model uncertainty with prior edge inclusion probabilities 2/(q − 1) for GU and 2/(p − 1) for GV. Repeat explorations suggest stability of the marginal likelihood estimation using smaller Monte Carlo sample sizes, and we use 2000 draws within each step of the model search. The add/delete Metropolis-within-Gibbs was run for 20 000 iterates starting from empty graphs. Results are essentially replicated starting at the full graphs. The most probable graphs visited, (ĜU, ĜV), are shown in Fig. 2; these are local modes and also have greater posterior probability than the true graphs also displayed, and this model was first visited after 2614 Markov chain steps. The edges in (ĜU, ĜV) generally have higher posterior edge inclusion probability than those not included; the lowest probability included edge has probability 0.52, while the highest probability excluded edge has probability 0.59. Thus, graphs discovered by highest posterior probability and by aggregating high probability edges are not dramatically different. The modal ĜU is sparser than the true GU, reflecting the difficulties in identifying very weak signals; for example, the modal graph lacks an edge corresponding to the true Ω1,6 = 0.05, and the posterior probability of that edge is naturally low. One measure of inferred sparsity is the posterior mean of the proportion of edges in each graph; these are about 28%, 59.6% for GU, GV, respectively. Additional posterior summaries and exploration of the posterior samples suggest clean convergence of the simulation analysis and the Metropolis–Hastings steps over graphs had good empirical acceptance rates of about 26%, 9% for GU, GV, respectively.
Fig. 2.
True graphs in the simulated data example together with graphs of highest posterior probability identified from the analysis.
6. Dynamic matrix-variate graphical models for time series
Using the theory and methods for matrix normal models developed above, we are now able to extend our ideas to matrix time series involving two covariance matrices and associated graphical models. In the notation below, the work of Carvalho & West (2007a, 2007b) is the special case of vector data with q = 1, U fixed, and inference on (V, GV) only.
A q × p matrix-variate times series Yt follows the dynamic linear model
for t = 1, 2, . . ., where (a) Yt = (Yt,ij), the q × p matrix observation at time t; (b) Θt = (Θt,ij), the qs × p state matrix comprised of q × p state vectors Θt,ij each of dimension s × 1; (c) ϒt = (ωt,ij), the qs × p matrix of state evolution innovations comprised of q × p innovation vectors ωt,ij each of dimension s × 1; (d) νt = (νt,ij), the q × p matrix of observational errors; (e) Wt is the s × s innovation covariance matrix at time t; (f) for all t, the s-vector Ft and s × s state evolution matrix Gt are known. Also, ϒt follows a matrix-variate normal distribution with mean 0, left covariance matrix U ⊗ Wt and right covariance matrix V. In terms of scalar elements, we have q × p univariate models with individual s-vector state parameters, namely,
| (4) |
for each i, j and t. Each of the scalar series shares the same Ft and Gt elements, and the reference to the model as one of exchangeable time series reflects these symmetries. In the example below, Ft = F and Gt = G, as in many practical models, but the model class includes dynamic regressions when Ft involves predictor variables. This form of model is a standard specification (Quintana & West, 1987; West & Harrison, 1997) in which the correlation structures induced by U and V affect both the observation and evolution errors; for example, if uij is large and positive, vector series Yt,i★ and Yt, j★ will show concordant behaviour in movement of their state vectors and in observational variation about their levels. Specification of the entire sequence of Wt in terms of discount factors (West & Harrison, 1997) is also standard practice, typically using multiple discount factors related to components of the state vector and their expected degrees of random change in time, as illustrated in the example below. The innovations here concern graphical modelling and inference on (U, V). The key theory, conditional on U, V, concerns the conjugate sequential learning and forecasting as data is processed, as follows.
Theorem 1. Define Dt = {Dt−1, Yt} for t = 1, 2, . . ., with D0 representing prior information. With initial prior (Θ0 | U, V, D0) ∼ N (m0, U ⊗ C0, V) we have, for all t:
posterior at t – 1: (Θt−1 | Dt−1, U, V) ∼ N (mt−1, U ⊗ Ct−1, V);
prior at t: (Θt | Dt−1, U, V) ∼ N (at, U ⊗ Rt, V) where at = (In ⊗ Gt)mt−1 and Rt = Gt Ct−1 G′t + Wt;
one-step forecast at t – 1: (Yt | Dt−1, U, V) ∼ N (ft, Uqt, V) with forecast mean matrix ft = (In ⊗ F′1 Gt)mt−1 and scalar qt = F′t Rt Ft +1; and
posterior at t: (Θt | Dt, U, V) ∼ N (mt, U ⊗ Ct, V) with mt = at + (Iq ⊗ At)et and Ct = Rt − At A′t qt where At = Rt Ft/qt and et = Yt − ft.
Proof. This stems from the theory of multivariate models applied to vec(Yt) (West & Harrison, 1997). The main novelty here concerns the separability of covariance structures. That is: for all t, the distributions for state matrices have separable covariance structures; for example, (Θt |Dt, U, V) is such that cov{vec(Θt) | Dt, U, V} = V ⊗ U ⊗ Ct; the sequential updating equations for the set of qs × p state matrices are implemented in parallel based on computations for the univariate component models, each of them involving the same scalar qt, s–vector At and s × s matrices Rt, Ct at time t.
Suppose now that U and V are constrained by graphs GU and GV, with priors as in equation (3) and sparsity priors over the graphs. Given data over t = 1, . . ., n, the sequential updating analysis on (GU, GV) leads to the full joint density
marginalized with respect to all state vectors. The one-step forecast error matrices et are conditionally independent matrix normal variates. Apart from the scalars qt, this is essentially the framework of § 2. Thus, with a small change to insert the qt, we are able to directly fit and explore dynamic graphical models using the analysis for random samples with embedded sequential updating computations.
7. A macro-economic example
An example concerns exploration of conditional dependence structures in macroeconomic time series related to US labour market employment. The data are Current Employment Statistics for the eight US states, New Jersey, New York, Massachusetts, Georgia, North Carolina, Virginia, Illinois and Ohio. We explore these data across nine industrial sectors: construction; manufacturing; trade, transportation and utilities; information; financial activities; professional and business services; education and health services; leisure and hospitality; and government. In our model framework, we have q = 8, p = 9 and monthly data over several years. Then U characterizes the residual conditional dependencies among states while V does the same for industrial sectors, in the context of an overall model that incorporates time-varying state parameters for underlying trend and annual seasonal structure in the series. Trend and seasonal elements are represented in standard form, the former as random walks and the latter as randomly varying seasonal effects. Specifically, in month t, the monthly employment change in state i and sector j is Yt,ij, modelled as a first-order polynomial/seasonal effect model (West & Harrison, 1997) with the state vector comprising a local-level parameter and 12 seasonal factors, so that the state dimension is s = 13.
The univariate models of equation (4) have state vectors Θt,ij = (μt,ij, ϕt,ij)′, where μt,ij is the local level and ϕt,ij = (ϕt,ij,k, ϕt,ij,k+1, . . ., ϕt,ij,11, ϕt,ij,0, . . ., ϕt,ij,k−1) contains current monthly seasonal factors, subject to 1′ϕt,ij = 0 for all i, j and t. Further, Ft = F (13 × 1) and Gt = G (13 × 13) for all t, where F′ = (1, 1, 0, . . ., 0). The state matrix G and the sequence of state evolution covariance matrices Wt (13 × 13) are
with the latter having entries as follows. The univariate Wt,μ and 12 × 12 matrix Wt,ϕ are defined via discount factors δl and δs and the corresponding block components of Ct as Wt,μ = Ct−1,,μ (1 − δl)/δl and Wt,ϕ = PCt−1,ϕ P′(1 − δs)/δs for each t. The discount factor δl reflects the rate at which the levels μt,ij are expected to vary between months, with 100( − 1)% of information on these parameters decaying each month. The factor δs plays the same role for seasonal parameters. We use δl = 0.9, δs = 0.95 to allow more adaptation to level changes than seasonal factors (West & Harrison, 1997); results, in terms of graphical model search and structure, are substantially similar using other values in appropriate ranges. In application, we can estimate discount factors and also extend the model to allow changes in discount factors to model change-points and other events impacting the series, based on monitoring and intervention methods (Pole et al., 1994; West & Harrison, 1997). Such considerations are secondary to our purposes in using this model for illustration of computational model search analysis for (U, V, GU, GV), but practically very germane. Model completion uses initial, vague priors with m0 = 0, the 104 × 9 matrix, and C0 = 100I13. The constraint that 1′ϕt,ij = 0 is imposed by transforming m0 and C0 as discussed in West & Harrison (1997).
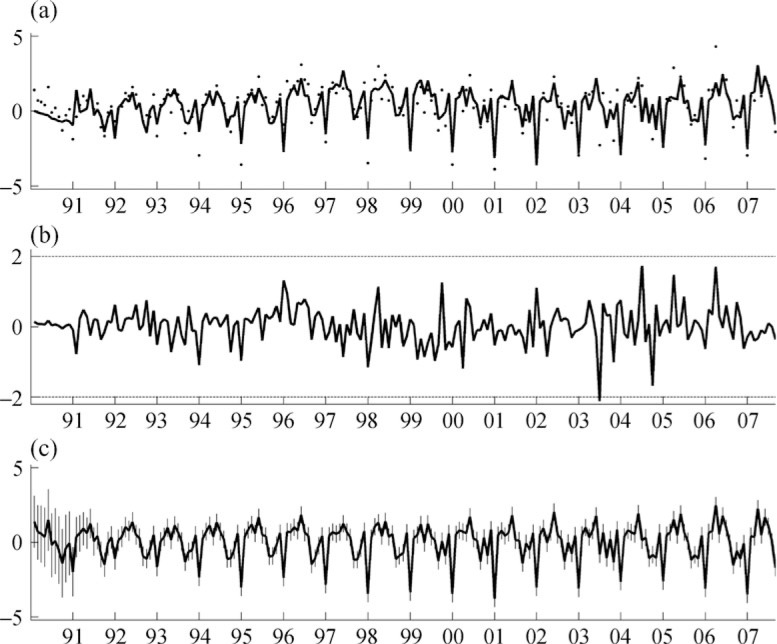
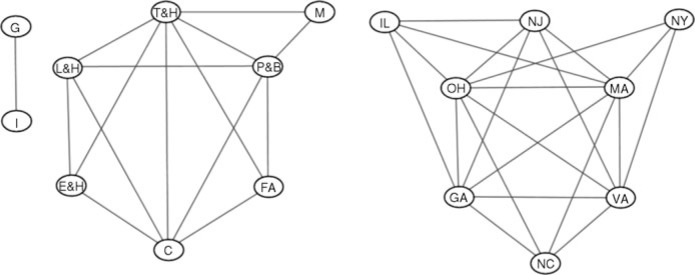
Model fitting estimates the movements in trend and seasonality, sequentially generating matrix series et whose row and column covariance patterns relate to (U, V). The North Carolina financial activities data and some aspects of the sequential model fit are graphed in Fig. 3. Priors for (U, V) use B = 5I8, D = 5I9 and b = d = 3, reflecting the range of residual variation, sparsity-encouraging priors with prior edge inclusion probabilities 2/(q − 1) for GU and 2/(p − 1) for GV. The add/delete Metropolis-within-Gibbs sampler was run for 20 000 steps. Two chains were run: one starting at empty graphs and one at full graphs. The most probable model identified, (ĜU, ĜV), is shown in Fig. 4. This, and the acceptance rates of graphs, were insensitive to the starting points. Beginning with empty graphs, the most probable model visited was found after 401 steps; its log posterior probability is −27 695.40, the sparsity of (ĜU, ĜV) in terms of percentage of edges included is (72.4%, 42.1%) and the acceptance rates are (7.3%, 11.9%). Beginning with full graphs led to a most probable model with log posterior probability −27 695.43 after 2194 steps, sparsity (73.7%, 41.9%) and acceptance rates (7.7%, 12.2%). Posterior edge inclusion probabilities are also consistent between the two runs; see Table 1. Further, the most probable graphs sit in a region of graphs of similar sparsity and posterior probability and the posterior is dense around this mode; see Fig. 5.
Fig. 3.
One of the 72 time series in the econometric example, plotted over 1990–2007. (a): Dots are monthly changes in employment of North Carolina financial activities and the line joins the corresponding one-step ahead forecasts over time. (b): Corresponding standardized one-step ahead forecast errors et/ √ qt. (c): Corresponding on-line estimated seasonal pattern with 95% point-wise credible intervals indicated by vertical bars at each month.
Fig. 4.
Highest posterior probability graphs that illustrate aspects of inferred conditional dependencies among industrial sectors and among states in analysis of the econometric time series data.
Table 1.
Posterior edge inclusion probabilities in graphical model analysis of the matrix econometric time series data
| NJ | NY | MA | GA | NC | VA | IL | OH | ||
|---|---|---|---|---|---|---|---|---|---|
| NJ | 1 | 0.05 | 1.00 | 0.55 | 0.01 | 1.00 | 1.00 | 1.00 | |
| NY | 1 | 1.00 | 0.19 | 0.00 | 1.00 | 0.00 | 0.59 | ||
| MA | 1 | 0.98 | 0.96 | 1.00 | 1.00 | 1.00 | |||
| GA | 1 | 0.93 | 0.89 | 0.75 | 1.00 | ||||
| NC | 1 | 1.00 | 0.06 | 1.00 | |||||
| VA | 1 | 0.31 | 1.00 | ||||||
| IL | 1 | 1.00 | |||||||
| OH | 1 | ||||||||
| C | M | T&U | I | FA | P&BS | E&H | L&H | G | |
| C | 1 | 0.02 | 1.00 | 0.16 | 0.75 | 1.00 | 0.99 | 1.00 | 0.06 |
| M | 1 | 1.00 | 0.28 | 0.02 | 0.98 | 0.01 | 0.03 | 0.01 | |
| T&U | 1 | 0.02 | 1.00 | 1.00 | 0.93 | 1.00 | 0.02 | ||
| I | 1 | 0.06 | 0.34 | 0.02 | 0.01 | 0.55 | |||
| FA | 1 | 1.00 | 0.00 | 0.04 | 0.02 | ||||
| P&BS | 1 | 0.02 | 1.00 | 0.00 | |||||
| E&H | 1 | 0.75 | 0.02 | ||||||
| L&H | 1 | 0.01 | |||||||
| G | 1 |
US states: NJ, New Jersey; NY, New York; MA, Massachusetts; GA, Georgia; NC, North Carolina; VA, Virginia; IL, Illinois; OH, Ohio. Industrial sectors: C, industrial construction; M, manufacturing; T&U, trade, transportation & utilities; I, information; FA, financial activities; P&BS, professional & business services; E&H, education & health services; L&H, leisure & hospitality; G, government.
Fig. 5.
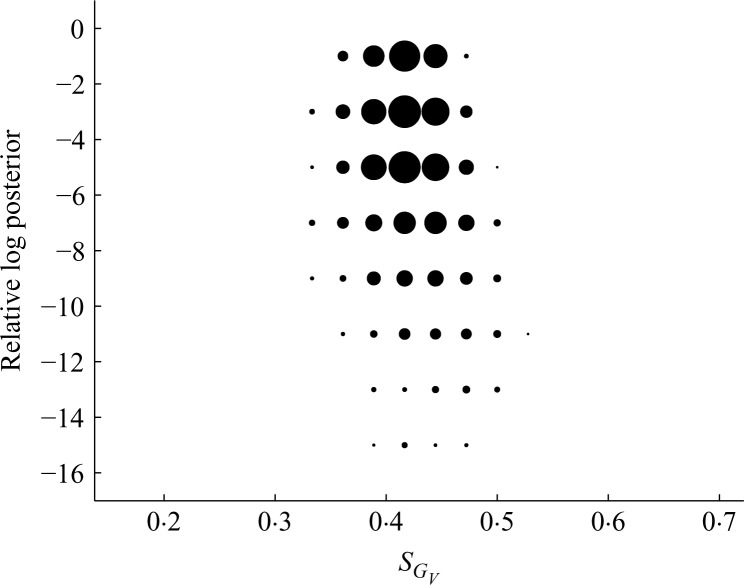
Summary of posterior on sparsity of GV in the econometric example. Circled areas are proportional to the fraction of posterior sampled graphs at several levels of posterior probability plotted against levels of sparsity SGV measured as the proportion of edges included.
Graphs with high probability in the region of the mode seem to reflect relevant dependencies in the econometric context. There are strongly evident conditional independencies particularly among subsets of the industrial sectors; see Table 1. Further, the posterior indicates overall sparsity levels through posterior means of about 73% for the proportion of edges included in GU and about 42% in GV. Figure 5 further illustrates aspects of the posterior over sparsity for GV.
8. Further comments
We have introduced Bayesian analysis of matrix-variate graphical models in random sampling and time series contexts. The main innovations include new priors for matrix normal graphical models, use of the parameter expansion approach, inference via Markov chain Monte Carlo for a specific graphical model, evaluation of marginal likelihoods over graphs using coupled candidate’s formula approximations, and the extension of graphical modelling to matrix time series analysis.
On the use of parameter expansion, Roy & Hobert (2007) and Hobert & Marchev (2008) provide theoretical support for the method in Gibbs samplers; in our models, this approach induces tractable and computationally accessible posteriors, leads to good mixing of Markov chain simulations, and is theoretically fundamental to the new model/prior framework in addressing identification issues directly and naturally.
On model identification, an alternative approach might use unconstrained hyper-inverse Wishart priors for each of (U, V) and run the Markov chain Monte Carlo simulation on the unconstrained parameters, similar to a strategy sometimes used in multinomial probit models (McCulloch et al., 2000). It can be argued that this is computationally less demanding than using our explicitly constrained prior and that inferences can be constructed from the simulation output by transforming to constraint-compatible parameters (U υ11, V/υ11). We had considered this, and note that posterior simulation analysis is marginally faster than under the explicitly identified model; in empirical studies, however, we find the computational benefit to be of negligible practical significance. Importantly, this approach relies on a proper prior for the effectively free, unidentified parameter υ11, and is sensitive to that choice. More importantly, the implied prior on (U υ11, V/υ11) is nonstandard and difficult to interpret, and raises questions in prior elicitation and specification; for example, the implied margins for variances are those of ratios of inverse gamma variates and difficult to assess compared to the traditional inverse gamma, and there are now dependencies in priors on left and right covariance matrices. Perhaps most important are the resulting effects on approximate marginal likelihoods; in examples we have studied, the approach yields very different marginal likelihoods and the impact of the marginal prior on the unidentified υ11 plays a key role in that. In contrast, and though very slightly more computationally demanding, the direct and explicitly constrained hyper-inverse Wishart prior is easy to interpret, specify and, with results from Carvalho et al. (2007), implement; synthetic examples have verified the resulting efficacy of the simulation and model search computations.
Our use of candidate’s formula to provide different approximations to marginal likelihoods over graphs can be extended to multiple such approximations. We have explored other constructions, and found no obvious practical differences in the resulting estimates in simulated examples. This is an area open for theoretical investigation and in other model contexts. This also offers a route to extending the analysis here to nondecomposable graphical models.
Our examples are in modest dimensional problems where local move Metropolis–Hastings methods for the graphical model components of the analysis can be expected to be effective, building on experiences in multivariate models (Jones et al., 2005). To scale to higher dimensions, alternative computational strategies such as shotgun stochastic search over graphs (Dobra et al., 2004; Jones et al., 2005; Hans et al., 2007) become relevant. A critical perspective is to define analysis that will rapidly find regions of graphical model space supported by the data. It is far better to work with a small selection of high-probability models than a grossly incorrect model on full graphs, and as dimensions scale the latter quickly becomes infeasible. Shotgun stochastic search and related methods reflect this and offer a path towards faster, parallelizable model search. There is also potential for computationally faster approximations using expectation-maximization style and variational methods (Jordan et al., 1999).
An interesting class of matrix graphical structures arises under autoregressive correlation specifications for the two covariance matrices. This generates a class of Markov random field models that is of potential interest in applications such as texture image modelling. With the matrix data representing a spatial process on a rectangular grid, taking covariance matrices U and V as those of two stationary autoregressive processes provides flexibility in modelling patterns separately in horizontal and vertical directions. We have experimented with examples that suggest potential for this direction in applying the new theory and methods we have presented.
Acknowledgments
The authors are grateful for the constructive comments of the editor and two referees on the original version of this paper. Research reported here was partially supported under grants from the U.S. National Science Foundation and the National Institutes of Health. Any opinions, findings and conclusions or recommendations expressed in this work are those of the authors and do not necessarily reflect the views of the NSF or NIH. Matlab code implementing the analysis and the Graph-Explore visualization software are freely available at www.stat.duke.edu/research/software/west.
References
- Besag J. A candidate’s formula: a curious result in Bayesian prediction. Biometrika. 1989;76:183. [Google Scholar]
- Carvalho CM, Massam H, West M. Simulation of hyper-inverse Wishart distributions in graphical models. Biometrika. 2007;94:647–59. [Google Scholar]
- Carvalho CM, West M. Dynamic matrix-variate graphical models. Bayesian Anal. 2007a;2:69–98. [Google Scholar]
- Carvalho CM, West M. Dynamic matrix-variate graphical models—a synopsis. In: Bernardo JM, Bayarri MJ, Berger JO, Dawid AP, Heckerman D, Smith AFM, West M, editors. Bayesian Statistics. Oxford: Oxford University Press; 2007b. pp. 585–90. VIII, [Google Scholar]
- Chib S. Marginal likelihood from the Gibbs output. J Am Statist Assoc. 1995;90:1313–21. [Google Scholar]
- Dawid AP. Some matrix-variate distribution theory: notational considerations and a Bayesian application. Biometrika. 1981;68:265–74. [Google Scholar]
- Dawid AP, Lauritzen SL. Hyper-Markov laws in the statistical analysis of decomposable graphical models. Ann Statist. 1993;21:1272–317. [Google Scholar]
- Dobra A, Jones B, Hans C, Nevins J, West M. Sparse graphical models for exploring gene expression data. J Mult Anal. 2004;90:196–212. [Google Scholar]
- Dutilleul P. The MLE algorithm for the matrix normal distribution. J Statist Comp Simul. 1999;64:105–23. [Google Scholar]
- Finn JD. A General Model for Multivariate Analysis. New York: Holt, Rinehart and Winston; 1974. [Google Scholar]
- Galecki A. General class of covariance structures for two or more repeated factors in longitudinal data analysis. Commun. Statist. A. 1994;23:3105–19. [Google Scholar]
- Gelman A. Parameterization and Bayesian modeling. J Am Statist Assoc. 2004;99:537–45. [Google Scholar]
- Gelman A. Prior distributions for variance parameters in hierarchical models. Bayesian Anal. 2006;3:515–34. [Google Scholar]
- Giudici P. Learning in graphical Gaussian models. In: Bernado JM, Berger JO, Dawid AP, Smith AFM, editors. Bayesian Statistics. Oxford: Oxford University Press; 1996. pp. 621–28. 5, [Google Scholar]
- Giudici P, Green PJ. Decomposable graphical Gaussian model determination. Biometrika. 1999;86:785–801. [Google Scholar]
- Gupta AK, Nagar DK. Matrix Variate Distributions Monographs and Surveys in Pure & Applied Mathematics. London: Chapman & Hall; 2000. p. 104. [Google Scholar]
- Hans C, Dobra A, West M. Shotgun stochastic search in regression with many predictors. J Am Statist Assoc. 2007;102:507–16. [Google Scholar]
- Hobert JP, Marchev D. A theoretical comparison of the data augmentation, marginal augmentation and PXDA algorithms. Ann Statist. 2008;2:532–54. [Google Scholar]
- Huizenga HM, de Munck JC, Waldorp LJ, Grasman R. Spatiotemporal EEG/MEG source analysis based on a parametric noisecovariance model. IEEE Trans Biomed Eng. 2002;49:533–9. doi: 10.1109/TBME.2002.1001967. [DOI] [PubMed] [Google Scholar]
- Jones B, Carvalho CM, Dobra A, Hans C, Carter C, West M. Experiments in stochastic computation for high-dimensional graphical models. Statist Sci. 2005;20:388–400. [Google Scholar]
- Jordan M, Ghahramani Z, Jaakkola T, Saul L. An introduction to variational methods for graphical models. Mach Learn. 1999;37:183–233. [Google Scholar]
- Lauritzen SL. Graphical Models. Oxford: Clarendon Press; 1996. [Google Scholar]
- Liu C, Rubin DB, Wu YN. Parameter expansion to accelerate EM: the PX-EM algorithm. Biometrika. 1998;85:755–70. [Google Scholar]
- Liu JS, Wu YN. Parameter expansion for data augmentation. J Am Statist Assoc. 1999;94:1264–74. [Google Scholar]
- Mardia KV, Goodall CR. Spatial-temporal analysis of multivariate environmental monitoring data. In: Patil GP, Rao CR, editors. Multivariate Environmental Statistics. Amsterdam: Elsevier; 1993. pp. 347–85. [Google Scholar]
- McCulloch RE, Polson NG, Rossi PE. Bayesian analysis of the multinomial probit model with fully identified parameters. J Economet. 2000;99:173–93. [Google Scholar]
- Mitchell MW, Genton MG, Gumpertz ML. A likelihood ratio test for separability of covariances. J Mult Anal. 2006;97:1025–43. [Google Scholar]
- Naik DN, Rao SS. Analysis of multivariate repeated measures data with a Kronecker product structured covariance matrix. J Appl Statist. 2001;29:91–105. [Google Scholar]
- Pole A, West M, Harrison PJ. Applied Bayesian Forecasting and Time Series Analysis. New York: Chapman-Hall; 1994. [Google Scholar]
- Quintana JM, West M. Multivariate time series analysis: new techniques applied to international exchange rate data. Statistician. 1987;36:275–81. [Google Scholar]
- Roy V, Hobert JP. Convergence rates and asymptotic standard errors for MCMC algorithms for Bayesian probit regression. J. R. Statist. Soc. B. 2007;69:607–23. [Google Scholar]
- Theobald DL, Wuttke DS. Empirical Bayes hierarchical models for regularizing maximum likelihood estimation in the matrix Gaussian procrustes problem. Proc Nat Acad Sci. 2006;103:18521–7. doi: 10.1073/pnas.0508445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Harrison PJ. Bayesian Forecasting and Dynamic Models. 2nd ed. New York: Springer; 1997. [Google Scholar]
- Whittaker J. Graphical Models in Applied Multivariate Statistics. Chichester, UK: John Wiley and Sons; 1990. [Google Scholar]