Abstract
Mycolic acids are attractive diagnostic markers for tuberculosis (TB) infection because they are bacteria-derived, contain information about bacterial species, modulate host–pathogen interactions and are chemically inert. Here, we present a novel approach based on mass spectrometry. Quantification of specific precursor → fragment transitions of approximately 2000 individual mycolic acids (MAs) resulted in high analytical sensitivity and specificity. We next used this tool in a retrospective case–control study of patients with pulmonary TB with varying disease burdens from South Korea, Vietnam, Uganda and South Africa. MAs were extracted from small volume sputum (200 µl) and analysed without the requirement for derivatization. Infected patients (70, 19 of whom were HIV+) could be separated from controls (40, 20 of whom were HIV+) with a sensitivity and specificity of 94 and 93%, respectively. Furthermore, we quantified MA species in lung tissue of TB-infected mice and demonstrated effective clearance of MA levels following curative rifampicin treatment. Thus, our results demonstrate for the first time the feasibility and clinical relevance of direct detection of mycobacterial lipids as biomarkers of TB infection.
Keywords: diagnostic marker, lipidomics, mass spectrometry, Mycobacterium tuberculosis, mycolic acids
INTRODUCTION
The number of mycobacterial infections has increased dramatically over the past two decades due to, among other factors, the HIV/AIDS epidemic (Harries & Dye, 2006; Iademarco & Castro, 2003; Saltini, 2006). Infection with Mycobacterium tuberculosis (MTB) remains a major global health threat, with over nine million new cases and close to two million deaths annually. Detection of active tuberculosis (TB) remains a serious problem in areas where TB is present and in pre-clinical and clinical trials. Furthermore, there are currently no markers available that reliably reflect drug response and clearance of bacterial remnants, for example from lesions in the lung.
Prior to the introduction of sequence-based techniques, one clinically useful method for mycobacterial speciation was high-performance liquid chromatography (HPLC) analysis of unique mycolic acids (MAs) (Butler & Guthertz, 2001). MAs, which are categorized according to the functional groups present in their meromycolate chain, form a very complex family of lipids. In fact, the now almost-forgotten discipline of ‘chemotaxonomy’ was a cornerstone of most pre-genetic speciation schemes for the Corynebacterium-Mycobacterium-Nocardia (CMN) family of Actinomycetes (Wayne et al, 1996).
MAs, α-alkyl-β-hydroxy branched chain fatty acids (for a molecular structure, see Fig 1B), are major envelope components of mycobacteria and targets of therapeutic intervention in TB. They are attractive diagnostic markers for mycobacterial infections for several reasons. (1) They are cell wall-associated bacterial molecules and are not synthesized in the human body. Thus, the presence of MAs in host tissue is indicative of bacterial infection. (2) The chemical diversity of MAs can be used for taxonomy. (3) MAs are involved in first-line recognition of bacteria during host–pathogen interactions, contributing to bacterial survival inside macrophages. For example CD1 receptors bind MAs and stimulate subpopulations of T cells, thus modulating the immune response to infection. (4) MAs represent a substantial mass fraction of bacterial cell walls and provide a stable and durable shell. Their hydrophobic nature renders low permeability to the cell wall and protects bacterial cells from dehydration. The MA biosynthetic machinery is thus an important target for antimycobacterial drugs (Barry et al, 1998; Ehrt & Schnappinger, 2007; Tonge, 2000). In fact, it is the chemical stability of MAs that permitted early methods for taxonomy and epidemiology of TB infections. Unfortunately, analysis of MAs is complicated by their hydrophobic properties and heterogeneous chemistry.
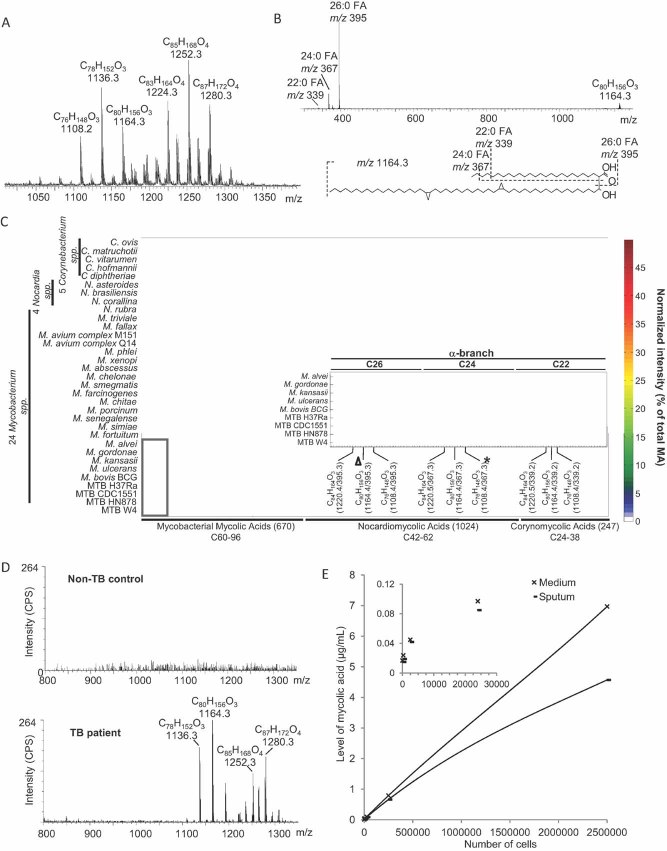
Figure 1. MS and fingerprints of MAs.
MAs from bacterial cell walls were extracted and analysed by ESI/MS in negative mode.
- ESI/MS spectrum of MAs derived from MTB Beijing strain.
- Product ion analysis (MS/MS) of m/z 1164, indicating the presence of C22:0 (m/z 339), C24:0 (m/z 367) and C26:0 (m/z 395) fatty acyls in the alpha-branch. See Figs S1–4 of Supporting information for additional ESI/MS of MAs derived from Corynebacteria, Nocardia and Mycobacteria. Based on these ESI/MS and MS/MS results, we developed a method for targeted analysis using MRM. Precursor/product ion pairs (i.e. MRM transitions) selective for 670 MAs of Mycobacteria, 1024 MAs of Nocardia and 247 MAs of Corynebacteria were used to profile 33 bacterial strains. The signal intensity for each MA species was normalized to the total signal intensity of all MAs measured in each strain (percent of total MA signal), and the resulting data is expressed in the heat plot format.
- The inset shows an enlarged section of the α-MAs in nine mycobacterial strains (box). The triangle indicates a major α-MA C80H156O3 with C26 as the alpha-branch. The asterisk indicates the α-MA C76H148O3 with C24 as the alpha-branch. The full list of all 1942 MAs measured here, along with their MRM transitions and alpha-branch fatty acyl compositions, is shown in Table S1 of Supporting information.
- Single stage mass spectra of sputum from a non-TB control and from a TB patient (ESI/MS). Ion counts are indicated as counts per second (CPS).
- LOD of MA in sputum and in media. Serial dilutions of MTB were spiked into control sputum and medium. After MA extraction, the MA were resuspended in 120 µl of mobile phase and quantified by MRM and normalization to internal MA standard C32. Ion responses for both medium and sputum are shown using a non-linear polynomial regression curve.
Here, we report the development of a rapid and sensitive diagnostic strategy that involves targeted quantification of 1942 MAs, expanding the current database of 203 MAs (http://lipidbank.jp/) by approximately 10-fold. Our approach is based on electrospray ionization tandem mass spectrometry (ESI/MS) and does not require derivatization of MAs (Butler & Guthertz, 2001; Laval et al, 2001). Various bacterial strains of the CMN family of Actinomycetes were grown in vitro and their MAs harvested by alkaline hydrolysis of delipidated cells. Using these complex lipid mixtures, we established a comprehensive list of MA precursor ions and corresponding fragment ions for multiple reaction monitoring (MRM). This experimental filter, which is both highly specific and sensitive, was next used to determine limits of detection of MA in vitro and further to assess which MAs are detectable in clinical samples. We show that quantification of MAs from sputum, in particular C26 α-MAs, leads to diagnostic performances comparable to or better than conventional acid-fast bacillus (AFB) smear staining. We also demonstrate that this analytical tool can serve as a diagnostic marker for TB infection and drug efficacy in laboratory animals.
To our knowledge, this is the first study to demonstrate the feasibility of direct measurement of MAs for TB case detection in a comprehensive clinical study and for assessment of drug efficacy in a laboratory setting. The approach is safe and fast, as no culturing is required. These results are thus a promising advance in the development of novel independent markers for TB infection and drug efficacy.
RESULTS
Targeted analysis of MAs using ESI/MS
We first used HPLC/ESI/MS (Shui et al, 2007) to profile MAs in 33 CMN organisms grown in vitro (see, e.g. MTB Beijing strain in Fig 1A). Differences in MA profiles between genera were easily observed (Figs S1–4 of Supporting information), and allow for their identification. We next used tandem mass spectrometry (MS/MS) and collision-induced dissociation (CID) to further characterize MAs in the 33 strains, revealing isobars with different lengths of alpha-branch fatty acids and of meromycolates (Fig 1B; Fig S5A and B of Supporting information and Table S1 of Supporting information). For example fragmentation of m/z 1164, which corresponds to an α-MA with a molecular composition of C80H156O3 which is present in many species of mycobacteria including MTB, yielded product ions corresponding to alpha-branch fatty acids with different chain lengths: trace amounts of m/z 339 (C22:0), m/z 367 (C24:0) and m/z 395 (C26:0) as the predominant acyl species (Fig 1B). Based on such product ion analysis, we established conditions for targeted analysis by MRM. In MRM, a precursor ion of interest (e.g. m/z 1108) is selected in the first mass analyser of a tandem mass spectrometer and fragmented in the collision cell, and a characteristic product ion (e.g. the acyl chain at m/z 367) is then selected in the second mass analyser. For example an α-MA with an m/z of 1108 and a C24:0 fatty acyl as the alpha-chain could thus be selectively monitored using the transition pair 1108/367 (Fig 1C, asterisk). A total of 1942 such MRM transitions specific for individual MA species were defined (Table S1 of Supporting information) and used to determine characteristic MA fingerprints of 33 CMN organisms (Fig 1C). Synthetic C32 MA (Laval et al, 2001) with a MRM transition of 495/255 was used as an internal standard to determine limits of detection and to quantify individual levels of various α-, keto- and methoxy-MAs extracted from mouse lung tissue and human sputum samples (see below). This MRM-based approach provides quantitative analysis that is very rapid (∼2 min analytical time per sample, though this does not take into account sample preparation, see discussion) and sensitive [limit of detection, LOD ∼ 1 pg of MA extracts, an ∼100-fold increase over existing methods (Shui et al, 2007)].
In a next step, we investigated if MAs can be extracted and analysed directly from sputum samples from TB patients. While no MA signal could be measured in sputum from non-TB control patients, a number of mycolates were extracted from TB patient's sputum (Fig 1D). Note that the lengths of MA extracted from bacterial cultures (Fig 1A) differ from MA extracted from sputum (D), with the major α-MA in culture being m/z 1136, but m/z 1164 in sputum. A similar trend of acyl lengthening can be observed for methoxy-MA m/z 1252 (culture) to m/z 1280 (sputum) and keto-MA m/z 1236 (culture) to m/z 1264 (sputum) (Fig 1A and D and not shown).
We further determined the minimum number of bacterial cells necessary for our MA detection approach, by performing a spiking experiment with a serial dilution of MTB cells added to non-TB sputum as well as culture medium. In sputum, around 10,000 colony forming units (CFU) were sufficient to detect bacteria based on their MA signal (signal-to-noise ratio, S/N = 3). A linear increase in MA levels was observed with increasing numbers of bacterial cells, for both, sputum and medium. In extracts from medium, MA signals were slightly higher than in extracts from sputum, which was probably due to MA extraction efficiency or ion suppression effects caused by the complex matrix of this body fluid (Fig 1E).
Quantitative analysis of MAs as a diagnostic marker for tuberculosis infection
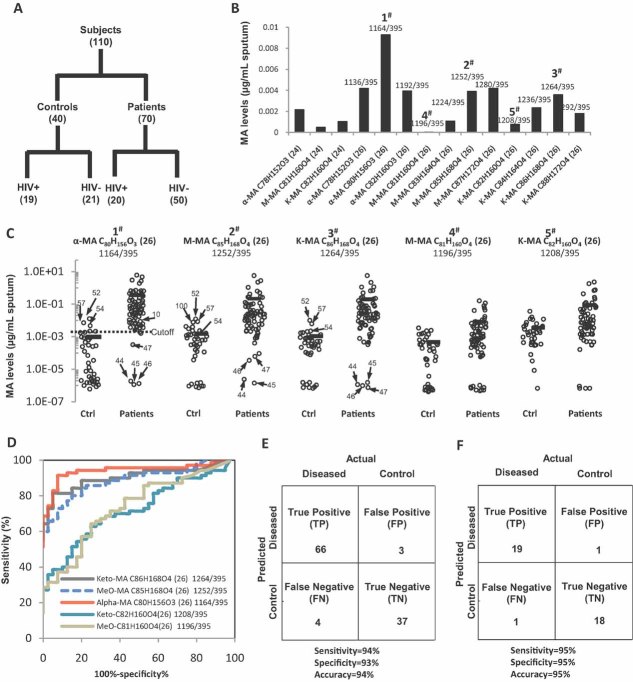
Using a blinded format, we conducted a retrospective, multicenter, case–control study of 70 patients with pulmonary TB with varying disease burdens and 40 non-TB controls (individuals with clinical symptoms of TB who were diagnosed as non-TB by culture). For both groups, the HIV status and bacterial burden were known (Fig 2A; for details, see demographic information in Table S2 of Supporting information). Strikingly, robust α-, keto- and methoxy-MA signals were detected in as little as 200 µl of sputum from TB patients (Fig 2B). This was too small of a sample volume for detecting MAs with alternative methods (Shui et al, 2007). Further, the concentration of major MAs detected in sputum obtained from TB-infected individuals was significantly (∼100 times) higher than in samples from non-TB controls (Fig 2C, see also discussion for false positives and negatives). As shown in Fig 2E, we classified 66 out of the 70 TB patients correctly as TB positive (‘true positive’) and 37 out of the 40 non-TB controls correctly as non-TB (‘true negative’). Three were falsely classified as TP positive (‘false positive’) and four were falsely classified as non-TB (‘false negative’). Using these data, we calculated a statistical sensitivity of 94% and a specificity of 93% (sensitivity: number of true positives divided by the sum of true positives and false negatives; specificity: number of true negatives divided by the sum of true negatives and false positives) and even slightly better values for the HIV positive individuals alone (Fig 2F).
Figure 2. MAs in sputum are diagnostic for TB infection in humans.
- A. Diagram illustrating the case–control set-up and number of TB patients (n = 70) and non-TB controls (n = 40) analysed. Samples originated from four different countries (see Table S2 of Supporting information for detailed demographic information).
- B. Sputum samples (200–500 µl) were extracted using organic solvents and levels of individual MA molecular species were determined using MS in MRM mode. Major MA species for α-MA (alpha-), M- (methoxy-) and K- (keto-) MA with their respective molecular composition are plotted, with (24) and (26) indicating the number of carbon atoms in their respective alpha chain.
- C. Individual MA molecular species varied in their power to differentiate between non-TB controls and TB patients. Numbered dots highlighting false positive and false negative results correspond to patient IDs in Table S2 of Supporting information. Cutoff for MRM 1164/395 between non-TB control and patients is 0.0026 µg/ml sputum.
- D. ROC curve displaying the classifying performance (positive diagnostic likelihood ratio) of individual MA molecular species, expressed by its true positive rate (sensitivity) and false positive rate (1—specificity), with α-MA C80H156O3 providing the best accuracy.
- E,F. The predictive power of MA quantification is shown by confusion matrix for all 110 samples (E) and for the 39 HIV+ patients (F) alone. Sensitivity and specificity were calculated as described in the text.
Careful inspection of the data obtained for each of the 110 participants showed that individual molecular species varied in their power to differentiate between non-TB controls and TB patients. Receiver operating characteristic (ROC) curves were computed to compare classifying characteristics of individual MA species (Fig 2D). MAs with (i) C26 alpha-branches, rather than those with C24 and C22 branches (Fig 2C and D and data not shown) and (ii) α-MAs, rather than oxygenated MAs such as methoxy-MAs or keto-MAs (Fig 2D), were best suited for diagnostic purposes. α-MA C80H156O3 (m/z 1164/395. Fig 1B) provided the best accuracy in our study population, yielding an area under the ROC curve of 0.94 for classification as TB/non-TB (red line in Fig 2D). Thus, this sensitive targeted MS analysis allowed for robust detection of MAs from minimally processed, small-volume sputum extracts. The large number of MAs included in the list helped identify those MAs present in clinical samples.
We next evaluated the possibility of using the full profiles of mycobacterial MAs, rather than individual MAs such as C26 α-MA, for diagnostic classification. This approach was based on the assumption that MA profiles of bacterial species grown in vitro reflect those found in sputum extracts. We used Euclidean cluster analysis to group mycobacterial strains according to similarities in their MA profiles. Strikingly, all sputum profiles analysed (four representative samples are shown in Fig S6 of Supporting information) fell within the MTB cluster. We further analysed MAs from two clinical non-tuberculous mycobacterial (NTM) isolates (NTM isolate-1 and -2) and one corresponding sputum sample (NTM sputum-2). NTM isolate-1 was characterized as Mycobacterium avium complex (MAC) by sequencing the 16S rRNA and rpoB regions (data not shown). Interestingly, the MA profile of NTM isolate-1 was closest to those of MAC M151 and MAC Q14 in our analysis. NTM isolate-2 was characterized as Mycobacterium massiliense, a rapidly growing mycobacterium that is indistinguishable from Mycobacterium chelonae/Mycobacterium abscessus with partial 16S rRNA gene sequencing (Simmon et al, 2007). MAs from both M. chelonae and M. abscessus were profiled in this study and were found to be most similar to NTM isolate-2 and the corresponding sputum (NTM sputum-2). Thus, in addition to indicating TB infection (Fig 2), the above results demonstrate that MA profiles allow for classification of the infecting mycobacteria (Fig S6 of Supporting information).
HIV co-infection is a complicating factor in TB diagnostics using assays that rely on immunological readouts, such as interferon-γ release. However, an approach that directly monitors bacterial factors such as MAs should not be influenced by such complications. We examined the effect of co-infection with HIV by comparing the predictive power of MA signals for both the complete set of 110 samples (Fig 2E) and the 39 HIV-positive patients only (Fig 2F). Sensitivities and specificities were very comparable, indicating that our approach is not impaired by HIV co-infection.
MAs as markers for drug efficacy in a controlled laboratory experiment
The majority of the study subjects (those from the FIND studies) had not received anti-TB chemotherapy for at least 60 days prior to enrolment. Eight of the 10 Korean patients had received combination therapy with first- and second-line drugs for more than a month at the time of specimen collection, but remained sputum positive (two of these were MDR patients; see Table S2 of Supporting information for detailed demographics and antibiotic treatment regimen). We did not observe differences (either qualitative or quantitative) in MA profiles between individuals who received anti-TB chemotherapy and those who did not within this limited number of patients. Larger study populations and longitudinal sampling over extended periods of time will be required to monitor MA levels during drug treatment in humans.
To evaluate our method for monitoring drug efficacy, we measured MAs in lung tissue of mice infected with MTB. These animals are routinely used in pre-clinical studies for development of novel treatments against TB. One commonly used marker for drug efficacy is body weight, which is only indirectly related to TB infection. The signatures of major MAs in mouse lung (Fig 3A) were very comparable to those observed in human sputum samples (Fig 2B). The bacillary burden of these mice as assessed by CFU in their lungs was 15.2 × 106 on average (as assessed in 5 out of the 10 animals tested for MA levels). We also measured MA levels in TB-infected mice that had been treated with the bactericidal antibiotic rifampicin (30 mg/kg body weight for 4 weeks), after which the mice were cured of TB (no detectable CFU in the lungs) (Jayaram et al, 2003). Impressively, the MAs in lung tissue of treated mice were significantly reduced compared to untreated mice, indicating that MAs were effectively cleared at some point during the 4-week treatment (Fig 3B). Thus, direct monitoring of pulmonary MAs can be used to differentiate TB-infected mice from healthy ones.
Figure 3. MAs in lung tissue are a diagnostic marker for TB infection in mice.
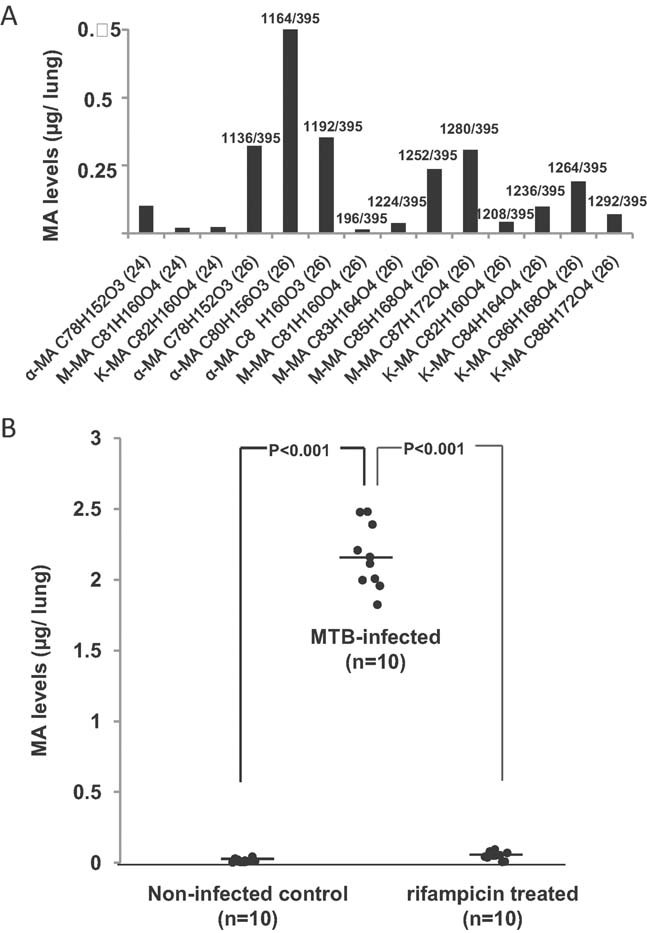
MAs were extracted from homogenized lungs of MTB-infected mice, spiked with synthetic C32 MA standard and the most abundant mycobacterial MA species were quantified using MRM.
- Levels of individual MA molecular species in the lung of one infected, untreated mouse 5 weeks post-infection.
- MA levels of non-infected control mice compared to MTB-infected and to rifampicin-treated mice. The average bacillary load in this infections was 15.2 million CFU/lung (n = 5).
DISCUSSION
Using a lipidomics approach, we identified a subclass of MAs, extracted directly from patients' sputum and mouse lungs, which can be used as a diagnostic marker for acute TB infection in humans and mice. The MA signal clearly differentiates these active TB samples from non-active or cured TB, as no MA (apart from three false positive samples) were detected in our non-TB control group (suspected TB patients) or in rifampicin-treated mice. Non-oxygenated MAs with C26 acyl moieties in their alpha-branch best classified active versus non-active TB. The key novelty of our approach is the rapid and direct measurement of distinct bacterial components with high sensitivity and selectivity in poorly defined heterogeneous biological material (sputum and lung tissue). Because no culturing was required, the method was quick and safe, and sample preparation was simple. Importantly, no chemical derivatization was needed, thus simplifying sample preparation and handling. In a case–control study spanning four ethnicities and varying disease burden, we demonstrated the feasibility of this approach for rapid differentiation of TB patients from non-TB patients, with sensitivity and specificity comparable to existing methods.
Our approach is highly synergistic with existing and alternative diagnostics. Our method provides speed, sensitivity and specificity over culture methods. Although the AFB smear stain is simple and reliable (i.e. ∼95% specificity) when used on good-quality specimens, many microorganisms are acid-fast, and thus NTM also lead to positive test results using AFB stains. Additionally, a major limitation of the AFB smear is its moderate sensitivity (∼60% on average). We compared the AFB smear in combination with bacterial culture to our new method (Table 1). Heavy burden cases (i.e. smear ++ and +++) were detected with equal accuracy by our method (100%, 15/15 smear ++ and 7/7 smear +++). Almost identical detection (97.5%) was seen with smear + cases (39/40). Thus, in these clear TB cases, the MA method is comparable in performance to the most widely used and most sensitive TB test currently available. It might offer advantages in specialized situations such as pediatric cases (where there is a very limited amount of sputum) or HIV positive/smear negative cases (where bacteria tend to be diluted in the sputum).
Table 1.
Diagnostic performance of our MA diagnostic tool compared to microscopy
| Smear − culture + | Smear scanty | Smear + | Smear ++ | Smear +++ | |
|---|---|---|---|---|---|
| Accuracy | 1/4 | 3/4 | 39/40 | 15/15 | 7/7 |
The number of TB cases accurately detected by our MA method was compared to the number of TB positive cases as determined by acid-fast bacilli (AFB) smear microscopy.
The more difficult cases are those in which the smear results are unclear (smear scanty) or negative (smear negative/culture positive). We were able to identify positive TB cases from 3/4 (75%) patients with smear scanty results and 1/4 (25%) smear negative/culture positive results (#10 in Fig 2C). Although our study population was relatively small, this result indicates that MA analysis is an important development that may lead to better detection of TB in a particularly critical subset of cases. Typically, these individuals are released without treatment and thus pose a risk for disease dissemination.
False negative detection with the MA approach included the remaining smear negative/culture positive cases (#44–47 in Fig 2C), and could be explained by limits in extraction efficiency and instrument sensitivity. To evaluate the predictive power of our method especially in these borderline cases, we analysed sputum samples from an additional 50 smear negative/culture positive subjects as replication cohort (see Table S2 of Supporting information for demographics). These subjects all produced two consecutive smear negative sputa, which were subsequently diagnosed as TB positive by liquid culture. Three out of these 50 subjects (6%) were detected positively with our MA method (subject #111, #114 and #128; data not shown). While this result is still superior to AFB staining, additional improvements in sensitivity are required. Approaches to improve our methodology would include enhancement of MA extraction efficiency (e.g. novel extraction approaches based on solid phases or supercritical fluid extraction) or increased analytical sensitivity (e.g. improved ionization sources and ion funnels).
We re-sampled and re-measured banked specimens of the three false-positive cases (#52, #54 and #57 in Fig 2C), but they remained false positive. We do not currently have a plausible explanation for this.
Even though C26 α-MA as a single marker seems sufficient to differentiate between active and non-active TB, the inclusion of all mycobacterial MA added specificity to our approach and allowed for additional classification based on MA profiles (Fig S6 of Supporting information). Patients infected with NTM could be clearly distinguished from those infected with TB using cluster analysis of MA patterns. Although detection of active TB remains a key goal in the development of novel diagnostics, differentiation between MTB and NTM infection is also important because markedly different chemotherapeutic regimens are used to treat these infections. Current technologies that differentiate between these infections and provide species identification rely on culture and DNA amplification of species-specific sequences, leading to a delay of up to 6–8 weeks before information needed for patient care is obtained (Bruijnesteijn van Coppenraet et al, 2004; Garcia-Quintanilla et al, 2002; Marris, 2007; Shrestha et al, 2003).
Biological clearance of MAs from lung lesions and their transport within the body remain poorly understood but are of great general interest. Distinct MAs have been associated with various biological functions, for example virulence, growth rate and drug resistance. MAs may vary by clade and the specific environmental conditions of the strain (Barry et al, 1998; Ehrt & Schnappinger, 2007; Rao et al, 2006; Riley, 2006; Tonge, 2000). The technique described here provides a powerful new approach to monitor subtle differences in MA structure that may be important for pathogenicity. Thus, in addition to potential development of new diagnostic tools (Kaufmann et al, 2005; Layre et al, 2009), the results from our study may be applicable to other areas including basic chemistry and biology of MAs (e.g. biosynthesis and transport) and their immunology (e.g. CD1-mediated activation of T cells). Recent and upcoming clinical trials in humans will require longitudinal testing of large numbers of specimens for drug efficacy and prediction of relapse. It will be particularly interesting to combine existing and other novel diagnostics (e.g. culture or molecular test such as the novel GeneXpert system (Boehme et al, 2010) with MA profiles in a longitudinal manner across different body fluids (i.e. sputum, urine and blood), as well as in connection with latent TB infections. These data will help us better understand responses to new drugs and clearance of mycobacteria at sites of infection (Warner & Mizrahi, 2006).
Although the MA approach requires specialized equipment, such analysis may be feasible in the future because of the increased technical capabilities of reference laboratories in areas with endemic TB infections. Unlike proteins and nucleic acids, MAs are inert and stable during sampling, even at tropical temperatures and during storage for extended periods of time. Importantly, given that TB epidemiology is primarily driven by the HIV/AIDS pandemic, with up to 50% of HIV positive patients succumbing to TB in some geographic areas, it is crucial that diagnostic tools accurately detect TB in both HIV negative and HIV positive individuals. Direct detection of bacteria-derived molecules circumvents the need for indirect readouts from immunological tests, which may be difficult to interpret in patients with multiple infections and compromised immune functions (i.e. elderly or HIV positive patients).
In summary, we have demonstrated that analysis of MAs by ESI/MS can be used as a potential diagnostic tool for rapid and sensitive identification of mycobacterial pathogens (including both MTB and NTM species). This method can be applied directly to human sputum and reflects the effects of chemotherapy, as shown in infected and rifampicin-treated mice, suggesting that it may be used to monitor treatment response and potentially to predict relapse. As a next step, the epidemiologic sensitivity and specificity should be evaluated, using a prospective validation cohort.
MATERIALS AND METHODS
Growth conditions
All strains were grown in Middlebrook medium (Difco Laboratories Ltd, West Molesey, UK) supplemented with 0.05% Tween 80 (v/v) (Sigma) either on 7H10 agar slants or in 7H9 broth, at 37°C; Mycobacterium ulcerans was grown at 30°C. Mycobacterium bovis BCG Pasteur (ATCC 35734) and MTB strains (H37Ra, CDC1551, HN878 and W4) were cultivated in broth under rotating conditions (125 rpm) until the mid-logarithmic phase. Effects of different growth conditions on MA profiles were evaluated by cultivating M. bovis BCG on agar as well as in Dubos media using the Wayne model of hypoxic dormancy and were found to be insignificant (Shui et al, 2007). The NTM strains MAC Q14, MAC M151, Mycobacterium gordonae, Mycobacterium fortuitum, Mycobacterium simiae, Mycobacterium kansasii, M. abscessus, M. chelonae and Mycobacterium xenopi were grown on agar and kindly contributed by the Department of Microbiology at National University Hospital, Singapore. Corynebacteria, Nocardia and all other mycobacterial species were cultivated as surface pellicles in broth and obtained from the laboratory of Mamadou Daffé and Marie Laneelle. NTM isolates-1 and -2 were obtained from expectorated sputum specimens (see below), grown on agar and provided as a kind gift by Kenneth Olivier (NIH, USA).
Animal experiments
All animal work was approved by the NITD Animal Welfare Committee (IUCAC 08-2008) and performed in compliance with national laws and institutional policies. Five-week-old female Balb/C mice were divided into three groups of 10 animals each. Two groups were infected by intranasal inoculation with 102–103 CFU of MTB Beijing W4. One of these groups was treated with rifampicin daily (30 mg/kg body weight, a dose that is curative and far below the LD50) (Jayaram et al, 2003). The second group was treated with vehicle only (PBS containing 0.25% carboxymethyl cellulose), and the third group was maintained as an uninfected, vehicle-treated control group. Five weeks post-infection, all mice were terminally bled under anaesthesia via retro-orbital sinus puncture. Lungs were homogenized in PBS containing 1% Triton X-100, and dilutions were plated for CFU analysis.
Clinical and demographic data of participants
The 110 participants analysed in this study represent at least four ethnic backgrounds originating from different geographical areas: the FIND study sites in the high-TB incidence countries of Vietnam (n = 82), South Africa (n = 2) and Uganda (n = 16), and the NIH study site in the mid-TB incidence region of South Korea (n = 10) at the National Masan Tuberculosis Hospital. All FIND samples were acquired under local IRB approvals and informed consent was obtained from all subjects. The Korean samples were collected under an IRB-approved clinical protocol to study multidrug resistant MTB (NCT00341601). Informed consent was obtained from all participants. From both sites, individuals were at least 18 years old (average 36), and the male-to-female ratio was 1:0.4.6 (see Table S2 of Supporting information for detailed demographic data). Inclusion for enrolment was based on the presence of one or more clinical symptoms of pulmonary TB (persistent cough for at least 3 weeks, abnormal chest radiography, fever, night sweats, weight loss or contact with an active TB case). Suspicion of TB was verified by positive AFB staining, solid culture and liquid culture using two consecutive sputum samples (see Table S2 of Supporting information for individual results). For culture-positive samples, speciation of the causative agent as MTB complex was performed using the Capilia method (Tauns Laboratories Inc., Japan). All individuals were tested for HIV, and positive cases were serologically confirmed. Participants from the FIND study sites had been without anti-TB treatment for at least 60 days prior to enrolment, whereas, eight individuals from the Masan hospital were on anti-TB chemotherapy at the time of specimen collection (Table S2 of Supporting information). All sputum samples were blinded for MA extraction and analysis. Sputum was further collected from two smear-positive patients with NTM infections at the NIH Clinical Center in the USA under an IRB-approved NIAID clinical protocol studying atypical mycobacterial disease (NCT00018044).
Sputum processing
No sputum specimen was concentrated. Expectorated sputum specimens (0.2–5 ml) from NIH study sites were digested and decontaminated by adding an equal volume of Sputasol solution (Oxoid Ltd, Basingstoke, UK), incubated at room temperature for 20 min with occasional vortexing, divided into aliquots and stored at −80°C until MA isolation. Sputum specimen from FIND study sites for MA extraction were neither digested nor decontaminated, but only homogenized by vortexing with glass beads for 1 min, divided into 0.5 ml aliquots and stored at −70°C until MA isolation.
For microscopy and culture using the MGIT system, sputum was decontaminated using NALC-NaOH. Staining for all smears was standard Ziehl–Nielsen (=acid-fast blue) staining. The smears were examined under bright field microscopy at a 400× magnification.
Isolation of MAs from bacteria, human sputum and mouse lung tissue
For MA isolation, bacteria, human sputum and mouse lung homogenates were treated with chloroform/methanol (2:1 v/v) and shaken overnight at 4°C to inactivate the pathogens and extract free lipids. After separating the phases with H2O and centrifuging at 9000 rpm for 2 min, the lower organic phase (containing free lipids) was removed. To reduce ion suppression by phospholipids and hydrolysed fatty acids, a second delipidation was carried out by repeating these steps. The upper aqueous phase was carefully removed, and the lower organic phase was collected and pooled with that from the first extraction. The intermediate layer was transferred to a fresh tube and dried. One volume of freshly prepared 20% tetrabutylammonium hydroxide was added to the dried material to hydrolyse the covalently linked MAs. The sample was vortexed thoroughly and heated for saponification in an oven at 105°C for 2 h. After cooling and acidification by adding ∼50 µl of 12 M HCl (desired pH ∼ 4–5), 2 ml of hexane were added. The sample was vortexed vigorously for 1 min and spun at 5000 rpm for 2 min. The upper hexane layer was transferred to a fresh screw-cap tube, and the extraction was repeated twice with an additional 2 ml of hexane each. The hexane layer from the re-extraction was pooled with that from the first extraction and dried using a stream of N2 gas.
The paper explained
PROBLEM
Tuberculosis (TB), which is caused by infection with Mycobacterium tuberculosis (MTB), remains a major global health threat, with over nine million new cases and close to two million deaths annually. Detection of active TB cases in endemic countries still relies on sputum smear microscopy, which only has an average sensitivity of 65% and which is highly dependent on the individual examiner. The more sensitive culture methodology on the other hand takes weeks, thus further delaying initiation of treatment and potential spread of pathogens. Better diagnostics are therefore urgently needed. Since a high percentage of patients are co-infected with HIV, diagnostics should be suitable for immunocompromised individuals. Furthermore, a specific molecular marker which reflects clearance of bacteria from lungs would be helpful to monitor treatment response in pre-clinical and clinical trials.
RESULTS
Using ESI/MS analysis of small amounts of human sputum (200 µl), we could demonstrate that mycobacteria-derived MAs serve as a reliable and direct marker for detection of acute TB. We used this tool in a well-controlled retrospective case–control study of patients with pulmonary TB with varying disease burdens from South Korea, Vietnam, Uganda and South Africa (n = 110). Infected patients (70, 19 of whom were HIV+) could be separated from controls (40, 20 of whom were HIV+) with a sensitivity and specificity of 94 and 93%, respectively. In an animal model of TB infection, our methodology was further able to distinguish active TB cases from cured ones.
IMPACT
MAs are an attractive diagnostic marker for TB infection because they are bacteria derived, contain information about bacterial species and are chemically inert. Our results demonstrate for the first time the feasibility and clinical relevance of direct detection of mycobacterial lipids as biomarkers for TB infection as well as infections caused by NTM. The developed methodology is robust, due to the stability of the analysed molecules and the independence of host factors, and also fast, giving reliable results within a day. Further we could show that MAs can serve as a reliable marker of infection in drug efficacy studies, featuring fast clearance from animal lungs after successful drug treatment.
HPLC/ESI/MS and tandem ESI analysis of MAs
HPLC/ESI/MS analysis of MAs was carried out as described (Shui et al, 2007). Briefly, a Waters XTerra column (1 mm × 150 mm) was used to separate lipids. Chloroform/methanol (1:1 v/v) with 5% of 300 mM piperidine (final concentration 15 mM) was used as the mobile phase for isocratic elution. The mass spectra were acquired from m/z 400 to 1500 on a Waters Micromass QTOF. Typically, 2 µl of sample were injected for analysis. MA profiles for Fig 1D and supporting information figures were obtained by combining QTOF MS spectra during the MA elution period during column separation, as described previously (Shui et al, 2007). Tandem MS of peaks from HPLC/ESI/MS for all 33 strains analysed was carried out with collision energies ranging from 40 to 80 volts.
Multiple reaction monitoring (MRM)-based MS analysis of MAs
An Applied Biosystems Triple Quadrupole/Ion Trap mass spectrometer (4000Qtrap, Foster City, California, USA) was used for quantification of individual MAs. Extracted MAs were resuspended in chloroform/methanol/200 mM piperidine (1:1:0.1 by volume) and infused into the mass spectrometer with the same solution as the mobile phase at a flow rate of 8 µl/min and measured in the negative ESI mode. Based on tandem MS of ions obtained from HPLC/ESI/MS and precursor ion scans of fatty acyl head groups, comprehensive sets of MRM transitions were set up for quantitative analysis of MAs (Table S1 of Supporting information). The signal intensity of each MRM value was normalized to the total MA counts or an internal standard for quantitative comparisons.
Statistical analysis
Concentrations of MAs extracted from human sputum and mouse lung tissues from different disease states were compared statistically. MA measurements were found to be distributed non-normally when employing a Shapiro–Wilk test. Therefore, the nonparametric Mann–Whitney U-test was used to compare TB-infected individuals with non-TB controls, and the Dunn's post hoc test was used to compare TB-infected, rifampicin-treated mice and uninfected mice. Statistical analysis was carried out using GraphPad Prism.
Isotopic correction
Due to the overlapping nature of pseudo-molecular ion peaks of the species of interest with the M + 2 isotope peak from another species that has a 2 Da lower mass, isotopic correction was required (Han & Gross, 2005). Consider any MRM transition, 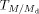 , where M is the m/z of the parent and Md is the m/z of the daughter. Let the peak intensity of this MRM transition be
, where M is the m/z of the parent and Md is the m/z of the daughter. Let the peak intensity of this MRM transition be 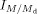 . To perform isotopic correction, the correction factor z must be calculated to obtain the corrected intensity
. To perform isotopic correction, the correction factor z must be calculated to obtain the corrected intensity 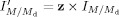 . Assuming that m, the total number of carbons in the transition
. Assuming that m, the total number of carbons in the transition 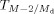 , is much larger than 1 (Han & Gross, 2005), the correction factor z is calculated as follows:
, is much larger than 1 (Han & Gross, 2005), the correction factor z is calculated as follows:
Clustering (Euclidean)
Hierarchical clustering was used to group samples that were similar in MA profiles. The algorithm aims to group samples with the smallest Euclidean distances. If we consider each sample as an object, a Euclidian distance metric is calculated between each pair of objects. The clustering algorithm looks for the pair of objects that is separated by the shortest distance and groups them together to form a new object. The distances between such newly formed objects are then recalculated to form a new distance metric. These steps are performed iteratively until all objects are merged to become a single object. To display the results, a dendrogram consisting of U-shaped lines is drawn. The height of each U represents the distance between the objects. Hierarchical clustering was performed using the pdist () function provided in MatLab (R2006a, The MathWorks, Natick, MA, USA) with Euclidean distance metric.
Acknowledgments
We thank Sindhu Ravindranand and Hui Chien Tay (NITD) for help with animal experiments and Xueli Guan (NUS) for technical assistance with figures. We are grateful to Kenneth Olivier (NIH) for providing NTM specimens. Special thanks are also due to Eloise Valli and Catharina Boehme (FIND) for their excellent help in managing the clinical samples from the FIND repository. This work was supported by the National Research Foundation (under CRP Award No. 2007-04), the Academic Research Fund (R-183-000-160-112), the Biomedical Research Council of Singapore (R-183-000-211-305), the National Medical Research Council (R-183-000-224-213) and partly by the Intramural Research Program of the NIH, NIAID. Work in the laboratory of MD was supported in part by a grant (SysteMTb HEALTH-F4-2010-241587) from the European Community.
Supporting information is available at EMBO Molecular Medicine online.
The authors declare that they have no conflict of interest.
Author contributions
GS, AKB, IAJ, HML and MWB did lipid extraction, mass spectrometry, data collection and analysis; MH and VD planned the animal work. ML, MD, LEV, SHH, JSL, SYE, HKK, SZZR, ALTM, CEB and GM performed the coordination and collection of bacterial strains and clinical samples; GHC and MWB carried out bioinformatics and statistical analysis; GS developed the MRM methods; MRW, GS and AKB conceived the project, directed scientifically and wrote the paper.
For more information
More general information on TB can be found on the World Health Organization (WHO) website:
Specific strategies to fight infectious diseases such as TB are summarized by the ‘Stop TB partnership’:
a partnership housed by the WHO
The not-for-profit ‘Foundation for Innovative New Diagnostics’ (FIND) has useful information regarding the urgent need to develop and implement novel TB diagnostics:
http://www.finddiagnostics.org/
The ‘Novartis Institute for Tropical Diseases’ (NITD) is a drug discovery research institute focusing on neglected tropical diseases including TB:
http://www.nibr.com/research/developing_world/NITD/
More information of lipid research can be found on the Wenk laboratory website: http://www.lipidprofiles.com
Supplementary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Barry CE, III, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, Yuan Y. Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruijnesteijn van Coppenraet ES, Lindeboom JA, Prins JM, Peeters MF, Claas EC, Kuijper EJ. Real-time PCR assay using fine-needle aspirates and tissue biopsy specimens for rapid diagnosis of mycobacterial lymphadenitis in children. J Clin Microbiol. 2004;42:2644–2650. doi: 10.1128/JCM.42.6.2644-2650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler WR, Guthertz LS. Mycolic acid analysis by high-performance liquid chromatography for identification of Mycobacterium species. Clin Microbiol Rev. 2001;14:704–726. doi: 10.1128/CMR.14.4.704-726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S, Schnappinger D. Mycobacterium tuberculosis virulence: lipids inside and out. Nat Med. 2007;13:284–285. doi: 10.1038/nm0307-284. [DOI] [PubMed] [Google Scholar]
- Garcia-Quintanilla A, Gonzalez-Martin J, Tudo G, Espasa M, Jimenez deAnta MT. Simultaneous identification of Mycobacterium genus and Mycobacterium tuberculosis complex in clinical samples by 5′-exonuclease fluorogenic PCR. J Clin Microbiol. 2002;40:4646–4651. doi: 10.1128/JCM.40.12.4646-4651.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- Harries AD, Dye C. Tuberculosis. Ann Trop Med Parasitol. 2006;100:415–431. doi: 10.1179/136485906X91477. [DOI] [PubMed] [Google Scholar]
- Iademarco MF, Castro KG. Epidemiology of tuberculosis. Semin Respir Infect. 2003;18:225–240. doi: 10.1053/s0882-0546(03)00074-4. [DOI] [PubMed] [Google Scholar]
- Jayaram R, Gaonkar S, Kaur P, Suresh BL, Mahesh BN, Jayashree R, Nandi V, Bharat S, Shandil RK, Kantharaj E, et al. Pharmacokinetics–pharmacodynamics of rifampin in an aerosol infection model of tuberculosis. Antimicrob Agents Chemother. 2003;47:2118–2124. doi: 10.1128/AAC.47.7.2118-2124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann SH, Cole ST, Mizrahi V, Rubin E, Nathan C. Mycobacterium tuberculosis and the host response. J Exp Med. 2005;201:1693–1697. doi: 10.1084/jem.20050842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval F, Laneelle MA, Deon C, Monsarrat B, Daffe M. Accurate molecular mass determination of mycolic acids by MALDI-TOF mass spectrometry. Anal Chem. 2001;73:4537–4544. doi: 10.1021/ac0105181. [DOI] [PubMed] [Google Scholar]
- Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J, Mori L, Stenger S, De LG, Puzo G, et al. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chem Biol. 2009;16:82–92. doi: 10.1016/j.chembiol.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Marris E. From TB tests, just a ‘yes or no’ answer, please. Nat Med. 2007;13:267. doi: 10.1038/nm0307-267. [DOI] [PubMed] [Google Scholar]
- Rao V, Gao F, Chen B, Jacobs WR, Jr, Glickman MS. Trans-cyclopropanation of mycolic acids on trehalose dimycolate suppresses Mycobacterium tuberculosis-induced inflammation and virulence. J Clin Invest. 2006;116:1660–1667. doi: 10.1172/JCI27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley LW. Of mice, men, and elephants: Mycobacterium tuberculosis cell envelope lipids and pathogenesis. J Clin Invest. 2006;116:1475–1478. doi: 10.1172/JCI28734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltini C. Chemotherapy and diagnosis of tuberculosis. Respir Med. 2006;100:2085–2097. doi: 10.1016/j.rmed.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Shrestha NK, Tuohy MJ, Hall GS, Reischl U, Gordon SM, Procop GW. Detection and differentiation of Mycobacterium tuberculosis and nontuberculous mycobacterial isolates by real-time PCR. J Clin Microbiol. 2003;41:5121–5126. doi: 10.1128/JCM.41.11.5121-5126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui G, Bendt AK, Pethe K, Dick T, Wenk MR. Sensitive profiling of chemically diverse bioactive lipids. J Lipid Res. 2007;48:1976–1984. doi: 10.1194/jlr.M700060-JLR200. [DOI] [PubMed] [Google Scholar]
- Simmon KE, Pounder JI, Greene JN, Walsh F, Anderson CM, Cohen S, Petti CA. Identification of an emerging pathogen, Mycobacterium massiliense, by rpoB sequencing of clinical isolates collected in the United States. J Clin Microbiol. 2007;45:1978–1980. doi: 10.1128/JCM.00563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonge PJ. Another brick in the wall. Nat Struct Biol. 2000;7:94–96. doi: 10.1038/72458. [DOI] [PubMed] [Google Scholar]
- Warner DF, Mizrahi V. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin Microbiol Rev. 2006;19:558–570. doi: 10.1128/CMR.00060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Good RC, Bottger EC, Butler R, Dorsch M, Ezaki T, Gross W, Jonas V, Kilburn J, Kirschner P, et al. Semantide- and chemotaxonomy-based analyses of some problematic phenotypic clusters of slowly growing mycobacteria, a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int J Syst Bacteriol. 1996;46:280–297. doi: 10.1099/00207713-46-1-280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.