Abstract
Early studies have shown how aberrantly expressed microRNAs are a hallmark of several diseases like cancer. MicroRNA expression profiling was shown to be associated with tumour development, progression and response to therapy, suggesting their possible use as diagnostic, prognostic and predictive biomarkers. Moreover, based on the increasing number of studies demonstrating that microRNAs can function as potential oncogenes or oncosuppressor genes, with the goal to improve disease response and increase cure rates, miRNA-based anticancer therapies have recently been exploited, either alone or in combination with current targeted therapies. The advantage of using microRNA approaches is based on its ability to concurrently target multiple effectors of pathways involved in cell differentiation, proliferation and survival. Here, we review our current knowledge about the involvement of microRNAs in cancer, and their potential as diagnostic, prognostic and therapeutic tools.
Keywords: biomarkers, diagnostics, human cancer, microRNAs, therapeutic impact
Discovery, biogenesis and mechanisms of action
Approximately 20 years ago, investigators first determined that components of the genome traditionally considered nonfunctional had, in fact, gene regulatory capacity.
MicroRNAs, initially discovered in 1993, when a small RNA encoded by the lin-4 locus was associated to the developmental timing of the nematode Caenorhabditis elegans by modulating the protein lin-14 (Lee et al, 1993), have then been revealed as essential part of the uncoding genome, playing a crucial role through a complicated gene regulation in all the most important processes and in different species, including vertebrates (Lagos-Quintana et al, 2001; a list of miRNA databases is reported in Table 1).
Table 1.
miRNA databases
| Name | Website | Reference |
|---|---|---|
| miRNA map | http://mirnamap.mbc.nctu.edu.tw/ | Hsu et al (2006) |
| miRBASE | http://mirbase.org/ | Griffiths-Jones et al (2008) |
| microRNA | http://www.microrna.org/microrna/home.do | Betel et al (2008) |
| coGemiR | http://www.cogemir.tigem.it/ | Maselli et al (2008) |
| miRGEN | http://www.diana.pcbi.upenn.edu/miRGen.html | Alexiou et al (2010) |
| deepBase | http://www.deepbase.sysu.edu.cn | Yang et al (2010) |
MicroRNAs are transcribed for the most part by RNA polymerase II as long primary transcripts characterized by hairpin structures (pri-microRNAs), and processed into the nucleus by RNAse III Drosha into 70–100 nts long pre-microRNAs (Lee et al, 2004). Drosha is a highly conserved 160 kDa protein containing two RNAse III domains and one double-strand RNA-binding domain. Drosha forms a huge complex, 500 kDa in Drosophila melanogaster (D. melanogaster) and 650 kDa in Homo sapiens, called Microprocessor and containing the co-factor Di George syndrome critical region 8 (DGCR8), also known as Pasha in D. melanogaster and C. elegans.
An alternative miRNA biogenesis pathway, called miRtron pathway, has been discovered among diverse mammals, drosophila and nematodes (Berezikov et al, 2007; Okamura et al, 2007; Ruby et al, 2007): miRtrons are regulatory RNAs, which get processed to form pre-miRs using the splicing machinery without Drosha-mediated cleavage.
The originated precursor molecules are exported by an Exportin 5-mediated mechanism to the cytoplasm (Yi et al, 2003), where an additional step mediated by the RNAse III Dicer, which acts in complex with the transactivating response RNA-binding protein (TRBP), generates a dsRNA approximately 22 nts long, named miRNA/miRNA*, including the mature miRNA guide, and the complementary passenger strand, the miRNA* (star miRNA; many publications refer to the two strand pair as miR-3p/miR-5p, referring to the direction of the functional miRNA). Whereas one of the two strands is selected as guide strand according to thermodynamic properties, the complementary one is usually subjected to degradation. The so-called miRNA* was initially thought to be the strand subjected to degradation, instead more recent evidence suggests that it does not simply represent a non-functional bioproduct of miRNA biogenesis, but it can be selected as a functional strand and play significant biological roles (Bhayani et al, 2012).
Dicer are very large enzymes (∼200 kDa) conserved among the species and containing different domains: a double strand RNA-binding domain (dsRBD), two RNAse III catalytic domains, one PAZ domain, which binds the 3′-end of small RNAs, and other domains with ATPasic and RNA-helicasic activity. Dicer recognizes the double strand region of the pre-miRNA in association with different proteins: RDE-4 (RNA interference; RNAi defective 4) in C. elegans, R2D2 e FMR1 (fragile X mental retardation syndrome 1 homolog) in D. melanogaster and members of the Argonaut family in other species. In particular these proteins are not needed for the endonucleasic activity of Dicer but they play a role in stabilizing the complex Dicer-miRNA. In mammalians the Argonaut 2 (AGO2) protein complex, characterized by RNAse H activity, cooperates in the Dicer-mediated processing of some pre-miRNAs, yielding to another intermediate processing product, called AGO2-cleaved precursor miRNA (ac-pre-miRNA; Krol et al, 2010).
Completed the processing steps, the mature single stranded miRNA product is then incorporated in the complex known as miRNA-containing ribonucleoprotein complex (miRNP), miRNA-containing RNA-induced silencing complex (miRgonaute or miR-RISC), a ribonucleoproteic complex containing Argonaute proteins, of which AGO1 and 2 have been the most extensively studied, the two miRNA strands and several additional factors, including the trans-activator RNA binding protein (TRBP).
Glossary
AntagomiR
Class of chemically engineered oligonucleotides able to silence endogenous miRNAs. This definition was used for the first time by Krutzfeldt and collegues (Krutzfeldt et al, 2005), who developed a pharmacological approach for silencing miRNAs in vivo, designing chemically modified, cholesterol-conjugated single-stranded RNA analogues, complementary to miR-122.
Biomarker
Any parameter, molecule or protein, which can be used as indicator of a particular physiological or diseased state, disease progression, outcome and response to therapies.
Dicer
Endoribonuclease belonging to the RNase III family that cleaves double-stranded RNA (dsRNA) and pre-microRNA (miRNA) into short dsRNA fragments.
Drosha
RNase III enzyme responsible for initiating the processing of microRNA into the nucleus, where long primary transcripts characterized by hairpin structures (pri-microRNAs) are cleaved to generate 70–100 nts long pre-miRNAs.
Epigenetic regulation
Regulation of gene expression or cellular phenotypes caused by mechanisms that do not involve chromosomal aberrations or changes in the DNA sequence hence the name epi- (Greek: επí- over, above and outer) genetics.
LNA-anti-miR
Antisense RNA oligonucleotide modified to increase stability and specificity. In details, the ribose moiety of an LNA nucleotide is modified with an extra bridge connecting the 2′-oxygen and 4′-carbon, which ‘locks’ the ribose in the 3′-endo conformation. LNA nucleotides can be mixed with DNA or RNA residues in the oligonucleotide whenever desired.
miRNome
The miRNA profiling describing the expression of all microRNAs in a specific biological context.
miR-mask
Single-stranded 2′-O-methyl-modified (or other chemically modified) antisense oligonucleotide fully complementary to predicted miRNA binding sites in the 3′-UTR of a specific target mRNA. The miR-mask is thus able to cover up the access of the miRNA to its binding site on the target mRNA, so as to impair its inhibitory function.
miR-Risc
Ribonucleoproteic complex (RNA-induced silencing complex) where the mature single stranded miRNA product is incorporated and which guides its inhibitory function on the target mRNA.
miRNA sponge
miRNA inhibitory transgene expressing an mRNA containing multiple tandem binding sites for an endogenous miRNA, and thus able to stably interact with the corresponding miRNA and prevent the association with its endogenous targets.
MetastmiR
MicroRNA playing a crucial role in the metastatic process.
Oncogene
Gene causing or contributing to tumour occurrence or progression. Oncogenes are generally mutated forms of normal cellular genes (proto-oncogenes). A gene capable, when activated, of transforming a cell.
OncomiRNA
MicroRNA playing an oncogenic role by targeting oncosuppressor molecules.
Tumour suppressor gene
Tumour suppressor gene (also called anti-oncogene) is a gene able to protect a cell from uncontrolled proliferation and other aberrant events leading to cancer development. When this gene is mutated to cause a loss or reduction in its function, the cell can progress to cancer, usually in combination with other genetic changes.
Tumour suppressor miRNA
MicroRNA playing an oncosuppressive role by targeting oncogenes.
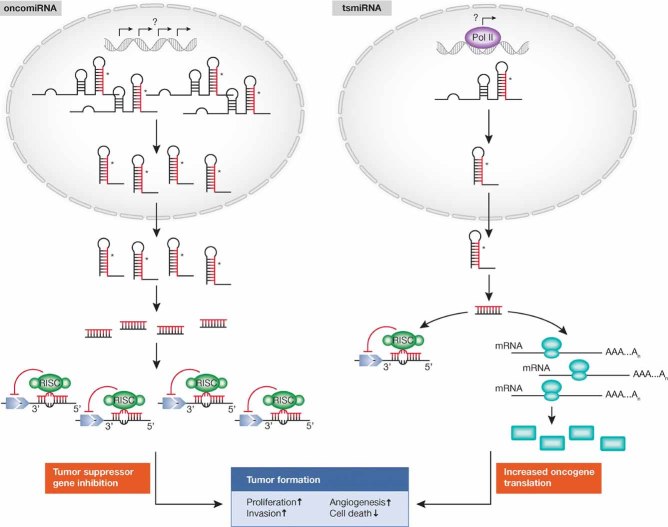
As a part of this complex, the mature miRNA is able to regulate gene expression at post-transcriptional level, binding for the most part through partial complementarity to target mRNAs, and mainly leading to mRNA degradation or translation inhibition.
More in detail, guided by the base pairing between the non-coding RNA and the target mRNA, miRNA-RISC-mediated gene inhibition can be split into three processes: (i) site-specific cleavage; (ii) enhanced mRNA degradation; and (iii) translational inhibition. The initial process, commonly defined as RNAi and restricted to miRNAs with a perfect or near-perfect match to the target RNA, is a very rare event in mammals, exclusively Ago2 dependent. Instead, the other two processes are more commonly associated with mismatched miRNA/target sequences that are the most likely scenario in mammals. The combination of these two processes is commonly defined as a non-cleavage repression, and can be carried out by any of the four mammalian Ago proteins (Su et al, 2009). Interestingly, recent genetic (Brodersen et al, 2008) and biochemical (Filipowicz et al, 2008) studies have also established that plant miRNAs can translationally repress their target mRNA, despite the near-perfect base pairing between the miRNA and target mRNA sequence.
However, the exact mechanism through which miRNAs can impair translation is still debated.
Moreover, even though it is known that microRNAs mainly recognize complementary sequences in the 3′-untranslated regions (UTRs) of their target mRNAs, more recent studies have reported that they can also bind to the 5′-UTR or the ORF (Lytle et al, 2007; Moretti et al, 2010; Ørom et al, 2008; Qin et al, 2010) and, even more surprisingly, they can upregulate translation upon growth arrest conditions (Vasudevan et al, 2007).
Finally, whereas the 5′-end of the microRNA (the so-called ‘seed site’) has always been considered the most important for the binding to the mRNA, recently the target sites have been further divided into three main classes, according to grade and localization of the complementarity (Brennecke et al, 2005): the dominant seed site targets (5′ seed-only), the 5′ dominant canonical seed site targets (5′ dominant) and the 3′ complementary seed site targets (3′ canonical).
Considering the different rules regulating the interaction between a microRNA and its target mRNA, it is not surprising that each miRNA has the potential to target a large number of genes (Betel et al, 2008; Friedman et al, 2009; Krek et al, 2005; Lewis et al, 2005). Equally, roughly 60% of the mRNAs share one or more sequences that are evolutionarily conserved and predicted to interact with miRNAs. Bioinformatics analysis predicts that 3′-UTRs of single genes are often targeted by several different miRNAs (Lewis et al, 2005). Many of these predictions have been validated experimentally, suggesting that miRNAs might cooperate to regulate gene expression (a list of computational tools for miRNA target prediction is reported in Table 2).
Table 2.
Computational tools for miRNA target prediction
| Name | Website | Reference |
|---|---|---|
| Targetscan | http://www.targetscan.org | Lewis et al (2005) |
| Pictar | http://www.pictar.org | Krek et al (2005) |
| RNA22 | http://cbcsrv.watson.ibm.com/rna22.html | Miranda et al (2006) |
| Tarbase | http://diana.cslab.ece.ntua.gr/tarbase/ | Sethupathy et al (2006) |
| PITA | http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html | Kertesz et al (2007) |
| microRNA | http://www.microrna.org/microrna/home.do | Betel et al (2008) |
| Diana-microT | http://diana.cslab.ece.ntua.gr/microT/ | Maragkakis et al (2009) |
| miRecords | http://mirecords.biolead.org/ | Xiao et al (2009) |
| Starbase | http://starbase.sysu.edu.cn/ | Yang et al (2011) |
To complicate the already intricate scenario, it has been recently reported that miRNAs can bind to ribonucleoproteins in a seed sequence and a RISC-independent manner and then interfere with their RNA binding functions (decoy activity; Beitzinger & Meister, 2010; Fig 1). Three studies have reported that miRNAs can also regulate gene expression at the transcriptional level by direct binding to the DNA (Gonzalez et al, 2008; Khraiwesh et al, 2010; Kim et al, 2008).
Figure 1.
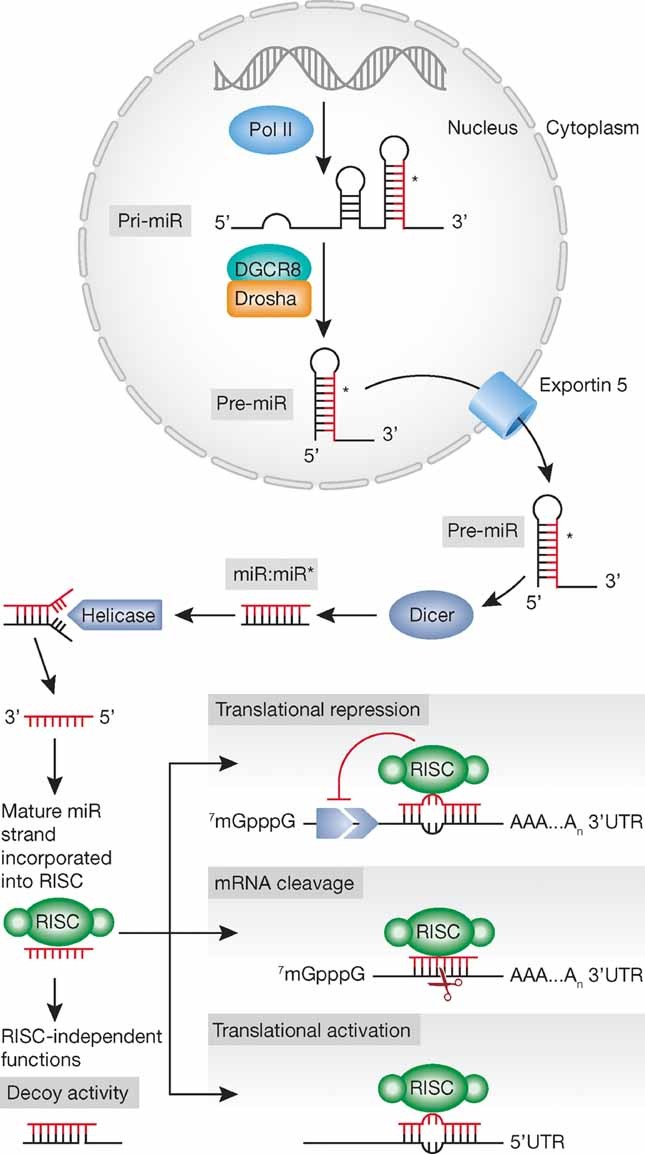
Biogenesis and mechanisms of action: an overview.
Overall, these data show the complexity and widespread regulation of gene expression by miRNAs, which should be taken into consideration when developing miRNA-based therapies.
MicroRNA expression in human cancer: diagnostic applications
The first evidence of the involvement of microRNAs in human cancer derived from studies on chronic lymphocitic leukemia (CLL), particularly in an attempt to identify tumour suppressors at chromosome 13q14, frequently deleted in CLL. Dr. Croce's group reported that rather than containing a protein coding tumour suppressor gene, this critical region contains in fact two microRNA genes, miR-15a and miR-16-1, expressed in the same polycistronic RNA. This result provided the first evidence that microRNAs could be involved in the pathogenesis of human cancer as the deletion of chromosome 13q14 caused the loss of these two microRNAs (Calin et al, 2002). Indeed, study of a large collection of CLLs showed knock down or knock out of miR-15a and miR-16-1 in approximately 69% of CLLs.
Following these initial observations, the same group mapped all the known microRNA genes and found many of them located in chromosomal loci prone to deletions or amplifications, as was found in many different human tumours (Calin et al, 2004). Indeed, chromosomal regions encompassing microRNAs involved in the negative regulation of a transcript encoding a known tumour suppressor gene can be amplified in cancer development. This amplification would result in the increased expression of the microRNA and consequent silencing of the tumour suppressor gene. Conversely, microRNAs repressing oncogenes are often located in fragile loci, where deletions or mutations can occur and result in reduced microRNA levels and overexpression of the target oncogene (Fig 2).
Figure 2.
MicroRNAs as oncogenes or tumour suppressor genes.
Therefore, alterations of microRNA expression are not exceptional but rather the rule in human cancer. Next, as an attempt to establish whether microRNA profiling could be used for tumour classification, diagnosis and prognosis, different platforms to assess the global expression of microRNA genes in normal and diseased tissues were developed (Calin & Croce, 2006): after an extensive use of custom-made (Liu et al, 2004) and then commercial miRNA microarrays, and bead-based flow cytometric miRNA analysis methods (Lu et al, 2005), the last generation of large-scale profiling method is represented by the high-throughput deep sequencing (Creighton et al, 2009; Farazi et al, 2011).
Genome-wide profiling showed that miRNA expression signatures (miRNome) allowed different types of cancer to be discriminated with high accuracy (Lu et al, 2005; Volinia et al, 2006) and the tissue of origin of poorly differentiated tumours to be identified. By contrast, mRNA profiles were highly inaccurate indicators of tissue or cancer type.
Indeed, miRNA mis-expression patterns are more accurate in identifying the origin of tumours that are otherwise difficult to be determined as the tumour spread to multiple metastatic sites, suggesting that tumours more clearly maintain a unique ‘tissue miRNA expression profile’. One study, for example, developed a classifier of 48 miRNAs from a sample of 336 primary and metastatic tumours, and was able to use this classifier to accurately predict the tissue origin in 86% of a blind test set, including 77% of the metastatic tumours (Rosenfeld et al, 2008). Given that cancers of undefined origin account for approximately 4% of all malignancies and are associated with poor prognosis (Oien & Evans, 2008), the continued development of miRNA classifiers has foreseeable benefits in aiding clinical diagnosis and subsequent treatment.
Another major issue in clinics is clearly represented by the need of biomarkers for an early diagnosis, extremely important considering that survival and prognosis of patients depends on the stage of the tumour at the time of detection, with an early diagnosis usually been associated with the best prognosis. MicroRNAs have revealed a great potential as new potential early diagnosis biomarkers: overexpression of miR-205 and miR-21 in ductal adenocarcinoma, for example has been reported to precede phenotypic changes in the ducts, thus suggesting the possibility to use them for an early detection of this neoplasm (du Rieu et al, 2010).
Furthermore, miRNAs are certainly more stable due to their small size as compared to long mRNAs, allowing expression profiling from fixed tissues or other biological material, and thus supporting their possible use as novel, minimally invasive and robust biomarkers. Certainly, miRNAs can be reliably extracted and detected from frozen and paraffin-embedded tissues, from blood (either total blood, plasma or serum; Mitchell et al, 2008; Schwarzenbach et al, 2011), circulating exosomes (Taylor & Gercel-Taylor, 2008), and from different biologic fluids like urine (Hanke et al, 2010), saliva (Michael et al, 2010; Park et al, 2009) and even sputum (Xie et al, 2010; Yu et al, 2010). Moreover, the profile of circulating miRNAs from individuals affected by different neoplasias was shown to reflect the pattern observed in the tumour tissues, suggesting the attractive possibility of using circulating miRNAs as easily detectable tumour biomarkers (Lawrie et al, 2008), especially for early diagnosis (Heneghan et al, 2010; Huang et al, 2010; Xing et al, 2010). More recently, by analysing plasma samples of lung cancer patients collected 1–2 years before the onset of disease, Sozzi's group (Boeri et al, 2011) has found microRNA signatures with strong predictive diagnostic and prognostic potentials.
miRNA profiles can distinguish not only between normal and cancerous tissue and identify tissues of origin, but they can also discriminate different subtypes of a particular cancer, or even specific oncogenic abnormalities. Gene expression profiling has already demonstrated its effectiveness at subtyping various cancers, however, miRNA profiles are equally discriminatory and can even be more informative, as expression changes can provide insights into the multitude of gene permutations observed in various cancer subtypes: links have indeed been made between misregulated miRNAs and the target genes that are affected, thus unraveling some of the unique gene networks involved (O'Day & Lal, 2010). miRNAs, for example, are differentially expressed between basal and luminal breast cancer subtypes (Blenkiron et al, 2007; Sempere et al, 2007), and can specifically classify estrogen receptor (ER), progesterone receptor (PR) and HER2/neu receptor status (Iorio et al, 2005; Lowery et al, 2009; Mattie et al, 2006). Some of the miRNAs associated with luminal or basal subtype, respectively, reflect their epithelial and myoepithelial origins. For instance, miR-200 family associates with the luminal subtype, and it is not surprising since they directly target the EMT regulators ZEB1 and ZEB2 (Gregory et al, 2008), whereas miR-145 and miR-205, preferentially expressed in normal myoepithelial cells, are dramatically reduced in basal-like triple negative tumours (ER−/PR−/HER2−), suggesting that this expression change might be a consequence of disease progression in this subtype (Sempere et al, 2007). Other examples are the differential expression of microRNAs according to specific histotypes of ovarian carcinoma (Iorio et al, 2007) and the ability of miR-205 expression to discriminate squamous from non-squamous non-small cell lung carcinoma (Lebanony et al, 2009).
Beside the expression profile studies based on microarray platforms, many other methods for detecting microRNAs have been developed, as quantitative real-time polymerase chain reaction (RT-qPCR; Chen et al, 2005; Raymond et al, 2005), in situ hybridization (de Planell-Saguer et al, 2010; Nuovo et al, 2009; Obernosterer et al, 2007; Pena et al, 2009) and high throughput sequencing (Schulte et al, 2010). The most important disadvantage of microarray technologies resides in the non-quantitative nature of this method, which, therefore, requires further experimental validation. Real-time PCR is extremely sensitive and accurate, however it is a more expensive and low-throughput method. In situ hybridization, based on the detection of specific miRNAs by hybridizing a complementary strand (probe) to the sequence of interest in morphologically preserved tissue sections or cell preparations, is certainly highly sensitive and allows the analysis at a single cell or subcellular level. However, it represents a technically challenging, semi-quantitative and low throughput method. Finally, whereas all these techniques are restricted to detection and profiling of previously identified miRNA sequences, sequence-based methods allow the identification of unknown microRNAs. Indeed, initially used to detect microRNAs expressed at a low level, and extremely expensive and time consuming, since 2007 deep sequencing methods, which rely on next generation sequencing machines, fast and accurate, have led to the discovery of new microRNAs. Recently, these methods have been used to reveal the differential expression of miRNAs in ovarian cancer (Wyman et al, 2009) and in favourable versus unfavourable neuroblastoma (Schulte et al, 2010).
In summary, despite the encouraging reports, to be successful at narrowing down the biology of a specific tumour to enable more accurate diagnoses, which would be of immense benefit to both doctors and patients, the accuracy of the method is of vital importance. And, similar to all big innovations, also for the use of microRNA signatures in clinics there are some kinks that need to be addressed. Reported lack of consistency between different studies, for example, certainly gives rise to some concern. Such differences typically arise from sample selection or preparation, experimental design and/or data analysis (Xu & Wong, 2010). Indeed, the use of different controls for data normalization can explain some of the observed variability across studies (Peltier & Latham, 2008). Another possibility that must be considered is the dynamic and immediate regulation in miRNA levels in stress response (Marsit et al, 2006) and in hypoxia (Kulshreshtha et al, 2007); thus, time of collection and processing could impact miRNA levels.
Nevertheless, experimental evidence reported up to date is certainly encouraging and promising, and even though a more comprehensive validation is the primary concern, this is unlikely to represent an obstacle to the development of miRNAs in diagnostics.
MicroRNAs as prognostic and predictive biomarkers
Being able to discriminate tumour origins, subtypes, oncogenic mutations and cancer predisposition, and regulating the most important cellular processes, it is logical to hypothesize that miRNAs might be able to predict also cancer prognosis and/or response to specific therapies. Recently, several groups have reported success in utilizing miRNAs as prognostic markers to predict cancer outcome, thereby addressing this issue. After the first evidence in CLL, where a unique microRNA signature was associated with prognostic factors and disease progression in CLL (Calin et al, 2005) and lung cancer, where miR-155 overexpression and let-7a downregulation were able to predict poor disease outcome (Yanaihara et al, 2006), several other reports have supported the significance of microRNAs as prognostic biomarkers. For example in gastric cancer a robust 7-miRNA signature can predict overall survival and relapse-free survival (Li et al, 2010). Similarly, low miR-191 and high miR-193a levels were associated with a significantly shorter survival time as measured by Kaplan–Meier curves in melanomas (Caramuta et al, 2010).
Although predicting survival might be important in a more general sense, the prediction of response to specific therapies is of far greater clinical value. For instance, low levels of miR-26 are an independent predictor of poor survival in patients suffering from hepatocellular carcinoma (HCC); notably, however, patients with low miR-26 responded well to interferon-α treatment, resulting in improved survival (Ji et al, 2009). Therefore, miR-26 expression might be a useful biomarker to discriminate patients who could benefit most from an interferon-α therapy. The other way around is also true, as several miRNAs have been correlated with a poor response to specific treatments. In various cancers, increased miR-21 expression is an indicator of poor outcome (Dillhoff et al, 2008; Rossi et al, 2010; Schetter et al, 2008) and it is also sufficient to predict poor response to adjuvant chemotherapy in adenocarcinomas (Schetter et al, 2008).
High levels of miR-125b in breast cancer predict poor response to taxol-based treatments in vitro (Zhou et al, 2010), and a similar finding has been reported for miR-21 in pancreatic cancer patients treated with gemcitabine (Giovannetti et al, 2010).
Beside the use as predictive biomarkers, the correlation between microRNA expression and response to specific therapies has also suggested their promising potential as therapeutic adjuvant, even though this hypothesis mostly derives from in vitro studies of gain or loss of function, where candidate miRNAs are initially identified in tumour cell lines with different degrees of resistance to specific therapeutic drugs and then targeted in order to overcome drug resistance. One of the first reports describing the involvement of microRNAs in chemoresistance was performed in cholangiocarcinoma cell lines (Meng et al, 2006), where inhibition of miR-21 and miR-200b increased sensitivity to gemcitabine. This first evidence was followed by many other studies (Chen et al, 2010a; Kovalchuk et al, 2008; Xia et al, 2008). Beside chemotherapy, microRNAs can also improve the responsiveness to targeted therapies: overexpression of miR-221 and miR-222 is responsible for resistance to anti-estrogenic therapies, as Tamoxifen (Miller et al, 2008; Zhao et al, 2008), and Fulvestran (Rao et al, 2011), whereas ectopic expression of oncosuppressor miR-205 is able to improve the responsiveness to tyrosin kinase inhibitors through direct targeting of HER3 (Iorio et al, 2009).
These associations highlight not only the importance of microRNAs as predictive biomarkers, but also the possibility to use them as an alternative approach for tackling drug resistance.
Mechanisms of miRNAs deregulation: from genetic abnormalities to epigenetic regulation
Regulation of microRNA expression can be exerted through several mechanisms, which result to be altered in human diseases, including cancer: chromosomal abnormalities, as suggested by the evidence that microRNAs are frequently located in regions of the genome involved in alterations in cancer (Calin et al, 2004), and confirmed by several studies (Calin et al, 2002; Tagawa & Seto, 2005; Zhang et al, 2006); mutations, as the inherited mutations in the primary transcripts of miR-15a and miR-16-1 responsible for reduced expression of the two microRNAs in vitro and in vivo in CLL (Raveche et al, 2007); polymorphisms (SNPs), as described in lung cancer (Hu et al, 2008).
In addition to structural genetic alterations, microRNA expression can be also modulated as a consequence of defects in the microRNA biogenesis machinery, as supported by changes in microRNA levels consequent an altered Drosha or Dicer activity in different tumour types (Merritt et al, 2008; Nakamura et al, 2007; Thomson et al, 2006). Moreover, it has been reported that also deregulation of different cofactors can affect miRNA expression with important biological implications: DGR8 knock out mice arrest early in development and have defects in ES cell proliferation and differentiation (Wang et al, 2007); Lin-28, originally discovered as a heterochronic gene regulating developmental timing in worms, is able to block let-7 biogenesis and it is activated in many human tumours (15%), being particularly associated with less differentiated cancers (Viswanathan et al, 2008).
The deregulated microRNA expression in cancer can also be due to epigenetic changes, as altered DNA methylation. An extensive analysis of genomic sequences of miRNA genes have shown that approximately half of them are associated with CpG islands, suggesting that they could be subjected to this mechanism of regulation (Weber et al, 2007). Several reports have indeed shown that aberrant methylation can result in aberrant microRNA expression in cancer: by treating T24 bladder cancer cells and human fibroblasts with DNMT inhibitor 5-Aza-2′-deoxycytidine, Saito and colleagues (Saito et al, 2006) reported a strong upregulation of miR-127, a microRNA characterized by a CpG island promoter able to target the proto-oncogene BCL-6, found silenced in several cancer cells. With the same approach of unmasking epigenetically repressed microRNAs inducing chromatin-remodelling by drug treatment, hypermethylation and down-modulation of miR-9-1 has been described in breast cancer (Lehmann et al, 2008), as well as the clustered miR-34b and miR-34c in colon cancer (Toyota et al, 2008).
Conversely, upregulation of putative oncogenic microRNAs can result from DNA hypomethylation, as shown in lung adenocarcinoma for let-7a-3 (Brueckner et al, 2007) or epithelial ovarian cancer for miR-21 (Iorio et al, 2007).
A differential methylation approach was used to identify epigenetically regulated microRNAs by profiling DNMT1- and DNMT3b-deficient colorectal cancer cells: out of 18 upmodulated microRNAs in comparison to WT cells, the only unmethylated in normal tissue but hypermethylated and silenced in tumour, was miR-124a, which embedded in a large CpG island and is able to target cyclin D kinase 6, thus mediating the phosphorylation of RB tumour suppressor gene (Lujambio et al, 2007).
Methylation is not the only epigenetic modification that can affect microRNAs expression: Scott et al (Scott et al, 2006) showed that in SKBR3 breast carcinoma cells, an extensive and rapid alteration of microRNA levels followed histone deacetylase inhibition.
Epigenetic drugs, such as DNA demethylating agents and histone deacetylase inhibitors, are able to reverse an aberrant methylation or acetylation status, thereby raising the intriguing possibility of also restoring the expression of tumour suppressor microRNAs, and reverting a tumoural phenotype.
To complicate the scenario, however, microRNAs themselves can regulate the expression of components of the epigenetic machinery, creating a highly controlled feedback mechanism: miR-29 family can directly target the de novo DNA methyltransferases DNMT-3A and -3B, while indirectly, through regulation of the transactivator Sp1, maintain the DNA methyl transferase DNMT1. Interestingly, introduction of miR-29s into lung cancers and AMLs was shown to result in reactivation of silenced tumour suppressors and inhibition of tumourigenesis (Fabbri et al, 2007; Garzon et al, 2009). Loss of miR-290 cluster in Dicer-deficient mouse embryonic stem cells leads to the upmodulation of the RBL-2, which repressed DNMT3a, DNMT3b and DNMT1 (Benetti et al, 2008; Sinkkonen et al, 2008); miR-1, involved in myogenesis and related diseases, directly targets HDAC4 (Chen et al, 2006).
Finally, miRNA dysregulation can result from increased or decreased transcription activity of a transcription factor at the promoter. The miR-34a, miR-34b and miR-34c family of miRNAs, for instance, was shown to be directly induced by the tumour suppressor p53 and to be partially responsible of the phenotype induced by this oncosuppressor (Chang et al, 2007; He et al, 2007).
Vice versa, oncogenes can also affect microRNA expression, and a clear example is represented by the oncoprotein MYC, which is able to both induce oncogenic microRNAs, as the miR-17-92 cluster, and negatively regulate transcription of tumour suppressor miRNAs, such as let-7 (Chang et al, 2009) and miR-29 family members (Mott et al, 2010).
Nevertheless, despite the advances in our understanding of the mechanisms causing miRNA deregulation, the daunting task still remains the elucidation of the biological role of miRNAs in the initiation and in the development of cancer.
MicroRNAs as oncogenes or oncosuppressor genes: functional evidence
Over time, cancers have developed sophisticated networks of biological activities allowing them to develop and, in some cases, evade treatment. This complex program relies on the communication between multiple cell types, including the primary tumour as well as the stromal cells. Hanahan and Weinberg described six essential features of cancer progression: self-sufficiency in growth signals, insensitivity to anti-growth signals, apoptosis evasion, limitless replicative potential, sustained angiogenesis and tissue invasion and metastasis (Hanahan & Weinberg, 2000). Dysregulated miRNAs may function as either tumour suppressors or oncogenes in cancer by targeting each one of these features (Fig 3).
Figure 3.
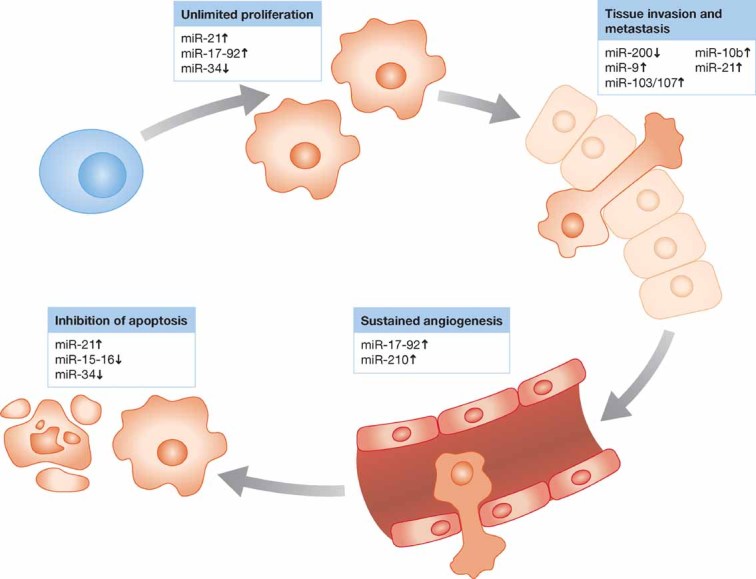
MicroRNAs targeting the hallmarks of cancer.
Gain- and loss-of-function experiments, together with target prediction analyses, have provided insights into the role of miRNAs in carcinogenesis.
Computational algorithms for target identification (Table 2), mainly based on the free binding energy between a miRNA and a putative target mRNA sequence, are by definition prediction tools, which need an experimental validation. The validation method most commonly used is represented by a reporter assay, where the co-transfection of the miRNA of interest and the tested 3′-UTR cloned downstream a luciferase gene reduces the reporter activity, inhibitory effect reverted by mutating the miRNA binding sequence of the target mRNA. However, even though this method suggests a physical and functional interaction between a miRNA and its target, it does not prove it directly. To this aim, more rigorous pull-down assays have been designed, as immunoprecipitation of labelled miRNA/mRNA complexes and consequent target identification by RT-PCR and sequencing (Hsu et al, 2009), or immunoprecipitation with Ago2 antibody, thus isolating the ternary, presumably functional, miRNA/mRNA/Ago2 complex (Chi et al, 2009).
However, the rules of miRNA/target mRNA regulation are even more complicated than expected. A very recent report by Pandolfi's group (Salmena et al, 2011) has introduced the revolutionary concept that miRNA effect on mRNA containing common miRNA recognition elements (MREs) can be affected by ceRNAs (competing endogenous RNAs): RNA transcripts, both protein coding and non-coding, can compete for miRNA binding, thus co-regulating each other.
Gain-of-function approaches have shown that miRNAs acting as tumour suppressors target oncoproteins with crucial roles in various cancer pathways, such BCL2 (targeted by miR-15a–miR-16-1; Cimmino et al, 2005), RAS (regulated by let-7; Johnson et al, 2005), myeloid cell leukaemia sequence 1 (BCl-2-related, MCL1, and targeted by miR-29; Garzon et al, 2009; Mott et al, 2007) and MYC (targeted by let-7; Sampson et al, 2007). Vice versa, to assess the biological effects of oncogenic miRNAs, often overexpressed in cancer cells, in vitro silencing was carried out using antisense oligonucleotides. For example miR-21 expression has been reported at high levels in breast (Iorio et al, 2005), glioblastomas (Ciafre et al, 2005), pancreas (Bloomston et al, 2007) and colon cancer (Schetter et al, 2008) among others. Chan and colleagues blocked miR-21 expression in glioblastoma cell lines and reported an increased activation of caspases and apoptosis (Chan et al, 2005). Additional studies showed that miR-21 anti-apoptotic effects occurs by targeting the tumour suppressors phosphatase and tensin homologue (PTEN) and programmed cell death 4 (PDCD4) (Frankel et al, 2008; Meng et al, 2007). More recently, Slack's group (Medina et al, 2010) has shown that mice conditionally expressing miR-21 develop a pre-B malignant lymphoid-like phenotype, thus demonstrating that miR-21 is a genuine oncogene.
miR-17-92 cluster and miR-155, both discovered to be overexpressed in lymphoproliferative disorders, including lymphomas and leukaemia (Garzon et al, 2008; He et al, 2005), were the first examples of miRNAs with oncogenic activity validated in engineered animal models. Infection of murine haematopoietic stem cells with a retrovirus carrying the miR-17-92 cluster accelerated the development of lymphomas in Myc transgenic mice (He et al, 2005). Transgenic mice overexpressing miR-17-92 cluster in B cells were discovered to develop lymphoproliferative disease and autoimmunity (Xiao et al, 2008). Lymphocytes higher rate of proliferation and lower rate of activation-induced cell death in these mice were partially accredited to the direct targeting of the anti-apoptotic genes Bim and Pten by miR-17-92 cluster. Moreover, Ventura and colleagues showed that mice deficient for miR-17-92 cluster die shortly after birth with lung hypoplasia and a ventricular septal defect (Ventura et al, 2008). Finally, Mu and colleagues showed that deletion of the complete miR-17-92 cluster slows down Myc-induced oncogenesis (Mu et al, 2009).
In contrast, miR-155 overexpression in the lymphoid compartment was sufficient to cause cancer without any other cooperative mutation or Myc expression. miR-155 transgenic mice developed polyclonal lymphoid proliferation followed by acute lymphocytic lymphoma or leukaemia (Costinean et al, 2006). These data were the first to report that the dysregulation of a single miRNA can lead to malignancy. Further, it was one of the first, and still of the few, microRNA engineered animal models, which, through knock out or transgene introduction, provide the genetic demonstration of the causative involvement of a specific microRNA in a biological phenomenon.
In addition to classical tumour suppressor or oncogene functions, miRNAs have been implicated also in cell migration and metastasis. The highly expressed miR-10b in metastatic breast cancer, positively regulates cell migration and invasion (Ma et al, 2007). Elegant experiments confirmed that overexpression of miR-10b in non-metastatic breast cancer cells initiates invasion and metastasis. The authors showed that these effects are mediated by direct targeting of HOXD10 by miR-10b, facilitating the overexpression of the well-known pro-metastatic gene RHOC. Besides, recently, the same lab reported that silencing miR-10b can inhibit metastasis in a breast cancer mouse model, thereby highlighting the therapeutic potential of targeting metastasis-associated miRNAs (Ma et al, 2010). Another study showed that miR-126 and miR-335 act as negative regulators of tumour invasion and metastasis in human breast and lung cancer (Tavazoie et al, 2008).
MiR-34a, lost in several tumour types and involved into the network mediated by the well known ‘genome guardian’ p53 (He et al, 2007), inhibits migration and invasion by down-regulation of MET expression in human HCC cells (Li et al, 2009).
Because the ‘epithelial–mesenchymal transition’ (EMT) is thought to promote malignant tumour progression, several groups have recently reported that microRNAs are involved in this process. Indeed, members of miR-200 family of microRNAs and miR-205 have been shown to reduce cell migration and invasiveness targeting ZEB transcription factors, known inducers of EMT (Gregory et al, 2008) and PKCε in prostate cancer (Gandellini et al, 2009). The oncogenic miR-21 stimulates invasation, extravasation and metastasis in different tumour types, included colorectal cancer (Asangani et al, 2008) and breast cancer (Zhu et al, 2008), while oncosuppressor miR-205 has opposite effects, reducing invasion in vitro and suppressing lung metastasis in vivo (Wu et al, 2009).
Interestingly, it has been observed that primary tumours and metastasis from the same tissue show a similar pattern of microRNAs expression (Rosenfeld et al, 2008). miRNA profiling is a more accurate classifier than mRNA profiling, and thus has the potential to solve one of the most challenging issues in cancer diagnostic: the origin of metastasis of unknown primary tumours.
During metastatic process, neo-angiogenesis is the crucial step allowing cells to reach and disseminate through the systemic circulation. MicroRNAs can control tumour progression at this level, either promoting or inhibiting the proliferation of endothelial cells. In endothelial cells, miR-221 and miR-222 repress proliferative and angiogenetic properties of c-Kit, while hypoxic signals reduce expressions of miR-16, miR-15b, miR-20a and miR-20b, directly targeting VEGF, and supporting the angiogenic process (Poliseno et al, 2006). Angiogenesis can be also promoted by miR-210, activated by hypoxia and directly repressing endothelial ligand Ephrin A3 (Pulkkinen et al, 2008). miR-17-92 cluster sustains MYC angiogenic properties by repressing connective tissue growth factor (CTGF) and anti-angiogenic adhesive glycoprotein thrombospondin 1 (TSP1; Dews et al, 2006), also targeted by miR-27b and let-7f (Kuehbacher et al, 2007).
It is unlikely that miRNAs will be found responsible for a specific phenotype by aiming at a specific target. Instead, it is largely accepted that miRNAs engage in complex interactions with the machinery that controls the transcriptome and concurrently target multiple mRNAs. This is probably the most intriguing rationale supporting the idea of using microRNAs as anticancer drugs.
MicroRNAs as therapeutic targets or tools
One of the most appealing properties of miRNAs as therapeutic agents, and probably the most important advantage in comparison with approaches targeting single genes, is their ability to target multiple molecules, frequently in the context of a network, making them extremely efficient in regulating distinct biological cell processes relevant to normal and malignant cell biology.
One example was clearly reported by Chen and colleagues, who demonstrated that miR-181 has a critical role in the regulation of T cell receptor sensitivity targeting multiple phosphatases, effect that was not obtained silencing single component of the pathway by RNAi (Li et al, 2007). Moreover, O'Day and colleagues (O'Day & Lal, 2010) underlines how microRNAs altered in breast cancer regulate a network of interconnected molecules, where the central node is represented by Myc. The same group had already provided evidence that miR-24 inhibits proliferation through direct targeting of c-Myc, E2F1 and a number of related molecules (Lal et al, 2009).
The first indications of the feasibility and the efficacy of a microRNA-based therapy in cancer, using these small molecules as both targets and tools, came from preclinical models aimed to understand the biological role of a specific miRNA. Reintroduction of miR-15a/16-1, for example induces apoptosis in leukaemic MEG01 cells and inhibits tumour growth in vivo in a xenograft model (Calin et al, 2008), while silencing oncogenic miR-21 with antisense oligonucleotides generates a pro-apoptotic and anti-proliferative response in vitro in different cellular models, also reducing tumour development and metastatic potential in vivo (Si et al, 2007).
Moreover, microRNAs involved in specific networks, like apoptosis, proliferation or receptor-driven pathways, could likely affect the response to targeted therapies or chemotherapies, thus suggesting their possible use also as adjuvant tools.
In summary, there are two main strategies to target miRNA expression in cancer. Direct strategies involve the use of oligonucleotides or virus-based constructs to either block the expression of an oncogenic miRNA or to reintroduce a tumour suppressor miRNA lost in cancer. Indirect strategies involve the use of drugs to modulate miRNA expression by targeting their transcription and their processing.
Nevertheless, stability and effective delivery into target tissues remain a major hurdle for direct microRNA-based therapy. Indeed, the first challenging aspect in delivering a therapeutic RNA is represented by its exiting the circulatory system, transiting the cell membrane and escaping from endosomal vescicles into the cytoplasm. Moreover, the size of an unconjugated therapeutic RNA is 7–20 kDa, and molecules less than 50 kDa are filtered by the kidney and excreted. Finally, systemic delivery into the bloodstream is challenged by phagocytic immune cells, such as macrophages and monocytes, which remove complexed RNAs from the body. To overcome these issues, several modifications, some of which already developed for siRNAs, can be applied to miRNAs.
For reduction in mature microRNA levels (due to mutation or any other mechanisms like defects in processing machinery or aberrant transcription), the therapeutic approach could be to exogenously deliver synthetic double-stranded hairpin by complexing with lipids or delivery proteins. As reported by Tazawa et al (Tazawa et al, 2007), miR-34a transiently inhibits human colon cancer tumour progression when administered subcutaneously in complexes with atelocollagen, which was recently shown to be a very powerful system for efficient in vivo delivery of small interfering RNA molecules into tumours. Chen and colleagues have developed a LPH (liposome–polycation–hyaluronic acid) nanoparticle formulation modified with tumour-targeting single chain antibody fragment (scFv) for systemic delivery of miR-34a to lung metastasis of murine melanoma cells (Chen et al, 2010b).
Unmodified dsRNAs are vulnerable to nucleases in vivo, which limits the use of this class of compound to privileged local environments where local administration is possible. Using a conditional mouse lung cancer model, where conditional expression of oncogenic K-ras could be activated, Esquela-Kerscher et al showed that the intranasal administration of an adenovirus expressing let-7a RNA hairpin reduced tumour formation in vivo (Esquela-Kerscher et al, 2008). In 2009, Kota et al (Kota et al, 2009) used a systemic administration of miR-26a in a mouse model of HCC using adeno-associated virus (AVV) to inhibit cancer cell proliferation and induction of tumour-specific apoptosis. These results are consistent with previous findings made by that same group, which demonstrated that MYC-induced liver tumours result in concomitant downregulation of various microRNAs (Chang et al, 2008).
In short term experiments of cardiac hypertrophy, conducted by Carè et al (Care et al, 2007), overexpression of miR-133 by adenovirus delivery significantly reduced the expression of foetal genes and resulted in reduced size of left ventricular cardiac myocytes. For stable miRNA reintroduction, the expression can be enforced by a viral vector with Pol III promoter, including U6, H1 and tRNA (Kawasaki & Taira, 2003; Tiscornia et al, 2003; Xia et al, 2004), upstream an artificial short hairpin RNA (shRNA), which bypasses Drosha processing but it is cleaved and loaded into miRISC by Dicer. The advantage of Pol III promoters is to provide high expression of miRNAs from well-defined transcription start and termination sites; however, these promoters have no cell specificity. In addition, exceedingly high levels of shRNA expression increase the probability of off-target silencing, elicit non-specific effects such as interferon response and can also saturate Exportin5 pathway of endogenous miRNAs with fatal consequences (Grimm et al, 2006). Alternatively, the pri-miRNA can be expressed from an RNA Pol II promoter, leaving open the possibility for tissue-specificity or induced ectopic miRNA expression. Furthermore, most miRNAs are known to be downstream Pol II promoters, within known protein coding genes and expressed by Pol II activity (Lee et al, 2004). Therefore, strategies using Pol II-directed synthesis of shRNA that mimic the natural miRNA synthesis could be an efficient therapeutic approach.
However, it is important to underline the possible hazards in reintroducing a microRNA with a viral system. Depending on the nature of the system, the delivered material can be integrated into the host DNA or remain episomal: retroviral and lentiviral vectors integrate their DNA into the host genome, whereas adenoviral vector replicates as autonomous unit. Because the site of integration is unpredictable, there is always a risk of insertional mutagenesis and activation of proto-oncogenes. Another drawback of retroviral vectors is that their use is limited to actively dividing cells. Adenoviral vectors, which could be used in principle for both dividing and quiescent cells, are in fact mainly recommended for stable expression in non-dividing cells, to prevent dilution of the adenoviral genome (which remains not integrated into the host genome). Another limitation of using adenoviruses is the strong immunological response that they potentially induce, even though new species are being tested to reduce these effects (reviewed in Nayak & Herzog, 2010).
To achieve miRNA loss-of-function, chemically modified anti-miR oligonucleotides (AMOs) have been developed (Weiler et al, 2006). The most important property of these oligonucleotides is their specificity and high binding affinity to RNA and a number of them has being pursued in clinical studies. miRNA downregulation has been achieved using 2-O-methyl oligonucleotides (Hutvagner et al, 2004; Meister et al, 2004). miR-122 inhibition was obtained by treating mice with AMOs containing 2-O-methoxyethyl groups resulted in lower plasma cholesterol (Esau et al, 2006). Intravenous administration of cholesterol-conjugated AMOs against miR-16, miR-122, miR-192 and miR-194 resulted in a significant reduction of corresponding miRNA levels in liver, lung, kidney, heart, intestine, fat, skin, bone marrow, muscle, ovaries and adrenals (Krutzfeldt et al, 2005; Weiler et al, 2006).
Very interestingly, Ma and colleagues, after demonstratig the crucial role of miR-10b as metastmiR in breast cancer, have exploited a possible therapeutic application, reporting that systemic treatment of tumour-bearing mice with miR-10b antagomirs suppresses breast cancer metastasis (Ma et al, 2010).
Other modified AMOs are represented by locked nucleic acid (LNA)-oligonucleotides, able to inhibit exogenously introduced miRNAs with high specificity (Ørom et al, 2006).
The first clinical trial in human of LNA-anti-miR (a placebo-controlled, double-blind, randomized, single dose, dose-escalating safety study of SPC3649 in a total of 64 healthy male volunteers) has been conducted by Santaris to study SPC3649 (LNA-antimiR-122; http://ClinicalTrials.gov, Identifier: NCT00688012). miR-122 is an abundant miRNA in the liver. The hepatitis C virus (HCV) genome harbours two closely spaced miR-122 target sites in the 5′ non-coding region required for HCV replication. Kauppinen and colleagues showed that administration of LNA-anti-miR into mice resulted in a dose-dependent depletion of mature miR-122 (Elmen et al, 2008b). An efficacy study conducted by Elmen et al on non-human primates (Elmen et al, 2008a), showed a dose-dependent sequestration of mature miR-122 and a long lasting decrease of total plasma cholesterol.
Nevertheless, the same limitations encountered with the application of synthetic miRNA duplexes are encountered in the applications of antagomirs, namely their effective delivery into target tissues.
An interesting approach to overcome these problems is to target miRNA by saturating them with target mRNAs. Ebert et al (Ebert et al, 2007) developed miRNA inhibitory transgenes, called ‘miRNA sponges’, expressing an mRNA containing multiple tandem binding sites for an endogenous miRNA and thus able to stably interact with the corresponding miRNA and prevent the association with its endogenous targets. Both designed polymerase Pol II- and Pol III-driven miRNA sponges showed more efficiency for miRNA inhibition compared to standard 2′-O-Me antagomirs.
Lentiviral vectors have proven to be effective tool to ectopically express miRNAs using suitable transcriptional control units. It has been reported by Gentner et al (Gentner et al, 2009) that stable miRNA-223 knock down can be achieved in vivo by transducing bone marrow stem and progenitor cells with multiple target sequences from strong promoters and transplanting them into lethally irradiated congenic recipients. They showed that overexpressing miR-targets specifically affects the targeted miRNA rather than saturating the effector pathway. However, the need for strong promoters and multiple vector integrations to obtain a high miR-target expression could increase the risk of insertional mutagenesis in target cells, potentially confounding the identification of miRNA knock down phenotypes, and thus representing a potential limitation of this strategy.
miRNA-masking antisense oligonucleotides technology (miR-mask) is another strategy developed by Choi and colleagues (Choi et al, 2007). In contrast to miRNA sponges, miR-masks consist of single-stranded 2′-O-methyl-modified antisense oligonucleotides fully complementary to predicted miRNA binding sites in the 3′-UTR of a specific target mRNA. Although unwanted effects or off-target effects can be dramatically reduced with this approach, this may be a disadvantage for cancer therapy for which the targeting of multiple pathways might be desirable.
Su et al (Su et al, 2011) have applied a nanotechnologic approach to the use of anti-miRNAs: systemic delivery of a chemically stabilized anti-miR-122 complexed with interfering nanoparticles (iNOPs) effectively silences the liver-expressed miR-122 in mice, resulting in lowering of plasma cholesterol.
Beside targeted therapies and chemotherapies, microRNAs could also alter the sensitivity to radiotherapy, as recently reported by Slack's group (Weidhaas et al, 2007): a potential therapeutic use for anti-miR-34 as a radiosensitizing agent in p53-mutant breast cancer could be envisaged; in lung cancer cells, let-7 family can suppress the resistance to anticancer radiation therapy, likely through RAS regulation.
Evidence described up to date provides the experimental bases for the use of microRNAs as both targets and tools in anti-cancer therapy, but there are at least two primary issues to address to translate these fundamental research advances into medical practice: development of engineered animal models to study cancer-associated microRNAs, and improvement of miRNAs/anti-miRs in vivo delivery efficiency (Fig 4).
Figure 4.

Current approaches to in vivo miRNA targeting.
Moreover, all the strategies developed so far to modulate miRNA expression are designed to only modify one miRNA or a family of miRNAs. As miRNAs coordinate in cancer pathogenesis and the phenotypical effects result from multiple interactions between miRNAs and the transcriptome, it is reasonable to search for strategies that aim at re-programming aberrant miRNA networks in cancer.
A possible approach might be represented by the modulation microRNA expression by targeting components of the biogenesis machinery, or elements of the regulatory networks, as the epigenetic program. Demethylating agents as Decitabine and 5-azacytidine, for examples are currently approved for the treatment of myelodysplastic syndrome, although they have shown activity in many other malignancies, including AML (Galm et al, 2006). These drugs are known to inhibit DNA methyltransferases, resulting in tumour suppressor gene re-expression. As previously described, miRNAs have also been shown to be actively re-expressed after treatment with these drugs and to largely contribute to the therapeutic effects of these compounds. Even though it is tempting to suggest that many of the biological effects of these drugs may be mediated by the re-expression of non-coding RNAs, this still needs to be verified.
Moreover, it may sound surprising that reprogramming of the whole ‘microRNome’, including both oncomiRNAs and tumour suppressor miRNAs, can lead to a specific anti-tumoural effect: how is the balance shifted in favour of a specific effect? This might be due to the possibility that most microRNAs seem to exert a role as oncosuppressors, and consequently are mostly dowregulated in human neoplasia (Lu et al, 2005). Even though still debated and controversial, this hypothesis is supported by the evidence that Dicer or Drosha silencing promotes cellular transformation and tumourigenesis in vivo: conditional loss of Dicer1 in the lung tissues of mice enhances the development of lung tumours in a K-ras mouse model (Kumar et al, 2009). Finally, loss of Dicer and/or Drosha has also been inversely correlated with outcome in lung cancer (Karube et al, 2005), cancers of the ovarian epithelium (Merritt et al, 2008), and more recently in other tumour types as nasopharyngeal carcinoma (Guo et al, 2012), neuroblastoma (Lin et al, 2010) and breast (Grelier et al, 2009).
A few reports, however, describing a positive correlation between Dicer expression and poor outcome in colorectal cancer (Faber et al, 2011) and in prostate cancer (Chiosea et al, 2006), or the overexpression of Drosha in cervical cancer (Muralidhar et al, 2011), raise the important issue to validate this still debated question, and verify whether the effect of targeting the microRNA machinery might be tissue-related.
To make it even more complicated, recently Piccolo's group (Martello et al, 2010) described a microRNA family, miR-103-107, able to empower the metastatic potential targeting Dicer and thus attenuating the global microRNA biosynthetis, with a particular effect mediated by the downregulation of miR-200 family, and consequent switch to a more mesenchymal and aggressive phenotype.
Conclusions and future perspectives
The past decade has witnessed an explosion of research focused on small non-coding RNAs. From viruses to plants, to humans, these small RNA regulators of gene expression have been demonstrated to be involved in every biological system.
Cancer is defined by abnormal and uncontrolled cell division, a phenotype that arises from the misregulation of several genes. miRNAs are major regulators of gene expression, with roles in nearly every area of cell behaviour, development and survival; therefore, it is not surprising that miRNAs are actively altered in all types of cancers (Croce, 2009), acting as oncogenes or tumour suppressor genes.
However, although significant advances have been made for the future role of miRNAs in diagnostics, there have been far fewer reported successes in the development of miRNAs therapeutic strategies: the main issues that need to be addressed are certainly the discovery of the microRNAs playing a crucial role in the biology of a specific tumour type by altering a whole network of target proteins; the validation of the targets and the accurate prevision of the putative unwanted off target effects; the development of efficient methods of a specific drug delivery.
Referring to the use of microRNAs as the targeted therapy of the future is probably too optimistic and premature at this point; however, the number of discoveries, increasing so fast in the last few years, is certainly encouraging and promising.
Pending issues
A complete understanding of the complex mechanisms regulating the interaction between microRNAs and their targets, to unravel the networks regulated by these small molecules and exploit the possibility to modulate their activity with therapeutic purposes, is mandatory. To this aim, important issues to take into account are the potential tissue specificity, the possible side or off target effects and the existence of several other players able to affect miRNA function on a specific mRNA, as the competition for miRNA binding exerted by other coding and non-coding RNAs.
To have a clear, genetic demonstration of the involvement of a specific miRNA in the occurrence or progression of a specific tumour, engineered mouse models, as reported in few cases to date, need to be developed.
To use miRNAs as reliable diagnostic, predictive or prognostic biomarkers, the development or optimization of efficient, sensitive and reproducible detection methods are of primary importance. Indeed, reported lack of consistency between different studies certainly gives rise to some concern, and may arise from sample selection or preparation, experimental design and/or data analysis, suggesting the need of a more comprehensive validation.
Despite the increasing and encouraging body of evidence suggesting a possible use of microRNAs as therapeutic agents, either as tools or targets, there are several issues that need to be clarified and overcome to translate the success obtained in preclinical settings to the bedside. Indeed, whereas several chemical modifications have been developed to prevent degradation and increase the stability of a therapeutic RNA, either a miRNA or an antisense, and different methods have been exploited to increase the delivery efficiency, several aspects need to be further and carefully evaluated, as: an efficient and specific delivery, through systemic administration, to less accessible organs; the evaluation of possible side effects; the careful analysis of potential hazards by using viral particles.
The potential of microRNA expression to predict response to different therapies needs to be further investigated and validated: either the possibility to select subgroups of patients that most likely will present a better response to a specific treatment, and the possible use of microRNAs as therapeutic adjuvant tools to improve the response and overcome resistance, are both critically important aspects in clinics.
Indeed, microRNAs involved in specific networks, as the apoptotic, proliferation or receptor-driven pathways, could likely influence the response to targeted therapies or to chemotherapy, but unfortunately most part of the current reports relies on preclinical, generally in vitro, experiments, thus indicating that this represents still an open question.
Acknowledgments
CM Croce was supported by grants of the National Cancer Institute; Iorio MV was supported by Start Up AIRC Grant.
The authors declare that they have no conflict of interest.
References
- Alexiou P, Vergoulis T, Gleditzsch M, Prekas G, Dalamagas T, Megraw M, Grosse I, Sellis T, Hatzigeorgiou AG. miRGen 2.0: a database of microRNA genomic information and regulation. Nucleic Acids Res. 2010;38:D137–D141. doi: 10.1093/nar/gkp888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Beitzinger M, Meister G. Preview. MicroRNAs: from decay to decoy. Cell. 2010;140:612–614. doi: 10.1016/j.cell.2010.02.020. [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhayani MK, Calin GA, Lai SY. Functional relevance of miRNA* sequences in human disease. Mutat Res. 2012;731:14–19. doi: 10.1016/j.mrfmmm.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabro E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sultmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramuta S, Egyházi S, Rodolfo M, Witten D, Hansson J, Larsson C, Lui WO. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhu HH, Zhou LF, Wu SS, Wang J, Chen Z. Inhibition of c-FLIP expression by miR-512-3p contributes to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2010a;23:1457–1462. doi: 10.3892/or_00000784. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010b;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 2009;10:490–497. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Planell-Saguer M, Rodicio MC, Mourelatos Z. Rapid in situ codetection of noncoding RNAs and proteins in cells and formalin-fixed paraffin-embedded tissue sections without protease treatment. Nat Protoc. 2010;5:1061–1073. doi: 10.1038/nprot.2010.62. [DOI] [PubMed] [Google Scholar]
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, Tsongalis GJ, Suriawinata AA, Carrère N, Buscail L, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56:603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008a;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008b;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber C, Horst D, Hlubek F, Kirchner T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur J Cancer. 2011;47:1414–1419. doi: 10.1016/j.ejca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F, van Kouwenhove M, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galm O, Herman JG, Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20:1–13. doi: 10.1016/j.blre.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta P, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Pisano DG, Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7:2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial–mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, Léon-Goddard S, Rimokh R, Mikaelian I, Venoux C, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Guo X, Liao Q, Chen P, Li X, Xiong W, Ma J, Li X, Luo Z, Tang H, Deng M, et al. The microRNA-processing enzymes: Drosha and Dicer can predict prognosis of nasopharyngeal carcinoma. J Cancer Res Clin Oncol. 2012;138:49–56. doi: 10.1007/s00432-011-1058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, Warnecke JM, Sczakiel G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Kelly R, Newell J, Kerin MJ. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15:673–682. doi: 10.1634/theoncologist.2010-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135-9. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu RJ, Yang HJ, Tsai HJ. Labeled microRNA pull-down assay system: an experimental approach for high-throughput identification of microRNA-target mRNAs. Nucleic Acids Res. 2009;37:e77. doi: 10.1093/nar/gkp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Ménard S, Croce CM, Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snøve O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3'UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from non-squamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, Kreipe H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, London WB, Chang CH, Yu AL. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–7850. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, varez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Maselli V, Di Bernardo D, Banfi S. CoGemiR: a comparative genomics microRNA database. BMC Genomics. 2008;9:457. doi: 10.1186/1471-2164-9-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JL, Kurita S, Cazanave SC, Bronk SF, Werneburg NW, Fernandez-Zapico ME. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J Cell Biochem. 2010;110:1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhar B, Winder D, Murray M, Palmer R, Barbosa-Morais N, Saini H, Roberts I, Pett M, Coleman N. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J Pathol. 2011;224:496–507. doi: 10.1002/path.2898. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Canaani E, Croce CM. Oncogenic All1 fusion proteins target Drosha-mediated microRNA processing. Proc Natl Acad Sci USA. 2007;104:10980–10985. doi: 10.1073/pnas.0704559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010;17:295–304. doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuovo G, Lee EJ, Lawler S, Godlewski J, Schmittgen T. In situ detection of mature microRNAs by labeled extension on ultramer templates. Biotechniques. 2009;46:115–126. doi: 10.2144/000113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- O'Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oien KA, Evans TR. Raising the profile of cancer of unknown primary. J Clin Oncol. 2008;26:4373–4375. doi: 10.1200/JCO.2008.17.6156. [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom UA, Kaupinnen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Wong DT. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- Pulkkinen K, Malm T, Turunen M, Koistinaho J, Yla-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J, Jin Y. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS ONE. 2010;5:e9429. doi: 10.1371/journal.pone.0009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, Burow ME, Ivan M, Croce CM, Nephew KP. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30:1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, Fredrickson T, Landgraf P, Ramachandra S, Huppi K, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, Sampath D, Fabbri M, Lerner S, Barron LL, Rassenti LZ, et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–952. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Marschall T, Martin M, Rosenstiel P, Mestdagh P, Schlierf S, Thor T, Vandesompele J, Eggert A, Schreiber S, et al. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res. 2010;38:5919–5928. doi: 10.1093/nar/gkq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, rtus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Baigude H, McCarroll J, Rana TM. Silencing microRNA by interfering nanoparticles in mice. Nucleic Acids Res. 2011;39:e38. doi: 10.1093/nar/gkq1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Stresemann C, Brueckner B, Lyko F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001–1005. doi: 10.4161/cc.6.9.4209. [DOI] [PubMed] [Google Scholar]
- Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs implicated in human disease. Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O'Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS, Tewari M. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA–target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- Xu JZ, Wong CW. Hunting for robust gene signature from cancer profiling data: sources of variability, different interpretations, and recent methodological developments. Cancer Lett. 2010;296:9–16. doi: 10.1016/j.canlet.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. StarBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, Xiong W, Li G, Lu J, Fodstad O, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496–22507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]