Abstract
Endometriosis is found in 5–15% of women of reproductive age and is more frequent in relatives of women with the disease. Activation of KRAS results in de novo endometriosis in mice, however, activating KRAS mutations have not been identified in women. We screened 150 women with endometriosis for a polymorphism in a let-7 microRNA (miRNA) binding site in the 3'-UTR of KRAS and detected a KRAS variant allele in 31% of women with endometriosis as opposed to 5% of a large diverse control population. KRAS mRNA and protein expression were increased in cultured endometrial stromal cells of women with the KRAS variant. Increased KRAS protein was due to altered miRNA binding as demonstrated in reporter assays. Endometrial stromal cells from women with the KRAS variant showed increased proliferation and invasion. In a murine model, endometrial xenografts containing the KRAS variant demonstrated increased proliferation and decreased progesterone receptor levels. These findings suggest that an inherited polymorphism of a let-7 miRNA binding site in KRAS leads to abnormal endometrial growth and endometriosis. The LCS6 polymorphism is the first described genetic marker of endometriosis risk.
Keywords: endometriosis, epigenetics, KRAS, let-7, microRNA
INTRODUCTION
Endometriosis is a benign, invasive, estrogen-dependent disorder characterized by the presence of endometrial glands and stroma outside the uterus. It is found in 5–15% of women of reproductive age with more than 70 million affected worldwide (Bulun, 2009; Gao et al, 2006; Hemmings et al, 2004). Endometriosis has a dramatic effect on health and quality of life, causing chronic pelvic pain and infertility in up to 50% of women with the disease (Fourquet et al, 2010). The yearly medical costs and indirect economic impact total more than $22 billion in the United States alone (Practice Committee, 2006; Simoens et al, 2007).
Although multiple medical therapies are available, there is no established cure for endometriosis; all treatments suppress the growth of both endometriosis and normal endometrium through hormonal mechanisms; however, none target disease-specific pathways. Surgical intervention has proven to be an effective treatment; however, the estimated recurrence rate still remains over 50% at 5 years after laparoscopic surgery (Hadfleld et al, 1996). In the United States, the mean delay in diagnosis is approximately 11 years (Guo, 2009). There is an obvious need to understand the biological basis of the disease in order to devise specific treatments, allow early diagnosis and potentially provide a means of prevention.
While the etiology and pathogenesis of endometriosis remain an active area of investigation, there is a genetic predisposition with a sevenfold risk of endometriosis in women whose mother or sister has the disease (Moen & Magnus, 1993; Simpson & Bischoff, 2002). It has been previously shown that some genes are expressed differentially in eutopic endometrium of endometriosis patients compared to normal endometrium (Kao et al, 2003; Taylor et al, 1999). However, no specific gene responsible for the disease has been identified in humans (Moen & Magnus, 1993; Simpson & Bischoff, 2002). Two large genome-wide association studies (GWAS) have linked disease susceptibility to 7p15.2 (a region between NFE2L3 and HOXA10) and to 9p21 in the CDKN2BAS gene with odds ratios of 1.22 and 1.44, respectively (Painter et al, 2011; Uno et al, 2010). Another genome wide linkage study identified 10q26 as a locus responsible for susceptibility to endometriosis (Treloar et al, 2005). Although these loci identify genetic linkage, they have not identified genes demonstrated to be involved in the pathophysiology of endometriosis.
No human studies have linked or identified alterations in the gene responsible for the only known murine model of spontaneous endometriosis. Activation of an oncogenic KRAS gene in the murine ovarian surface epithelium results in the de novo formation of lesions with endometriotic morphology (Dinulescu et al, 2005). The authors of that study speculate that RAS pathway activation by different mechanisms may play an important role in endometriosis in humans. Moreover, a recent study demonstrated that activation of mutated KRAS in transplanted endometrium in mice triggered endometriosis formation and long-term survival of the lesions (Cheng et al, 2011). However, despite thorough mutational analyses of KRAS in human endometriosis by several groups, no activating mutations in the coding regions of this gene have been found (Amemiya et al, 2004; Otsuka et al, 2004; Vercellini et al, 1994; Zhao et al, 2006).
We hypothesized that other possible mechanisms that alter the regulation of KRAS gene expression may be involved in the pathogenesis of human endometriosis. MicroRNAs (miRNAs) are small non-coding RNAs that degrade or prevent translation of their target genes by binding to the 3′-untranslated regions (UTRs) of mRNAs (Calin et al, 2004; Carletti & Christenson, 2009; Esquela-Kerscher & Slack, 2006). Single nucleotide polymorphisms (SNPs) within miRNAs or miRNA binding sites can alter mRNA stability or translation and, thereby, result in various pathological processes including malignant transformation (Calin et al, 2004; Esquela-Kerscher & Slack, 2006; Yang et al, 2008). KRAS is known to be regulated in a miRNA-dependent manner (Johnson et al, 2005).
KRAS is a crucial target of let-7 miRNAs including let-7a–g and i (Johnson et al, 2005; Roush & Slack, 2008). KRAS is downregulated through 10 let-7 complementary sites (LCS) found in the 3′-UTR of the KRAS gene (Chin et al, 2008). One of these LCS (LCS6) is known to harbour a SNP (T→G in the fourth position, rs61764370), which modifies let-7 binding in lung cancer cells (Chin et al, 2008). The incidence of this SNP in the general population is 5.8% (Chin et al, 2008). This variant allele is associated with an increased risk of the development of non-small cell lung cancer in people with only a moderate smoking history and is also a marker of poor prognosis in oral cancer (Chin et al, 2008; Christensen et al, 2009). Similarly, this SNP has been identified in more than 25% of patients with ovarian cancer and is a marker of an increased risk of developing epithelial ovarian cancer especially in BRCA-negative families with hereditary breast and ovarian cancer syndrome (Ratner et al, 2010).
Here, we hypothesized that the Ras pathway might be activated by the presence of this previously identified SNP in LCS6 in the 3′-UTR of KRAS in patients with endometriosis. We demonstrate the increased prevalence of this variant allele in women with endometriosis. We show that the presence of this SNP results in elevated KRAS protein expression causing increased proliferation and invasion of human endometrial stromal cells (hESC).
RESULTS
Prevalence of the KRAS LCS6 variant in women with endometriosis
To determine the prevalence of the KRAS variant allele in women with endometriosis, we identified 150 subjects who provided DNA samples. Subjects were an average age of 32.9 years and endometriosis was diagnosed an average of 7 years prior to the study. Thirty-one percent had a family history of endometriosis. DNA suitable for analysis was obtained from 132 subjects. The study included women with a current diagnosis or history of endometrioma (n = 89) and/or peritoneal endometriosis (n = 43). Among those subjects with an endometrioma, 69% (n = 61) had co-existing peritoneal endometriosis. Staging of disease was made according to the American Society for Reproductive Medicine revised classification of endometriosis. Surgical diagnosis of severe (stage IV) and moderate (stage III) or minimal/mild (stage I/II) endometriosis was made in 56% (n = 74), 33% (n = 43) and 11% (n = 15) of subjects, respectively. 77% (n = 102) of patients had severe pain (pain which interfered with everyday activities) and/or dysmenorrhea. 27% (n = 36) of subjects were diagnosed with infertility. An irregular menstrual cycle was present in 35% (n = 46) of subjects.
Chin et al determined the allele frequencies of the LCS6 SNP using a collection of genomic DNA from 2433 healthy individuals from a global set of 46 populations (Chin et al, 2008). An extensive database of genetic variations in these samples can be found, along with the population descriptions, in ALFRED (Cheung et al, 2000). They found that <3% of the 4,866 chromosomes, or 5.8% of people tested, had the G allele (variant) at the LCS6 SNP site. The frequency of this allele varied across geographic populations, with ‘European’ populations exhibiting the variant allele most frequently (7.6% of the chromosomes tested); African populations less frequently (<2.0% of chromosomes tested) and ‘Asian’ and Native American populations infrequently (<0.4% of chromosomes tested; Chin et al, 2008).
Of the 132 women with endometriosis, 41 (31%) were found to carry a variant allele at LCS6 in the KRAS 3′-UTR that prevents let-7 miRNA inhibition of KRAS. In subjects with ovarian endometriomas, 23 of 89 had the variant LCS6 while 18 out of 43 women with peritoneal endometriosis carried this alternative allele (25.8 and 41.8%, respectively). 5.3% of patients were homozygous for the alternative allele (7 out of 132), which represents 17% of KRAS-variant-positive cases. There was no difference in the mean age of subjects with KRAS non-variant and variant alleles (Table 1). A total of 56 and 57% of subjects with the non-variant and variant KRAS alleles, respectively, were surgically diagnosed with severe (stage IV) endometriosis. There was no difference in the frequency of endometriomas or peritoneal endometriosis between the groups with the KRAS variant or the non-variant allele. Subjects with the alternative KRAS allele, however, more frequently had infertility (42% vs.18% in the group with variant and non-variant KRAS allele, respectively, p = 0.0033). In contrast, those with non-variant KRAS allele complained of severe pain, dysmenorrhea and dyspareunia more frequently than the group with the KRAS variant allele. Those symptoms were found in 77%, 96% and 69% vs. 42%, 42% and 28% of women with the non-variant and variant KRAS genes, respectively (p = 0.0001, p = 0.00001 and p = 0.0001, respectively). Irregular menstrual cycles were equally frequent in patients from each group. The subjects were ethnically diverse and included 66 Caucasian, 9 black and 57 hispanic subjects. The rate of the KRAS variant was not significantly different between ethnic groups.
Table 1.
Clinical characteristics of women with the non-variant and variant KRAS alleles
| Variant KRAS allele | Non-variant (wt) KRAS allele | p-Value | |
|---|---|---|---|
| Age | 34.1 | 31.3 | 0.35 |
| Stage IV disease | 57% | 56% | 1 |
| Years from diagnosis | 9 | 6 | 0.31 |
| Infertility | 42% | 18% | 0.003 |
| Irregular menses | 28% | 35% | 0.34 |
| Severe pain | 42% | 77% | 0.001 |
| Dysmenorrhea | 42% | 96% | 0.0001 |
| Dyspareunia | 28% | 69% | 0.001 |
Subjects with the non-variant KRAS allele had severe pelvic pain, dyspareunia and dysmenorrhea more frequently than patients with the alternative KRAS allele. Subjects with the variant KRAS allele were more likely to be infertile. Mean age and severity were not different. n = 41 subjects with KRAS variant and 91 subjects with the WT allele.
In summary, the KRAS variant that prevents let-7 miRNA inhibition of KRAS was significantly increased in women with endometriosis. The prevalence of the KRAS variant allele in our endometriosis patient cohort is significantly higher than expected in any existing geographic population, supporting the hypothesis that this variant allele is a marker of increased endometriosis risk.
Effect of the LCS6 variant allele on KRAS and let-7 expression
Quantitative polymerase chain reaction (after reverse transcription) (RT-PCR) was used to determine the level of KRAS mRNA in cultured hESC from women without endometriosis, women with endometriosis heterozygous for the variant allele, and women homozygous for the wild-type (WT) allele (n = 10 subjects per group). hESC from women without endometriosis were found to express approximately threefold lower levels of KRAS mRNA compared to both hESC from women with endometriosis carrying WT KRAS (KRAS/Actin: 0.001 ± 0.0002 vs. 0.003 ± 0.0004; p = 0.0007) and 10-fold lower mRNA levels compared to those carrying the variant KRAS allele (0.01 ± 0.002; p = 0.0001). KRAS mRNA was approximately threefold higher in hESC of subjects with the variant KRAS LCS6 compared to hESC from subjects with the non-variant allele (p = 0.0049; Fig 1a). Western blot analysis showed that hESC from women with the KRAS SNP had a 2.8-fold increase in KRAS protein compared to endometrium carrying the non-variant allele (Fig 1b).
Figure 1. KRAS expression in hESCs obtained from endometrium of 10 women without endometriosis, eutopic endometrium of 10 women with endometriosis carrying WT KRAS and 10 women with endometriosis carrying the variant allele of KRAS gene at the LCS6 site.
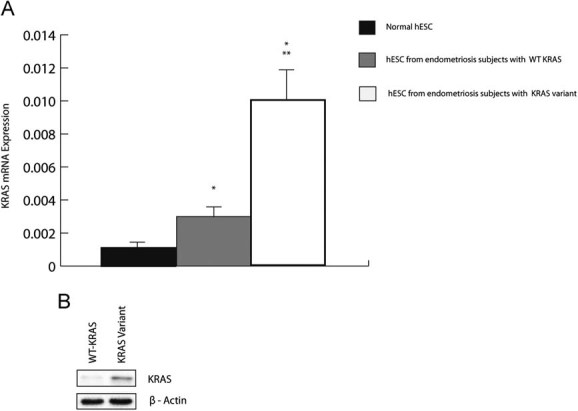
- q-RT-PCR results show comparatively low levels of KRAS mRNA in normal endometrium, increased KRAS mRNA in hESC from women with endometriosis and the WT KRAS allele (p = 0.0007) and highest expression of KRAS mRNA in hESC carrying the variant KRAS allele (p = 0.00018 when compared to normal hESC, p = 0.0049 when compared to hESC from endometriosis patients with WT allele). (*, difference is significant when compared to normal hESC; **, difference is significant when compared to hESC from women with endometriosis homozygous for the WT KRAS allele).
- Western blot results show a 2.8-fold increase in KRAS protein in hESC with the variant allele.
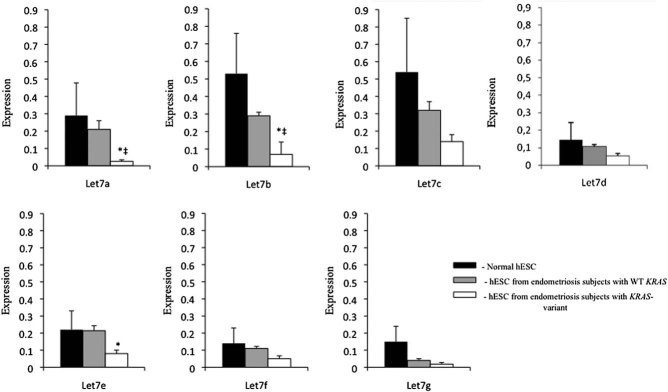
Let-7 is known to bind to the non-variant and not the variant LCS6 allele preventing KRAS protein synthesis. To assess the possibility of compensatory changes in let-7 miRNAs in the setting of the variant allele and elevated KRAS protein, we determined the level of let-7a–g miRNAs (Fig 2). Cultured endometrial stromal cells from women with endometriosis with the LCS6 variant in the KRAS gene showed lower levels of let-7a, b and e (0.26 ± 0.009, 0.07 ± 0.01 and 0.08 ± 0.02, respectively), compared to hESC from women with endometriosis with non-variant KRAS (0.21 ± 0.05, 0.29 ± 0.02, 0.21 ± 0.029, respectively; p = 0.0047, 0.003 and 0.05, respectively). let-7a and b were also lower in KRAS variant cells from endometriotic women compared to normal hESC (0.29 ± 0.188, 0.53 ± 0.23, respectively; p = 0.05 and 0.02, respectively). Compared to hESC isolated from normal endometrium, hESC from women with endometriosis exhibited lower levels of let-7 family miRNA. These alterations in let-7 in the presence of the KRAS variant allele could be a result of a feedback mechanism or due to another unidentified independent factor, but do agree with prior published findings of lower let-7 in KRAS variant-associated lesions.
Figure 2. Let-7 miRNA family expression in hESC from 10 subjects without endometriosis (normal controls), 10 subjects with endometriosis carrying WT KRAS and 10 subjects with endometriosis and the variant allele of KRAS gene.
q-RT-PCR results show a trend towards decreased transcript levels of all let-7 miRNAs in hESC from subjects with endometriosis compared to normal hESC from subjects without endometriosis (normal control). Endometrial cells from subjects with endometriosis with the LCS6 variant in the KRAS gene showed lower levels of let-7a, b and e compared to hESC from endometriosis with non-variant KRAS (*p = 0.0047, 0.003 and 0.05, respectively). let-7a and b were also lower in KRAS-variant cells from women with endometriosis compared to normal hESC (‡p = 0.05 and 0.02, respectively).
Effect of the LCS6 variant on the expression of KRAS using a luciferase reporter assay
To demonstrate that the increased level of KRAS protein seen in cultured hESC from subjects with the KRAS variant was in fact due to altered let-7 binding to the mutant LCS6, we introduced an siRNA construct designed to rescue let-7 activity by binding to the altered LCS6. Normal hESCs were co-transfected with a luciferase reporter construct carrying the variant LCS6, the siRNA or a siRNA negative control (Fig 3). Luciferase activity in the cells transfected with a reporter carrying the KRAS variant allele was approximately 30-fold greater than the activity from the reporter with the WT allele (relative luciferase activity: 0.1 ± 0.001 vs. 2.9 ± 0.05). The siRNA targeting the LCS6 variant RNA reduced the luciferase activity in the cells with the reporter containing the KRAS LCS6 variant by 70% (to 0.9 ± 0.13; p = 0.045).
Figure 3. The effect of siRNA mimicking let-7 action on luciferase expression from the reporter plasmid carrying WT or variant KRAS allele in Dual Luciferase Reporter assay.
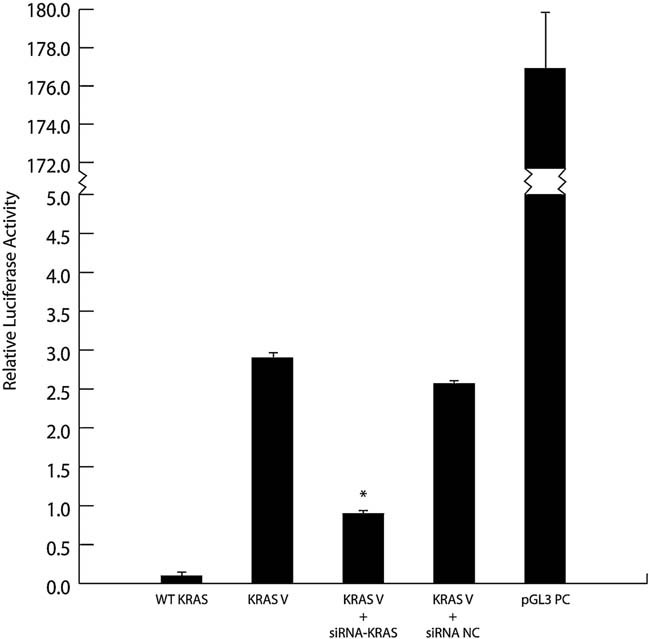
Normal hESC were co-transfected with pGL3 vector carrying the variant LCS6 of the KRAS gene and either siRNA modified to bind the variant LCS6 or a negative control RNA sequence. Luciferase activity from the reporter plasmid carrying the KRAS-variant allele was greatly increased compared to that from a reporter plasmid containing non-variant allele at baseline. There was a 70% reduction in luciferase activity when the KRAS-variant allele was co-transfected with siRNA designed to bind the LCS6 site (p = 0.045). No significant decrease in luciferase activity was obtained after co-transfecting this reporter plasmid with a control RNA sequence. The pGL3 control vector was used to assess transfection efficiency. Transfection with pGL3 carrying non-variant LCS6 of KRAS gene resulted in minimal luciferase activity likely due to inhibitory effects of endogenous let-7 miRNAs. Each experiment was carried out in duplicate in three separate experiments using cells from individual subjects. WT KRAS, hESC transfected with WT KRAS; KRAS V, hESC transfected with KRAS variant allele; KRAS V + siRNA, hESC co-transfected with KRAS variant allele and siRNA; KRAS V + siRNA NC, hESC co-transfected with KRAS variant allele and siRNA negative control; pGL3 PC, hESC transfected with pGL3 control vector.
Cell proliferation and invasion
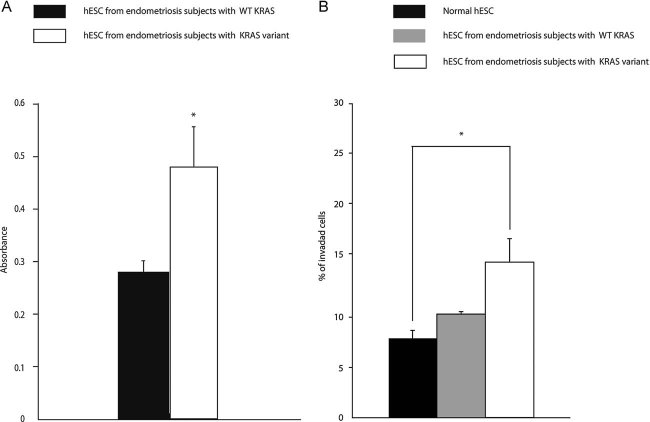
We assessed proliferation in the presence of the KRAS LCS6 variant allele using BrdU labelling. This assay showed a 71% increase of BrdU labelling in KRAS LCS6 variant hESC, indicative of an increased proliferation rate of endometrial cells from women with the variant allele compared to those with the non-variant allele (absorbance 0.48 ± 0.08 vs. 0.28 ± 0.02; p = 0.04; Fig 4a).
Figure 4. The effect of the KRAS variant allele on proliferation and invasion capacity of hESC.
- The effect of the KRAS variant allele on proliferation of hESC from women with endometriosis as determined by BrdU incorporation. There was a 71% increase (*p = 0.04) in the BrdU label in hESC with the variant allele (n = 5) versus WT KRAS LCS6 (n = 5). These results indicated an increase in cell proliferation rate of hESC containing the mutant KRAS LCS6.
- The effect of the KRAS variant allele on hESC invasion capacity. An invasion assay in which hESC from women with and without endometriosis and with the WT non-variant or the alternative KRAS allele was used to determine the ability to invade extracellular matrix. There was a significant increase in the invasion of hESC containing the variant allele (n = 9) compared to hESC without the variant allele (n = 6) (*p = 0.013). The difference between normal cells (n = 6) and cells from endometriosis with WT KRAS was not significant.
To determine the invasion capacity of these cells, we assessed the percentage of cells that invaded through ECM gel. The normalized absorbance (mean absorbance in sample wells minus mean absorbance in negative control wells) was 0.089 ± 0.006 for hESC from control subjects, 0.118 ± 0.019 for hESC from subjects with endometriosis carrying WT KRAS allele and 0.175 ± 0.026 for hESC from subjects with endometriosis carrying variant KRAS allele. Normalized absorbance in positive control wells (100% invasion) was 1.16 ± 0.028. The percent of invading cells was further calculated as 7 ± 0.5, 10 ± 1.8 and 15 ± 2.4% in samples from women without endometriosis, women with endometriosis and non-variant KRAS LCS6 and women with endometriosis and the variant KRAS LCS6 (Fig 4b). Invasion capacity of normal hESC and hESC from women with endometriosis without the variant were not significantly different, however, the cultured endometrial cells with the KRAS LCS6 variant allele invaded at a significantly increased rate compared to normal hESC (15% vs. 7%, respectively, p = 0.013).
The KRAS LCS6 variant allele in a murine endometriosis model
To evaluate differences in growth parameters in vivo, capacity for endometriotic lesion formation as well as histopathological and molecular characteristics of cultured endometrial stromal cells containing non-variant and variant alleles of the KRAS gene, a mouse model of endometriosis was used. We transplanted hESC obtained from subjects with and without the LCS6 variant under the kidney capsule of immune-deficient mice. Both non-variant and LCS6 variant cells formed endometriosis-like lesions with both glandular and stromal components (Fig 5a). The glandular component likely originated from progenitor stem cells in the culture as recently described (Cervello et al, 2011). Glandular cells are not identified in these cultures by the third passage, however, we cannot exclude a small number of contaminants (Taylor et al, 1998). Analysis of proliferation marker PCNA showed more cells (both epithelial and stromal) with stained nuclei in the lesion derived from LCS6 variant cells compared to those derived from non-variant cells. The percentage of stained nuclei in epithelium of lesions carrying variant KRAS allele was 54 ± 5% vs. 8 ± 1% in lesions created by non-variant hESC (p = 0.02). Stromal cells from lesions with the SNP exhibited 56 ± 5% of nuclei staining while in the lesions with WT KRAS, the percentage of positively stained nuclei was 34 ± 6% (p = 0.043; Fig 5b). These results suggest increased proliferation of cells harbouring the variant allele and were consistent with the results of the in vitro proliferation experiment. Apoptosis was assessed using cleaved caspase 3, and the amount of cleaved caspase 3 was equivalent in lesions derived from cells with either non-variant or LCS6 variant alleles (data not shown). Proliferation was increased without an increase in apoptosis in the LCS6 variant lesions.
Figure 5. Morphological and molecular features of endometriotic lesions containing the WT non-variant or variant alleles of the KRAS LCS6.
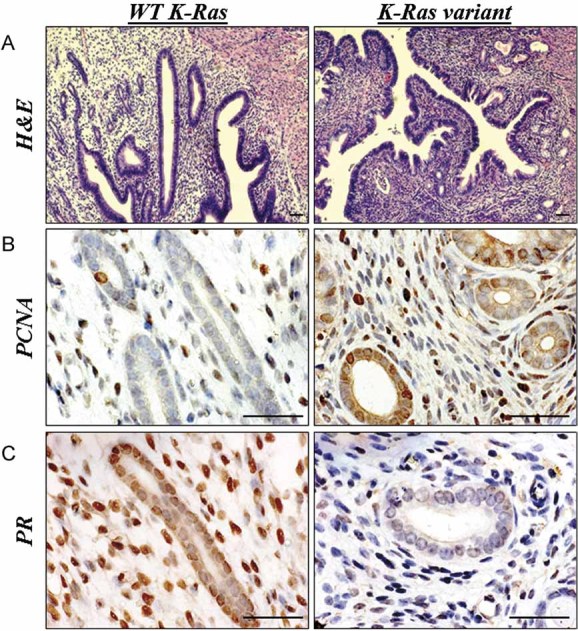
Cultured endometrial stromal cells were injected under the kidney capsule of immunodeficient mice.
- Morphological appearance of the lesions under the kidney capsule of immune deficient mice 1 month after transplantation of hESC either with or without the variant KRAS. In all cases the transplanted endometrial cells formed endometriosis lesions with both glandular and stromal components.
- Proliferation marker expression in endometriotic lesions in mice. Nuclear staining for PCNA was more prominent in epithelium and stroma of the lesions formed by KRAS variant positive cells (54 ± 5% and 56 ± 6% in epithelium and stroma, respectively), compared to those derived from normal cells (8 ± 4% and 34 ± 6% in epithelium and stroma, respectively; p = 0.02 and p = 0.043) indicating higher proliferation levels in these cells.
- PR expression in endometriotic lesions with WT non-variant or variant KRAS allele in mice. Lesions created by hESC carrying KRAS variant allele were characterized by a smaller number of nuclei stained positively for PR in both glandular and stromal cells. The epithelium of the lesions with variant cells was found to have only 35 ± 5% of nuclei positively stained compared to 75 ± 3%, in the lesions with WT KRAS (p = 0.02). Only 13 ± 8% of nuclei of stromal cells in KRAS variant lesions were found to express PR compared to 78 ± 7% in the non-variant lesions (p = 0.028). Scale bar represents 25 µm.
Progestins are commonly used to treat endometriosis and progesterone resistance has been described in this disease (Bulun et al, 2006; Cakmak & Taylor, 2010). To determine if alterations in the expression of sex steroid receptors were seen in the presence of the variant allele, we assessed estrogen receptor alpha (ERα) and progesterone receptor (PR) A and B. ERα levels were similar between lesions derived from cells with or without the variant LCS6 allele (data not shown). The number of nuclei positively stained for PR was decreased in the lesions with KRAS variant allele compared to the lesions created by WT hESC in both epithelium (35 ± 4% vs. 75 ± 3%, respectively, p = 0.02) and stroma (13 ± 8% vs. 78 ± 6%, respectively, p = 0.028; Fig 5c). The KRAS LCS6 variant cells retain the receptor for estrogen, which drives proliferation in endometrial cells; however, these cells show decreased PR, which drives differentiation.
DISCUSSION
In this study, we have identified a novel gene mutation associated with endometriosis. A variant SNP in the LCS6 let-7 miRNA binding site of the KRAS 3′-UTR was found in 31% of all cases of endometriosis, which is significantly higher than the 5.8% incidence observed in world populations. This variant allele is, therefore, potentially contributing to nearly one third of all endometriosis cases. Subjects with the non-variant KRAS allele more commonly presented with pain and dysmenorrhea than those with the variant allele; this may be due to the higher incidence of peritoneal endometriosis (as a proportion of total endometriosis), which is more likely to be symptomatic than are ovarian endometriomas. Subjects with the KRAS variant more often presented with infertility. Stage IV endometriosis, however, was equally common among subjects in each group.
This finding provides a mechanism for the pathogenesis of endometriosis in women with this functional mutation. Loss of the normal LCS6 let-7 binding site was associated with increased transcription and translation of KRAS. Ras proteins are crucial regulators of tyrosine kinase mitogenic and oncogenic activity. Effects of Ras activation include increased cell survival and proliferation (Khosravi-Far & Der, 1994; Schubbert et al, 2007). In this study, we demonstrate that the presence of the variant allele leads to a higher proliferation and invasion rate in endometrial cells. These properties may facilitate the invasion of endometrial cells into peritoneum and ovarian cortex. This mechanism supports the most accepted theory for the origin of endometriosis – retrograde menstruation and subsequent implantation and invasion of susceptible tissues (Giudice & Kao, 2004). The fact that only a portion of women develop this disease despite the nearly universal occurrence of retrograde menstruation could be explained by the presence of this allele.
Moreover, in the in vivo model of endometriosis, endometrial stromal cells harbouring the variant allele demonstrated a more aggressive behaviour. Cells containing the variant allele produced lesions that proliferated more and expressed lower levels of PR, which in turn may contribute to the diminished responsiveness to progesterone treatment. The behaviour of the variant cells in this model resembled those in mice with an activated KRAS gene that form endometriosis de novo (Dinulescu et al, 2005). Limitations of our model include the variability in hormone levels through the estrous cycle despite timing by vaginal cytology; however, this model also closely resembles the normal hormonal exposure seen in women. The activation of KRAS signalling through the LCS6 mutation explains the inability to find activating mutations in the coding regions of KRAS in humans with endometriosis. The mouse model of endometriosis can now be reconciled with the human disease, both caused by activating mutations disrupting regulation of the KRAS gene.
This variant in the let-7 binding site of KRAS has been established as a marker for predisposition to ovarian cancer (Ratner et al, 2010). This lends support to the theory that certain types of ovarian cancer may arise from endometriosis and helps to explain the increased risk of ovarian cancer in women with endometriosis (Nezhat et al, 2008). This variant SNP may be an early marker of those endometriosis patients with an increased risk of ovarian cancer, a hypothesis that will require additional validation.
Two large GWAS did not identify this SNP in women with endometriosis. The LCS6 polymorphism is not on the Illumina chip used in the larger European/US study. In addition, these studies confirmed the diagnosis of endometriosis by review of the medical records in a small minority of the subjects; and in the control group endometriosis status was not determined and can be expected to be approximately 10% in reproductive aged women. In contrast, all endometriosis subjects in this study were identified prospectively at the time of surgery and were thus clinically well annotated. Although GWAS studies are important for discovery of regions of the genome important in disease, they have not always proven to be useful in validation of functional markers due to the all-inclusive approach applied for cases and controls.
The KRAS pathway presents a potential therapeutic target for treatment of endometriosis. Our results demonstrate that synthetic small RNAs complementary to the variant allele will bind the LCS6 site and reduce reporter gene expression, suggesting a possible therapy for endometriosis. siRNA has been used as a drug because of its ability to induce specific, yet transient and reversible effects (Shim & Kwon, 2010).
In conclusion, a SNP in the let-7 miRNA binding site in 3′-UTR of the KRAS gene is a marker of endometriosis risk, explains the pathogenesis of endometriosis in the subgroup of patients with the SNP, provides a novel method of early endometriosis diagnostics, ovarian cancer prevention and offers potential treatment opportunities.
MATERIALS AND METHODS
Subjects and sample collection
From 2008 to 2010, a total of 150 DNA samples were collected from subjects diagnosed with endometriosis (ovarian or peritoneal) 132 of which were tested for the presence of K-Ras variant allele. DNA samples were received from subjects recruited at either Ponce School of Medicine and Health Sciences (PSMHS), Ponce, Puerto Rico (n = 48) or the Yale University School of Medicine (n = 102). The study included women with a current diagnosis or history of endometrioma (n = 89) and/or peritoneal endometriosis (n = 43). In all cases the diagnosis was made by biopsy; the lesion was surgically excised and the diagnosis of endometriosis was confirmed histologically. DNA was extracted from saliva, blood or tissue. Written informed consent was obtained from all participants. Approval for the experiments was obtained from the Yale University and the PSMHS Institutional Review Boards as well as from Yale University IACUC for mouse surgery protocol.
Evaluation of the LCS6 SNP in the 3′-UTR of the KRAS gene
DNA was isolated using the DNeasy Blood and Tissue kit or QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. For high-throughput genotyping, the isolated DNA samples were amplified using TaqMan PCR assays designed specifically to identify the T or G allele of the LCS6 SNP of KRAS gene (Applied Biosystems, Foster City, CA) as was described previously (Chin et al, 2008). Each assay was conducted in duplicate. Positive Results were confirmed by sequencing of the LCS6 region.
Tissue collection and cell culture
Endometrial biopsies were obtained from women with surgical and histological diagnosis of endometriosis. Biopsies were performed using the Pipelle catheter (CooperSurgical, Trumbull, CT) both from women positive for KRAS variant allele and women with WT KRAS gene. Endometrium from women without surgical evidence of endometriosis but with possible other benign gynecological conditions (e.g. fibroids, benign ovarian cysts) who tested negative for the presence of variant allele were used as controls. Briefly, endometrium was finely minced and cells were dispersed by incubation in HBSS containing HEPES (25 mm), 1% penicillin/streptomycin, collagenase (1 mg/ml, 15 U/mg) and DNase (0.1 mg/ml, 1500 U/mg) for 60 min at 37°C with agitation and pipetting. Endometrial cells were pelleted, washed and suspended in Ham's F12:DMEM (1:1) containing 10% FBS, 1% penicillin/streptomycin and 1% Amphotericin B. A mixture of endometrial cells (epithelial and stromal) were passed through a 40-µm sieve which allowed stromal cells to pass through while epithelial cells are retained on the sieve (Millipore, Billerica, MA). hESC were plated into 75 cm2 Falcon Tissue Culture flasks (BD Biosciences, Franklin Lakes, NJ). Cultured hESC at 3–5 passages were used for q-RT-PCR, proliferation, invasion, reporter assays and in the murine model. In each assay the number of passages was identical between the variant and control group.
As it has been previously shown by other investigators that stomal component of endometrium plays a crucial role in regulating endometrial homeostasis and controlling epithelial growth, hESC were chosen for use in all cell culture experiments (Arnold et al, 2001). Moreover it is well established that it is the defect in stromal cells which is responsible for defective estradiol metabolism in eutopic and ectopic endometrial tissue in patients with endometriosis (Cheng et al, 2007).
For all experiments hESC heterozygous for the variant allele were used and compare to either normal hESC or hESC from women with endometriosis and who were homozygous for WT KRAS allele.
Quantitative RT-PCR
To assess KRAS mRNA levels total RNA was extracted from primary cultured endometrial stromal cells using the Qiagen RNeasy isolation kit (Qiagen). The experiment included hECS from six normal subjects, six subjects with endometriosis carrying WT KRAS allele and six subjects carrying the variant KRAS allele. All samples were treated with RNase-free DNase (Ambion, Austin, TX) to remove the possibility of genomic DNA contamination. RNA samples were analysed by spectrophotometry to determine RNA concentration. mRNA (0.5 µg) was reverse-transcribed into cDNA using the iScript cDNA Synthesis Kit at 46°C for 40 min in a reaction mixture of 20 µl (Bio-Rad Laboratories, Hercules, CA). The resultant cDNA mixtures were stored at −20°C. Gene transcripts were amplified by real-time PCR using the Bio-Rad iCycler iQ system (Bio-Rad Laboratories). All primers were obtained from W. M. Keck Oligonucleotide Synthesis Facility, Yale University (Table 2). Real-time PCR was performed using the iQ SYBR Green Supermix Kit (Bio-Rad Laboratories). Reaction mixture included cDNA template (1 µg), forward and reverse primers, RNase-free water and the iQSYBRGreen Supermix, for a final reaction volume of 25 µl. The thermal cycling conditions were initiated by uracil-N-glycosylase activation at 50°C for 2 min and initial denaturation at 95°C for 10 min, then 40 cycles at 95°C for 15 s and annealing at 56.5°C for 30 s. Melting analysis was performed by heating the reaction mixture from 74 to 99°C at a rate of 0.2 C/s. Threshold cycle (Ct) and melting curves were acquired by using the quantitation and melting curve program of the Bio-Rad iCycler iQ system (Bio-Rad Laboratories). Only data with clear and single melting peaks were taken for further data analysis. Each reaction was performed in triplicate. The mRNA level of each sample was normalized to β-actin (ACTB) mRNA. Relative mRNA level was presented using the formula 2−ΔCt.
Table 2.
Primers used for quantitative RT-PCR
| Primers | Sequence |
|---|---|
| KRAS forward | tgaggactggggagggcttt |
| KRAS reverse | aggcatcatcaacaccctgtct |
| ACTB forward | gaagatcaagatcattgctc |
| ACTB reverse | aacgcaactaagtcatagtc |
| U6 forward | ctcgcttcggcagcaca |
| U6 reverse | aacgcttcacgaatttgcgt |
| let-7a | tgaggtagtaggttgtatagtt |
| let-7b | tgaggtagtaggttgtgtggtt |
| let-7c | tgaggtagtaggttgtatggtt |
| let-7d | agaggtagtaggttgcatagtt |
| let-7e | tgaggtaggaggttgtatagtt |
| let-7f | tgaggtagtagattgtatagtt |
| let-7g | tgaggtagtttgtacagtt |
For miRNA detection total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA). We employed the Poly (A) RT-PCR method using Invitrogen NCode miRNA First-Strand cDNA Synthesis MIRC-50 kit (Invitrogen). Conventional RT-PCR was used to assay miRNA levels with the specific forward primers to let-7a–g and the universal reverse primer complementary to the anchor primer. Anchor RT primer was used as the template for negative control and U6 small nuclear RNA was used as a control to determine relative miRNA expression. The real-time PCR profile was as described above, except for a melting temperature of 59°C.
Western blot analysis
Cultured endometrial stromal cells from five women with endometriosis carrying WT KRAS and five women with endometriosis with variant KRAS were lysed in Cell Lysis Buffer (Cell Signaling Technologies, Boston, MA) and centrifuged at 12,000 rpm for 2 min at 4°C, and the supernatant was collected. The protein content was quantified by the BCA assay method by using a protein assay kit (Bio-Rad Laboratories, Richmond, VA.). Aliquots (20 µg) were loaded onto 4–20% gradient polyacrylamide gel in MOPS buffer system (Invitrogen) and transferred to a nitrocellulose membrane by using a Transblot apparatus (Bio-Rad Laboratories) at 35 V overnight at 4°C. Subsequently, the membranes were incubated in blocking buffer (5% milk) for 1 h and then immunoblotted with mouse monoclonal KRAS antibody (1:200, ab55391, Abcam, Cambridge, MA) and Pan Actin antibody (1:25,000, 4968, Cell Signaling Technologies) overnight. After incubation with the primary antibody, the membranes were washed three times for 15 min with TBS [10 mM Tris–HCl (pH 7.4), 0.5 M NaCl] plus Tween 20 (0.2% v/v; TBST) and were incubated for 2 h in the corresponding HRP conjugated secondary antibody (1:2000; Invitrogen). The membranes were washed three times for 5 min in TBST. Proteins were detected with enhanced chemiluminescence (PerkinElmer, Waltham, MA). Quantification was performed using the ImageJ program.
Transfection and luciferase assay
pGL3 derivatives containing nearly the entire KRAS 3′-UTR (KRAS WT) and KRAS variant LCS6 were created as follows. KRAS WT includes 3910 bp of the KRAS 3′-UTR, which was amplified from human genomic DNA using the forward primer SMJ104 (ctagctagcatacaatttgtacttttttcttaaggcatac) and reverse primer LCJ5 (ctagctagctcaatgcagaattcatgctatccag). NheI restriction sites were included on the 5′-ends of the primers for convenient cloning. The product was first cloned into the TOPO cloning vector (Invitrogen) and then subcloned into pGL3 (Ambion) for use in subsequent luciferase assays. The luciferase reporter with the variant LCS6 KRAS 3′-UTR (KRAS mLCS6) was constructed through site-directed mutagenesis of KRAS WT using GeneTailor (Invitrogen). Normal endometrial stromal cells were plated in 12-well plates at 60% confluency. The adherent cells were co-transfected with 1 µg of luciferase reporter with variant K-Ras allele and a small interfering RNA (0.4 nM) designed to bind to the variant LCS6 K-Ras allele (ggacuggaguucacuacgugu). Qiagen AllStars Negative Control siRNA (0.4 nM) was used as negative control. To exclude the possibility that the endogenous levels of let-7 family miRNAs were not sufficient to down regulate KRAS a control set of cells was transfected with pGL3 basic derivative carrying WT regulatory region of K-Ras gene. pGL3 control vector was used as positive control of transfection efficiency. All cells were co-transfected with renilla vector (50 ng) using Fugene Transfection Reagent (Roche, Basel, Switzerland) using a DNA/Fugene transfection reagent volume ratio of 1:3. Luciferase activity was measured after 24 h of incubation according to the manufacturer's protocol using the Dual-Glow Luciferase assay system (Promega, Madison, WI). Firefly luciferase activity was normalized to renilla luciferase activity values for each sample. Experiments were performed three times in triplicate. The Mann–Whitney U-test was used for statistical analysis of the data.
Proliferation assay
Proliferation assay was performed with 5-bromo-2′-deoxy-uridine Labeling and Detection Kit III (Roche Applied Science, Germany). 0.5 × 105 of hESC with or without variant allele were plated into a 96-well plates. After culturing hESC for 48 h 10 µl of BrdU labelling solution was added into each well and incubated for 4 h. Then the culture medium containing the labelling solution was removed, cells were washed with serum containing wash medium and fixed with 200 µl of precooled 0.5 M ethanol in HCl for 30 min at −20°C. The cells were washed three times with serum containing medium and incubated with 100 µl of nucleases working solution per well for 30 min at room temperature in a water bath. Following three washes, 100 µl of of anti-BrdU-POD, Fab fragments and working solution were added. After 30 min of incubation at room temperature the antibody conjugate was removed and the cells were washed three times with washing buffer and incubated with 100 µl of peroxidase substrate per well. When positive samples showed distinctive green colour compared to negative control wells colorimetric analysis was performed using a microplate reader at 405 nm with a reference wavelength of approximately 490 nm (Bio-Rad Laboratories). The assay was performed three times in triplicate using hESC obtained from six different subjects in each group. The Mann–Whitney U-test was used for statistical analysis of the data.
Invasion assays
The invasion capacity from serum free towards serum containing medium through extracellular matrix gel and 8 µm pore membrane was analysed using the Millipore Colorimetric Migration Assay on 24-well plates (BD Falcon, Franklin Lakes, NJ, USA). Briefly, the membrane of each insert was covered with 100 µl of ECM gel (Sigma–Aldrich, St. Louis, MO) and the cells were kept in serum free medium (DMEM F12 + 1% penicillin/streptomycin + 1% Amphotericin B) for 24 h. 2 × 105 hESC from women without endometriosis and with endometriosis with WT or variant KRAS alleles were seeded into the inserts and incubated for 48 h. The lower chamber for this assay consisted of 24-well tissue culture plates (BD Falcon), which contained 500 µl of DMEM/F12 (1:1) with 10% FBS, 1% Pen/Strep and 1% Amphothericin B. For the control hESC (2 × 105) were plated directly into the lower chamber which represented 100% invasion. Invaded cells were stained, collected and lysed according to the manufacturer's instructions. Optical densities were read in triplicate at 560 nm using a Bio-Rad Laboratories (Hercules, CA, USA) plate reader. To determine the relative percent of invasion, results were compared to the 100% invasion control. Each experiment was performed three times in triplicate using specimens from six subjects without endometriosis, six subjects with endometriosis carrying WT KRAS allele and nine subjects with endometriosis carrying variant KRAS allele. The Mann–Whitney U-test was used to assess the significance of the difference in the acquired data.
Murine endometriosis model
Female 6–8-week-old immune-deficient mice (CB17SCID) were purchased from Charles River Laboratory. Experimental endometriosis was created in six mice. All surgeries and tissue collection were synchronized by use of vaginal cytology. All mice were operated on in their proestrous phase. In three mice endometriosis was created using cultured endometrial stromal cells from three different subjects with endometriosis and who tested positive for the KRAS variant SNP. In the other three mice endometriosis was created using cultured endometrial stromal cells from three different subjects with endometriosis but negative for the SNP (i.e. possessing the normal KRAS LCS6). Cells were cultured as described above, passaged 3–5 times, used at 100% confluency and harvested using 0.05% trypsin/EDTA. Cells were counted manually with a hemocytometer. The protocol for mouse kidney capsule cell transplantation was adopted and modified from Szot et al (Szot et al, 2007). Briefly, 1 × 106 cells were suspended in 20 µl normal saline and transferred to PE50 tubing system (BD Biosciences). The tubing was placed into 15 ml Falcon tubes and centrifuged at 1000 rpm to form a pellet. The tubing was maintained at 37°C until the procedure was performed. Each mouse was anaesthetised with intraperitoneal injection of xylazine/ketamine solution (100 and 10 mg/kg, respectively). Meloxicam was used for analgesia (0.2 mg/kg). The kidney was exteriorized and normal saline solution was injected in the peritoneal cavity to avoid dehydration. The kidney capsule was incised using a 27 gauge needle, the tubing containing the cell pellet inserted into the incision and the pellet was released under the kidney capsule. Thermocautery was used to close the kidney incision. The kidney was put back into the abdominal cavity and the abdominal wall was sutured. The skin was closed using a surgical stapler. After 4 weeks mice were sacrificed and the kidneys harvested, formalin fixed and paraffin embedded. Five micron sections were stained using haematoxylin and eosin (H&E) or used for immunohistochemistry (IHC) as described below. The experiment protocol was approved by Yale Institutional Animal Care and Use Committee.
The paper explained
PROBLEM
Endometriosis is a common, benign gynecological disorder, which is a frequent cause of chronic pelvic pain and infertility in 5–15% of reproductive age women. Although studied for many years, the exact pathogenesis as well as etiology of this disease remain unclear. It has been previously shown that activation of the KRAS gene caused de novo formation of endometriosis in mice, however, no activating mutations were found in the coding region of this gene in human endometriosis.
RESULTS
The authors hypothesized that there might be an activating mutation in the regulatory regions of KRAS gene causing excessive production of the protein with subsequent activation of the Ras pathway. 132 women with endometriosis were evaluated for a newly identified polymorphism in a miRNA let-7 binding site in the 3′-UTR of the KRAS gene, which has been previously associated with an increased risk of lung and ovarian cancer. Thirty-one percent of the subjects were found to carry this variant allele compared to only 5.8% of the general population. The presence of this mutation was found to be associated with higher KRAS mRNA and protein levels and lower let-7 levels in endometrial stromal cells of women with endometriosis as well as an increased proliferation rate and invasion capacity of these cells.
IMPACT
This study identified a novel gene mutation, which is associated with up to one third of endometriosis cases. This mutation potentially represents a new therapeutic target for endometriosis as well as the basis of a potential screening method for endometriosis risk.
Immunohistochemistry
IHC was conducted on formalin-fixed paraffin-embedded mouse kidneys containing transplanted hESCs that had either the variant or normal LCS6 allele of the KRAS gene. Sections were deparaffinized, and dehydrated through a series of xylene and ethanol washes. Each specimen was stained with H&E for histological evaluation. For IHC analysis, after a 5-min rinse in distilled water, an antigen-presenting step was performed by steaming the slides in 0.01 mole/l sodium citrate buffer for 20 min, followed by cooling for 20 min. Slides were rinsed for 5 min in PBS with 0.1% Tween 20 (PBST), and sections were circumscribed with a hydrophobic pen. Endogenous peroxidase was inactivated by incubation in 3% hydrogen peroxide for 5 min, followed by a 5-min PBST wash. After a preincubation with 2% normal goat or horse serum to block non-specific sites, sections were incubated with primary antibodies in a humidified chamber for 18 h at 4°C. Antibodies used were against PCNA, ERα, PR (Santa Cruz Biotechnology, Santa Cruz, CA) and Cleaved Caspase-3 (Asp175; Cell Signaling Technology, Beverly, MA). Slides were then incubated with the appropriate biotin conjugated secondary antibodies followed by avitin–biotin–horseradish peroxidase complex and diaminobenzidine tetrahydrochloride before counterstaining with Gill's haematoxylin (ABC kit; Vector Laboratories, Burlingame, CA). Negative control sections were processed in an identical manner but substituting primary antibodies with normal rabbit IgG. All negative control sections showed no colour reaction. The number of stained nuclei was counted separately in epithelium and stroma in five high-power fields on each slide by three-independent researchers and averaged for each experimental animal. The Mann–Whitney U-test was used to determine the significance of differences between experimental groups.
Statistical analysis
Chi-squared test was used to compare the frequencies of clinical symptoms among the groups of patients with non-variant and alternative KRAS allele. T-test was used to evaluate statistical significance of experiments used to asses KRAS mRNA and let-7 miRNA. The Mann–Whitney U-test was used to assess the significance of the differences in proliferation, invasion and luciferase activity and to compare IHC staining indices. Reported are mean ± standard error of the mean (SEM).
Acknowledgments
This study was funded by NIH Grants U54 HD052668 and R01 HD036887.
Conflict of interest statement: Dr. Weidhaas has patented IP around the KRAS variant. The other authors declare that they have no conflict of interest.
Author contributions
OG was involved with conception and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content and statistical analysis; RP, SP, EM, TP and EC were involved in acquisition, analysis and interpretation of data, drafting and revision of the manuscript and technical support; IF was involved in data acquisition and revision of the manuscript; JW was involved in conception and design of the study, acquisition of data, critical revision of the manuscript for important intellectual content and supervision; HST was involved in conception and design of the study, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, obtaining funding and supervision; HST had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Supplementary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Amemiya S, Sekizawa A, Otsuka J, Tachikawa T, Saito H, Okai T. Malignant transformation of endometriosis and genetic alterations of KRAS and microsatellite instability. Int J Gynecol Obstet. 2004;86:371–376. doi: 10.1016/j.ijgo.2004.04.036. [DOI] [PubMed] [Google Scholar]
- Arnold JT, Kaufman DG, Seppälä M, Lessey BA. Endometrial stromal cells regulate epithelial cell growth in vitro: a new co-culture model. Hum Reprod. 2001;16:836–845. doi: 10.1093/humrep/16.5.836. [DOI] [PubMed] [Google Scholar]
- Bulun SE. Mechanisms of disease endometriosis. N Engl J Med. 2009;360:268–279. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- Bulun SE, Cheng YH, Yin P, Imir G, Utsunomiya H, Attar E, Innes J, Julie Kim J. Progesterone resistance in endometriosis: link to failure to metabolize estradiol. Mol Cell Endocrinol. 2006;248:94–103. doi: 10.1016/j.mce.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Cakmak H, Taylor HS. Molecular mechanisms of treatment resistance in endometriosis: the role of progesterone-hox gene interactions. Semin Reprod Med. 2010;28:69–74. doi: 10.1055/s-0029-1242996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletti MZ, Christenson LK. MicroRNA in the ovary and female reproductive tract. J Anim Sci. 2009;87:29–38. doi: 10.2527/jas.2008-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervello I, Mas A, Gil-Sanchis C, Peris L, Saunders PT, Critchley HD, Simon C. Reconstruction of endometrium from human endometrial side population cell lines. PLoS ONE. 2011;6:21221. doi: 10.1371/journal.pone.0021221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YH, Imir A, Fenkci V, Yilmaz MB, Bulun SE. Stromal cells of endometriosis fail to produce paracrine factors that induce epithelial 17β-hydroxysteroid dehydrogenase type 2 gene and its transcriptional regulator Sp1: a mechanism for defective estradiol metabolism. Am J Obstet Gynecol. 2007;196:391.e1-7. doi: 10.1016/j.ajog.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Cheng CW, Licence D, Cook E, Luo F, Arends MJ, Smith SK, Print CG, Charnock-Jones DS. Activation of mutated KRAS in donor endometrial epithelium and stroma promotes lesion growth in an intact immunecompetent murine model of endometriosis. J Pathol. 2011;224:261–2269. doi: 10.1002/path.2852. [DOI] [PubMed] [Google Scholar]
- Cheung K, Osier M, Kidd J, Pakstis A, Miller P, Kidd K. ALFRED: an allele frequency database for diverse populations and DNA polymorphisms. Nucleic Acids Res. 2000;28:361–363. doi: 10.1093/nar/28.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;268:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, Marsit CJ, Kelsey KT. A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003–1007. doi: 10.1093/carcin/bgp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, Jacks T. Role of KRAS and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs: microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fourquet J, Gao X, Zavala D, Orengo JC, Abac S, Ruiz A, Laboy J, Flores I. Patients' report on how endometriosis affects health, work, and daily life. Fertil Steril. 2010;93:2424–2428. doi: 10.1016/j.fertnstert.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Outley J, Botteman M, Spalding J, Simon JA, Pashos CL. Economic burden of endometriosis. Fertil Steril. 2006;86:1561–1572. doi: 10.1016/j.fertnstert.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–1799. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- Guo S-W. Recurrence of endometriosis and its control. Hum Reprod Update. 2009;15:441–461. doi: 10.1093/humupd/dmp007. [DOI] [PubMed] [Google Scholar]
- Hadfleld R, Mardon H, Barlow D, Kennedy S. Delay in the diagnosis of endometriosis: a survey of women from the USA and then UK. Hum Reprod. 1996;11:878–880. doi: 10.1093/oxfordjournals.humrep.a019270. [DOI] [PubMed] [Google Scholar]
- Hemmings R, Rivard M, Olive DL, Poliquin-Fleury J, Gagné D, Hugo P, Gosselin D. Evaluation of risk factors associated with endometriosis. Fertil Steril. 2004;81:1513–1521. doi: 10.1016/j.fertnstert.2003.10.038. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kao LC, Germeyer A, Tulac S, Lobo S, Yang JP, Taylor RN, Osteen K, Lessey BA, Giudice LC. Expression profiling of endometrium from women with endometriosis reveals candidate genes for disease-based implantation failure and infertility. Endocrinology. 2003;144:2870–2881. doi: 10.1210/en.2003-0043. [DOI] [PubMed] [Google Scholar]
- Khosravi-Far R, Der CJ. The Ras signal transduction pathway. Cancer Metastasis Rev. 1994;13:67–89. doi: 10.1007/BF00690419. [DOI] [PubMed] [Google Scholar]
- Moen MH, Magnus P. The familial risk of endometriosis. Acta Obstet Gynecol Scand. 1993;72:560–564. doi: 10.3109/00016349309058164. [DOI] [PubMed] [Google Scholar]
- Nezhat F, Datta MS, Hanson V, Pejovic T, Nezhat C, Nezhat C. The relationship of endometriosis and ovarian malignancy: a review. Fertil Steril. 2008;90:1559–1570. doi: 10.1016/j.fertnstert.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Otsuka J, Okuda T, Sekizawa A, Amemiya S, Saito H, Okai T, Kushima M, Tachikawa T. KRAS mutation may promote carcinogenesis of endometriosis leading to ovarian clear cell carcinoma. Med Electron Microsc. 2004;37:188–192. doi: 10.1007/s00795-004-0252-5. [DOI] [PubMed] [Google Scholar]
- Painter JN, Anderson CA, Nyholt DR, Macgregor S, Lin J, Lee SH, Lambert A, Zhao ZZ, Roseman F, Guo Q, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Gen. 2011;43:51–54. doi: 10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine. Endometriosis and infertility. Fertil Steril. 2006;86:156–160. [Google Scholar]
- Ratner E, Lu L, Boeke M, Barnett R, Nallur S, Chin LJ, Pelletier C, Blitzblau R, Tassi R, Paranjape T, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–6515. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010;277:4814–4827. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- Simoens S, Hummelshoj L, D'Hooghe T. Endometriosis: cost estimates and methodological perspective. Hum Reprod Update. 2007;13:395–404. doi: 10.1093/humupd/dmm010. [DOI] [PubMed] [Google Scholar]
- Simpson JL, Bischoff FZ. Heritability and molecular genetic studies of endometriosis. Ann N Y Acad Sci. 2002;955:239–251. doi: 10.1111/j.1749-6632.2002.tb02785.x. [DOI] [PubMed] [Google Scholar]
- Szot GL, Koudria P, Bluestone JA. Transplantation of pancreatic islets into the kidney capsule of diabetic mice. J Vis Exp. 2007;9:404. doi: 10.3791/404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor H, Arici A, Olive D, Igarashi P. HOXA10 is expressed in response to sex steroids at the time of implantation in human endometrium. J Clin Invest. 1998;101:1379–1384. doi: 10.1172/JCI1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HS, Bagot C, Kardana A, Olive D, Arici A. HOX gene expression is altered in the endometrium of women with endometriosis. Hum Reprod. 1999;14:1328–1331. doi: 10.1093/humrep/14.5.1328. [DOI] [PubMed] [Google Scholar]
- Treloar SA, Wicks J, Nyholt DR, Montgomery GW, Bahlo M, Smith V, Dawson G, Mackay IJ, Weeks DE, Bennett ST, et al. Genomewide linkage study in 1,176 affected sister pair families identifies a significant susceptibility locus for endometriosis on chromosome 10q26. Am J Hum Genet. 2005;77:365–376. doi: 10.1086/432960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno S, Zembutsu H, Hirasawa A, Takahashi A, Kubo M, Akahane T, Aoki D, Kamatani N, Hirata K, Nakamura Y. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Gen. 2010;42:707–711. doi: 10.1038/ng.612. [DOI] [PubMed] [Google Scholar]
- Vercellini P, Trecca D, Oldani S, Fracchiolla NS, Neri A, Crosignani PG. Analysis of p53 and ras gene mutations in endometriosis. Gynecol Obstet Invest. 1994;38:70–71. doi: 10.1159/000292450. [DOI] [PubMed] [Google Scholar]
- Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- Zhao ZZ, Nyholt DR, Le L, Martin NG, James MR, Treloar SA, Montgomery GW. KRAS variation and risk of endometriosis. Mol Hum Reprod. 2006;12:671–676. doi: 10.1093/molehr/gal078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.