Abstract
A workshop on ‘The Biology of Intracellular Bacterial Pathogens’ was held last October in a venue of the International University of Andalusia (UNIA) located in the World Historic Heritage town of Baeza, in the South of Spain. This Workshop gathered leading scientists from around the world to discuss their latest findings related to the mechanisms that intracellular pathogens use to subvert and manipulate host cell functions. The workshop focused on novel aspects that imprint current research in this discipline, including the heterogeneous behaviour of the pathogen at the population level, the host determinants that modulate susceptibility to the infection, the search for new drugs to combat these particular types of infections and also cutting edge technologies based on new imaging approaches and the use of microfluidics. Discussion on these topics provided new insights into the biology of these pathogens and enriched the field with new ideas for understanding why colonization of the intracellular niche of eukaryotic cells is a preferred strategy used by important human pathogens.
Keywords: host response, intracellular pathogen
Together with viruses, intracellular bacterial and parasite pathogens constitute the largest group of microorganisms inflicting human health. Bacteria that colonize the interior of the eukaryotic cell cause many important infectious diseases such as tuberculosis, typhoid fever, tularaemia, Q-fever, brucellosis and pneumonia among others, with high morbidity and mortality in humans. Noteworthy, many of these pathogens are also prone to establish asymptomatic infections. This duality is in most cases poorly understood at the molecular level. A feature frequently evidenced in infections caused by intracellular bacteria and parasites is the sophistication and variety of mechanisms directed to manipulate processes central to the host physiology, including cytoskeleton dynamics, vesicular trafficking, release of defence molecules and sensing of pathogen molecules. Despite abundant information, in most cases we cannot ‘predict’ the outcome of a given pathogen-host interplay. Many factors, including population diversity and bistability in the responses (from both the pathogen and the host) seem to play decisive roles in this delicate battle. With these ideas in mind, Jorge Galán (Yale University, New Haven) and Francisco García-del Portillo (Centro Nacional de Biotecnología-CSIC, Madrid) organized this Workshop, to which more than 50 participants attended.
Cell biology of intracellular infections: balanced coexistence of the two partners
The two first sessions of the Workshop were dedicated to discuss at the cellular level different aspects of the interaction occurring between intracellular bacteria and the host. Guy Tran Van Nhieu (CIRB, College de France, Paris) described how the human pathogen Shigella flexneri manipulates epithelial cell physiology at the site of bacterial entry. S. flexneri, which is devoid of classical adhesins or invasins at its surface, invades epithelial cells in two phases (Fig 1). The first involves 'capture' of bacteria by nanometer thin micropodial extensions (NMEs) and retraction of the NME to bring bound bacteria in contact with the cell body. The second phase corresponds to the localized membrane ruffling linked to the activity of Shigella type three effectors. Local events spatially restricted to the site of bacterial entry include InsP3 signalling, Ca2+ release from intracellular stores, and Erk1/2 activation in the infected cell, which promote 'NME retraction' and bacterial internalization. Mitochondria located at entry sites also participate in bacterial invasion. Overall, these findings highlighted how pathogens can manipulate processes in specific areas of the cell. Raphael Valdivia (Duke University, North Carolina) described elegant subversion mechanisms of cytoskeletal structures such as actin and intermediate filaments by Chlamydia. These cytoskeleton proteins accumulate around the bacterial inclusion and were proposed to act as a ‘corset’ preventing leakage of pathogen components in the cytosol of the infected cell. Data were also presented on the effectiveness in controlling infection of peptides that block the action of the important Chlamydia protease CPAF, which cleaves intermediate filaments maintaining vacuole integrity and protects against caspase 1-mediated cell death. A new message of this talk was the apparent ‘altruistic’ behaviour of reticulate bodies (RB). Some of the RBs eventually lyse and release glycogen and other cellular components to the intra-inclusion lumen to provide RBs in the process of differentiation into infectious elementary bodies (EBs) with the necessary nutrients. The type of decisions directing the fate of some RBs to produce glycogen and others to produce enzymes required to metabolize such polymers remains an interesting subject of study for future research. The talk by Craig Roy (Yale University, New Haven) focused on the elegant mechanisms used by Legionella pneumophila to regulate the activities of host GTPases involved in vesicular trafficking. The effector DrrA, for example, mediates the recruitment and activation of the host GTPase Rab1 to the vacuole containing Legionella. Activation of Rab1 on the plasma membrane-derived vacuole is sufficient to promote the tethering and fusion of vesicles derived from the endoplasmic reticulum by a process that involves non-canonical SNARE interactions. Additionally, the effectors DrrA and AnkX post-translationally modify Rab1 and a related GTPase called Rab35 through the covalent attachment of AMP or phosphocholine residues respectively. This modulates the ability of these GTPases to conduct their normal cellular functions without preventing Rab1-mediated fusion of vesicles with the Legionella-containing vacuole. Thus, precise manipulation of host GTPases by Legionella is a strategy used to convert an endocytic vacuole into a specialized compartment that supports bacterial replication. Thomas F. Meyer (Max Planck Institute for Infection Biology, Berlin) showed recent data obtained upon sRNAi-based screenings designed to unravel host functions that are targeted by a pathogen, during infection. This work has been performed with two relevant human pathogens, influenza A virus (IAV) and Chlamydia trachomatis. Meyer's group discovered that ∼300 human cellular factors could support or antagonize IAV replication. In the case of C. trachomatis, host functions related to inflammation, cytoskeletal dynamics, fatty acid/lipid metabolism, and DNA damage response, were shown to be involved in the infection. Importantly, these studies reveal that a ‘host-directed therapy’ may be an alternative and effective strategy to combat intracellular infections, minimizing the emergence of microbial resistance. Francisco García-del Portillo (Centro Nacional de Biotecnología-CSIC, Madrid) reported data on how Salmonella controls intracellular proliferation within fibroblasts, a cell type in which this pathogen establishes a persistence state. Data obtained in non-growing intracellular bacteria revealed that the two type-III secretion systems that this pathogen has are temporally regulated in a strictly defined manner inside the fibroblast. Loss of this temporal control results in bacteria that either proliferate or are killed by the host cell. These data highlight how infection may proceed only when factors are put into game in a coordinated fashion by intracellular bacteria at precise times upon colonization of the host cell. William Sullivan (University of California, Santa Cruz) reported interesting aspects of the biology of Wolbachia, an insect and nematode endosymbiont that is inherited via the germ line (oocytes). Wolbachia is transmitted through invertebrates and causes important human diseases such as African River Blindness and Elephantiasis. Using Drosophila as a model, W. Sullivan showed that this bacterium during oogenesis utilizes the host microtubule network and the respective motors to first move to the anterior side of oocytes in a dynein-dependent manner and later to the posterior side using kinesin. Host factors such as Gurken (Grk) are required for Wolbachia proliferation. Both the mRNA and protein forms of this host factor influences Wolbachia titers. The Grk protein acts as a transforming growth factor homolog whereas grk mRNA serves as a platform to assemble a protein complex controlling dorsal/ventral determination in Drosophila. Interestingly, Wolbachia colocalizes with grkRNP complexes. Wolbachia and the host could establish a tight regulatory loop modulated by grkRNP function and the extent at which bacterial proliferate. This was a clear example of how intracellular bacterial pathogens and hosts can reach ‘balanced’ stages ensuring homeostasis in both partners. Pascale Cossart (Institut Pasteur, Paris) described the key role played by mitochondria in Listeria infection. This organelle ‘fragments’ upon bacterial infection in a process dependent on listeriolysin-O (LLO) and the uptake of extracellular Ca2+ (Fig 1). Manipulation of the mitochondria status using RNAi against host proteins Mfns and Drp1 affects bacterial infection. In addition, data were presented regarding the role of the Listeria secreted protein LntA in subverting nuclear responses. LntA interacts with BAHD1, a host protein inducing heterochromatin formation. LntA binding to BAHD1 would reprogram the infected cell to up-regulate interferon-lambda (INF-λ) responses and interferon-stimulated genes (ISGs). LntA production must be tightly regulated in vivo since Listeria virulence is attenuated in both ΔlntA bacteria and in variants that overproduce the protein. Altogether, these findings indicate that evolution has probably maintained LntA in pathogenic Listeria to finely tune the host immune response.
Figure 1. Representative phenomena discussed at the meeting.
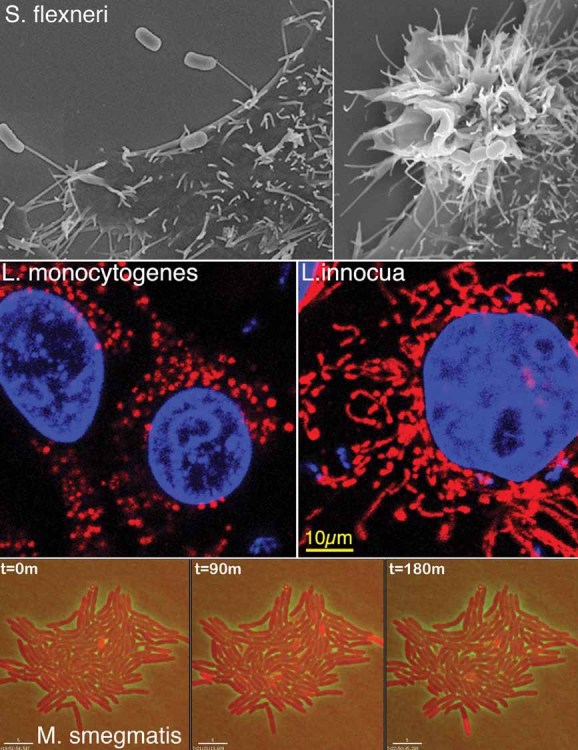
Upper panels, scanning electron micrographs of invasive Shigella flexneri captured by NMEs (left) or inducing membrane ruffling during cell invasion (right); middle panels, fragmentation of mitochondria occurring during Listeria monocytogenes invasion of HeLa epithelial cells (left), which is not observed upon entry of non-pathogenic L. innocua expressing InlB (right). Mitochondria were labelled with cytochrome-C antibody (red) and host cell nuclei and bacteria with Dapi (blue); lower panels, live imaging denoting dynamic expression of a KatG-dsRed fusion protein (red bacteria) in a subpopulation of Mycobacterium smegmatis as a function of time.
Regulation of intracellular infections: not all individuals are the same
This session was started by John McKinney (EPFL, Lausanne), who reported on the variability of responses exhibited by individuals of a Mycobacterium tuberculosis population when exposed to antibiotics (Fig 1). This phenomenon, which is starting to be analysed at the genetic and molecular level, has a tremendous impact in the fate of intracellular infections. Experiments performed in microfluidic devices coupled to time-lapse fluorescence microscopy revealed that M. tuberculosis populations are composed of individuals growing at different doubling rates, ranging from 6–7 h to 2–3 weeks. Some individuals of the population persist in the presence of the antibiotic and daughter cells derived by outgrowth of the survivors after antibiotic withdrawal show the same kinetics of cell death upon re-exposure to the antibiotic. Surprisingly, the growth rate of individual cells does not correlate with their fate (death or persistence) under antibiotic exposure. Therefore, non-genetic cell-to-cell phenotypic variation seems to underlie the response of M. tuberculosis to antibiotics and the dogma that slow-growing cells are predisposed to persist better in the presence of antibiotics is not universally true. Current effort is now focused on determining which bacterial functions are linked to the capability of M. tuberculosis to persist and withstand antibiotic treatment. One of these functions is the catalase-peroxidase KatG. Due to its NADH oxidase activity, this catalase modifies the antibiotic isoniazid, and the modified antibiotic is the active form that blocks mycolic acid biosynthesis. Increased KatG activity correlated with lower persistence rates. Interestingly, KatG expression was shown to be ‘pulsatile’—a feature postulated to be not specific for KatG and probably occurring in many if not all genes—and pulses of KatG expression were strongly correlated with ensuing cell death. Therefore, M. tuberculosis may be constantly taking decisions of the type ‘on-off’ in key functions linked to persistence, and fluctuations in gene expression at the single-cell level may explain the heterogeneity of cellular behaviour at the population level. This type of single-cell and population studies is certainly a major future area of new research. Importantly, fluctuations in gene expression are also observed in intracellular mycobacteria located inside macrophages. Eric Rubin (Harvard School of Public Health, Massachusetts) reported studies performed in M. tuberculosis addressing how mistranslation can be beneficial for the pathogen to get diversity maintaining the same genetic make-up. This intracellular pathogen lacks Gln(glutamine)-tRNA synthetase, and Gln is loaded in correct positions during translation due to an enzyme that modifies Glu (glutamic acid) in the Glu-tRNA. Using a mutated tRNA can drive amino acid misincorporation to high levels. Interestingly, increased mistranslation rates can result in phenotypic resistance to some antibiotics. Thus, a new concept was discussed on how the absence of some tRNA synthetase has been maintained during evolution of some intracellular bacterial pathogens to gain protein diversity in the population and ensure better adaptation to stress.
Manipulation of host responses by intracellular pathogens: again a game played by few individuals within the entire population
Jean Pierre Gorvel (Centre d'Immunologie de Marseille-Luminy, Marseille) reported on the ability of the important human pathogen Brucella abortus to signal to the host with a cyclic glucan made of 17–25 glucose units bound by β-1,2 linkages (CβG). CβG modulates lipid rafts organization at the Brucella-containing vacuole. Interestingly, Brucella replicates in dendritic cells and CβG behaves as a potent immune-activator of human and mouse dendritic cells. Since CβG is normally stored in the periplasm of producing bacteria, it was proposed that few intracellular Brucella could lyse and that the released CβG could subsequently activate DC maturation. CβG is produced in large amounts (1–5% of the bacterial dry weight) and lysis of few bacteria may be sufficient to alert the host for controlling the proliferation of the rest of the population and reach a host-pathogen equilibrium favouring the long-term infections that characterize many cases of brucellosis. Brett Finlay (University of British Columbia, Vancouver) described new findings on Salmonella SspH2, an effector protein of the type-III secretion system and an E3 ligase encoded in the pathogenicity island 2 (SPI-2) found to interact with a co-chaperone protein of the NOD1/NOD2 receptors. Such co-chaperone protein is also conserved in plants. Therefore, ‘intracellular sensing systems’ based on receptors of the Nod and NLR families might be subjected to fine-tuning by pathogen effector proteins. It would be interesting for future research to dissect the primary signals sensed by the pathogen leading to such counter-attack directed to ‘poison’ the NOD-NLR host defences. Wolf Dietrich-Hardt (ETH, Zurich) reported on the regulation of the Salmonella pathogenicity island 1 (SPI-1) required for epithelial cell invasion. Under invasion-inducing conditions, only 20–40% of the bacterial population express SPI-1. This is clearly a bistable phenotype since a) ON and OFF bacteria form two distinct sub-populations; b) both phenotypes remain stable even if shifted to conditions normally not favouring SPI-1 expression (“hysteresis”); c) new cultures grown from ON or OFF bacteria do again form both phenotypes. The reasons for this had remained unclear. Interestingly, ON-bacteria expressing SPI-1 suffer from two types of “cost”. First, they exhibit lower growth rates than OFF bacteria or commensals like E. coli. Second, they invade the gut mucosa, a tissue protected by efficient innate immune defences (NADPHox, NOS2, INF-γ, MyD88) killing approx. 90% of all invaders. These costs should represent a significant disadvantage to the entire Salmonella population. However, they are compensated by the growth advantage that the pathogen gains in the gut lumen against the competing microbiota. This advantage is fuelled by the inflammatory response of the intestinal mucosa, which favours pathogen growth in the gut lumen and disfavours the commensals. Thus, the inflammation is triggered by the SPI1-ON subpopulation, which fosters pathogen growth in the gut lumen and efficient transmission of bacteria that may never have expressed SPI-1. Bistable gene expression may have therefore evolved as a means to minimize the costs associated with the expression of certain virulence factors. Jonathan Howard (University of Cologne) discussed the anti-parasitic role of ‘immunity-related GTPases’ (IRG proteins). There are 23 IRG-encoding genes in mice. Loss of one of these, Irgm1 (LRG47), causes an unexplained lymphopenia and systemic loss of immune function, with consequent susceptibility to Salmonella, Listeria, M. tuberculosis, Toxoplasma, Leishmania and Trypanosoma. All other IRG proteins have so far been implicated exclusively in cell-autonomous resistance to Toxoplasma gondii and Chlamydia trachomatis. IRG proteins, such as Irga6 and Irgb6, locate at the parasitophorous vacuole surrounding T. gondii, damage the vacuolar membrane and inhibit parasite proliferation. At the vacuole membrane, IRG proteins can be inhibited by kinases and pseudokinases secreted by virulent T. gondii strains. Genes encoding the arsenal of IRGs are absent in higher primates, including humans, raising the question of why this potent mechanism against intracellular pathogens was abandoned in some vertebrates.
Drug discovery in intracellular infection models, symbiotic relationships and host susceptibility to intracellular infections
David Clarke (University College Cork, Ireland) described how bacteria of the genus Photorhabdus transit from a mutualistic lifestyle in nematodes to a pathogenic one in insect larvae. Of the three species known in this genus, only P. asymbiotica has been linked to infections in humans. Unlike the other species of the genus, which contain only one, P. asymbiotica contains two type-III secretion systems, which remain poorly characterized yet. Other bacterial products required for the pathogenic lifestyle include toxins and lipopolysaccharide (LPS). Interestingly, transition from pathogenicity to mutualism does not depend on quorum sensing systems or signalling molecules acting as second messengers. Instead, primary and secondary metabolism play a key role in this lifestyle change since malate dehydrogenase (Mdh) and fumarase (FumC) are absolutely required for this transition. Other bacterial functions seem also to be required for bacterial colonization of the nematode i.e. transmission. During transmission, Photorhabdus replicate inside nematode rectal gland cells. Indeed bacterial replication and nematode development are tightly coupled and regulated by the LysR-like regulatory protein HdfR. Overall, these observations illustrate how a bacterium can switch between different host-dependent lifestyles. David Russell (Cornell University, Ithaca) showed transcriptomic data relevant for the search of new drugs for combating M. tuberculosis infections. Data obtained from macrophages at different times post-infection were indicative of the different physiological states of the bacterium following invasion of the host cell. Global analysis of these responses indicates that intracellular M. tuberculosis faces stress linked to detoxification of C3 compounds such as propionyl-CoA. Overall the data indicate that the selective pressure(s) inside the macrophage are significantly different from those in broth culture. In collaboration with Vertex Pharmaceuticals, the lab has just completed a high-throughput screen of 340,000 compounds for small molecules active against intracellular M. tuberculosis. Of note is that approximately 50% of compounds identified exhibit preferential activity against intracellular and not extracellular bacilli. This represents a new class of compounds active against an intracellular infection. Lalita Ramakrishan (University of Washington, Seattle) discussed the importance of investigating the ‘host side’ in infections caused by intracellular pathogens and reported investigations on factors that may predispose humans to infections caused by M. tuberculosis. Data collected in the zebrafish model with M. marinum highlighted that the early granuloma acts not as a structure limiting pathogen dissemination but just the contrary. Moreover, the bacteria proliferating within the macrophages of these structures express several drug-efflux pumps that both enable their intracellular growth and induce antibiotic tolerance. Tumour necrosis factor (TNF) was confirmed to be a key defence molecule against mycobacteria and a new primary mechanism uncovered in the zebrafish for its protection. Genetic data first in the zebrafish and then in human populations showed that production of LTA4H, an enzyme involved in arachidonic acid metabolism, correlates to host susceptibility to intracellular infections by this pathogen. LTA4H production is tightly linked to synthesis of lipoxins A4 and leukotriene B4, which modulate TNF levels. High lipoxin A4 levels leads to low TNF production and increased bacterial growth and the reverse occurs for leukotriene B4. Importantly, the data accumulated reinforce the old assumption that a ‘balanced’ host response to M. tuberculosis is fundamental to control infection. Thus, human individuals having either low or high LTA-4H levels are equally susceptible to infections caused by this intracellular pathogen. This evidence alerts us to a scenario in which treatment with anti-inflammatory drugs on infected human individuals can in one group control the infection whereas in the other exacerbates the disease.
In conclusion, the Workshop unravelled great challenges for future research in the biology of these successful intracellular pathogens, especially in areas still under-investigated as population dynamics during the infection process from both the pathogen and host sides. Such variability is convincing the researchers in this field to consider disease, asymptomatic infection or pathogen clearance as different manifestations of a same game, in which a few players may dictate the most important rules. The understanding of these differential behaviours at a mechanistic level signifies a tremendous amount of work, which will be certainly rewarding considering that resulting information will help to prevent or treat the important diseases that intracellular bacterial pathogens cause in humans.
Acknowledgments
We thank all the conference presenters and participants for the high level of their presentations and the stimulating discussions. We also thank Guy Tran Van Nhieu, Fabrizia Stavru and John McKinney for the images shown in the figure of this report. We apologize to those whose work has not been cited due to space limitations.
The authors declare that they have no conflict of interest.


