Abstract
Although ∼50% of all types of human cancers harbour wild-type TP53, this p53 tumour suppressor is often deactivated through a concerted action by its abnormally elevated suppressors, MDM2, MDMX or SIRT1. Here, we report a novel small molecule Inauhzin (INZ) that effectively reactivates p53 by inhibiting SIRT1 activity, promotes p53-dependent apoptosis of human cancer cells without causing apparently genotoxic stress. Moreover, INZ stabilizes p53 by increasing p53 acetylation and preventing MDM2-mediated ubiquitylation of p53 in cells, though not directly in vitro. Remarkably, INZ inhibits cell proliferation, induces senescence and tumour-specific apoptosis, and represses the growth of xenograft tumours derived from p53-harbouring H460 and HCT116 cells without causing apparent toxicity to normal tissues and the tumour-bearing SCID mice. Hence, our study unearths INZ as a novel anti-cancer therapeutic candidate that inhibits SIRT1 activity and activates p53.
Keywords: apoptosis, Inauhzin, p53, SIRT1, small molecule inhibitor
INTRODUCTION
The vital importance of the tumour suppressor gene TP53 in preventing human cancer development and progression is not only demonstrated by the fact that its mutations are detected in 50% of all types of human cancers (Hollstein et al, 1991), but also emphasized by accumulating evidence that the functions and stability of the p53 protein are often abrogated via post-translational mechanisms in the rest of human cancers with wild-type (WT) TP53 (Brown et al, 2009; Kruse & Gu, 2009). Cancers need to frequently disarm p53, because it, once activated, triggers cell growth arrest, apoptosis, autophagy or senescence, which are detrimental to cancer cells (Vogelstein et al, 2000; Vousden & Prives, 2009), and impedes cell migration, metabolism or angiogenesis, which are favourable to cancer cell progression and metastasis (Vousden & Prives, 2009). These cellular functions of p53 are executed primarily via its transcription-dependent and independent activities (Vousden & Prives, 2009). However, because these functions are also deleterious to normally growing stem cells and developing tissues (Hong et al, 2009), higher eukaryotes have evolved an elegant feedback mechanism to monitor p53 level and activity (Eischen & Lozano, 2009).
Two chief monitor proteins of p53 are MDM2 (HDM2 in human; Wu et al, 1993) and MDMX (also known as MDM4; Shvarts et al, 1996). In a feedback fashion, they work together to directly inhibit the transcriptional activity of p53 (Gu et al, 2002) and mediate p53 degradation via ubiquitin-dependent proteolysis (Haupt et al, 1997; Kubbutat et al, 1997), as MDM2 possesses an E3 ubiquitin ligase activity (Honda et al, 1997) and its mRNA expression is stimulated by p53 (Barak et al, 1993; Wu et al, 1993), thus, keeping p53 level and activity marginally detectable in most of normal mammalian cells or tissues. This feedback regulation as firmly established in mouse models (Jones et al, 1995; Montes de Oca Luna et al, 1995) is subjected to tight regulation (Wade et al, 2010; Zhang & Lu, 2009). On one hand, a variety of cellular genotoxic or non-genotoxic stresses can reverse this feedback inhibition (Kruse & Gu, 2009) via post-translational modifications of either p53 or MDM2/MDMX, such as acetylation (Tang et al, 2008), phosphorylation (Banin et al, 1998; Maya et al, 2001; Shieh et al, 1997) and protein–protein interactions (Zhang & Lu, 2009; Zhang et al, 1998), to ultimately activate p53 that protects cells from transformation and neoplasia. Among the modifications, acetylation and ubiquitylation occur at a similar set of lysine residues within p53 and thus are mutually exclusive, that is that acetylation of p53 by p300/CBP prevents its degradation by MDM2 and activates its activity, whereas, MDM2 inhibits p53 acetylation by p300/CBP (Ito et al, 2001; Kobet et al, 2000; Li et al, 2002). Consistently, deacetylation of p53 by an NAD-dependent deacetylase, SIRT1 (Cheng et al, 2003; Luo et al, 2001; Vaziri et al, 2001) or a class I histone deacetylase, HDAC1 (Luo et al, 2000), facilitates MDM2-mediated p53 degradation and inactivates p53. On the other hand, cancers often hijack this feedback regulation to favour their own growth, as human breast cancers, osteosarcomas, lymphomas or leukaemia express high levels of MDM2 or MDMX through distinct mechanisms without p53 mutation (Onel & Cordon-Cardo, 2004). Also, deacetylases are often highly expressed in cancers (Jung-Hynes & Ahmad, 2009; Nosho et al, 2009; Tseng et al, 2009). For instance, SIRT1 is highly expressed in cancers largely due to the downregulation of a gene called hypermethylated-in-cancer-1 (HIC-1; Chen et al, 2005; Tseng et al, 2009; Wales et al, 1995). HIC-1 encodes a transcriptional repressor that inhibits the expression of SIRT1, but is frequently turned off via hypermethylation of its promoter in cancers (Fleuriel et al, 2009; Fukasawa et al, 2006; Hayashi et al, 2001), though it is a p53 target gene as well (Chen et al, 2005; Wales et al, 1995). In theory, this high level of deacetylases would readily maintain p53 in a deacetylated status in cancer cells, consequently favouring MDM2/MDMX-mediated degradation. Hence, this highly cancer-pertinent and well-defined p53–MDM2–MDMX pathway offers multiple molecule targets for screening small molecules as potential therapies for WT p53-harbouring cancers.
Indeed, several small molecules have been identified to target the p53 pathway (Brown et al, 2009). For instance, Nutlin-3, Rita and MI-219 can interfere with the p53–MDM2 binding (Issaeva et al, 2004; Shangary et al, 2008; Vassilev et al, 2004), consequently increasing p53 level and activity. Recently, Tenovins were reported to inhibit the activity of SIRT2 and SIRT1, inducing p53 acetylation and activity (Lain et al, 2008). These exciting studies not only consolidate the p53–MDM2 pathway as a valid target, but also provide multiple candidates for development into anti-cancer drugs, though their clinical significance is still under investigation. Since none of the potent inhibitors of the MDM2–p53 binding, such as Nutlin-3 or MI-219 (Shangary et al, 2008; Vassilev et al, 2004), could effectively affect the MDMX–p53 interaction, we were initially motivated to search for small molecules that could interfere with this interaction, hoping to complement the inhibitory effect of existing MDM2 inhibitors on cancer growth by performing a computational 3D structure-based search followed by a cell-based assessment of top candidates. From this two-step approach, however, we surprisingly uncovered a small molecule that suppresses SIRT1 activity and induces the acetylation, level and activity of p53, consequently and effectively repressing the growth of xenograft tumours derived from human lung and colon WT p53-containing cancer cells.
RESULTS
Identification of Inauhzin (INZ) as a potent activator of p53 with defined functional moieties
Comparison of the structures of the MDM2–p53 and MDMX–p53 complexes (Kussie et al, 1996; Popowicz et al, 2007) revealed that the N-terminal hydrophobic pocket of MDMX for p53 binding is much shallower than that of MDM2. This information explained why MDM2 inhibitors failed to affect MDMX–p53 binding and also prompted us to initiate a computational structure-based screening using the AutoDock computer program (Morris et al, 2008) for the docking of virtual compounds that could distinguish the p53 binding sites on MDM2 and MDMX. From our initial computational screening of half a million of commercially available compounds from the ChemDiv chemical library, we selected and purchased 50 top candidates. These compounds were tested in cell-based assays at 10 µM for their ability to induce p53 levels in human lung carcinoma H460 cells using an immunoblotting (IB) analyses. To our delight, one small molecule, 10-[2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)butanoyl]-10H-phenothiazine (abbreviated as INZ; Fig 1B), induced p53 levels as effectively as actinomycin D (ActD; 10 nM) and in a much more pronounced manner than did the rest of the compounds tested (Fig 1A and data not shown).
Figure 1. Identification of INZ as a novel p53 activator.
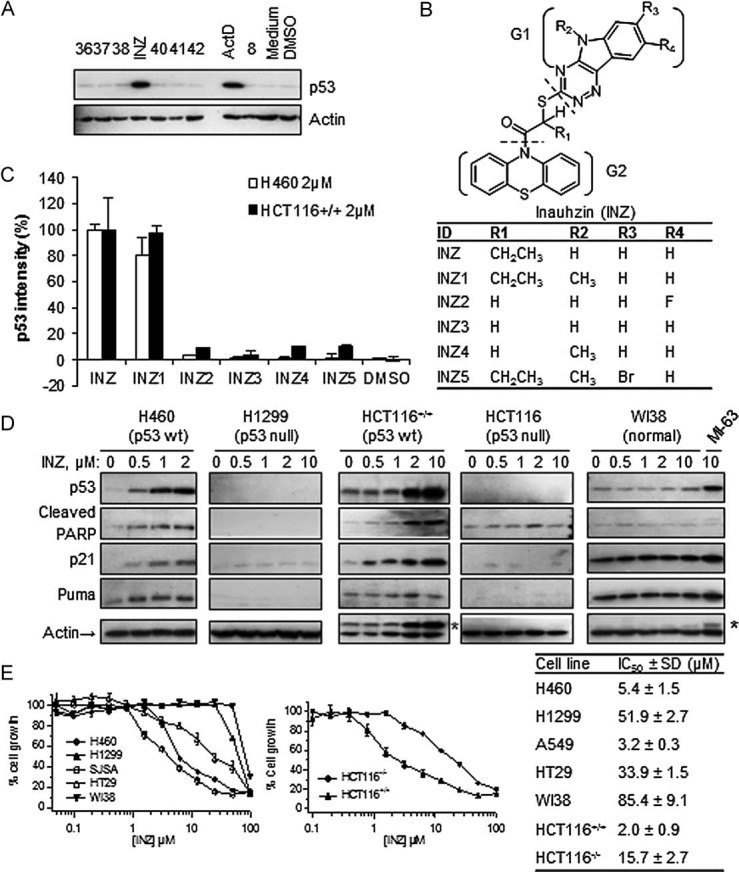
- Screening for compounds that increase p53 levels in cells as detected by IB. H460 cells were harvested for IB after being treated with each of the top 50 compounds (10 µM) from computational-throughput screening for 18 h as shown in a representative blot here (number denotes each compound; INZ). Fifty micrograms of total proteins was used per lane (true for the following figures unless indicated).
- Chemical structures of INZ and its analogues 1–5.
- Cellular activity of INZ analogues was measured using IB that detects p53 levels in H460 and HCT116 cells. The induction levels of p53 were normalized with actin and plotted as percentage of the level of p53 in the cells treated with INZ (mean ± SD, n = 3).
- Dose-dependent activation of p53 pathway by INZ. Cells were treated with INZ or a control compound MI63 for 18 h and harvested for IB with the antibodies as indicated. * Indicates residual bands from p53 antibody.
- Cell growth inhibitory activity was evaluated by WST cell growth assays. IC50 values are represented as mean ± SD (n = 3).
After confirming this effect of INZ in several different p53-containing human cancer cell lines (Fig 1D and Fig S1 of Supporting Information; data not shown), we investigated the relationship between the structure and p53 induction activity of this compound in cells. We were able to obtain 46 commercially available compounds, which are similar to INZ (Fig 1B and data not shown). The analysis of those compounds in p53 activation in H460 and HCT116 cells by IB (Fig 1C and data not shown) indicated that a unique structure scaffold might be required for the activity of INZ in cells. Both the triazino[5,6-b]indol (G1) and phenothiazine (G2) moieties are essential for p53 induction, as the analogues without either of them failed to induce p53 (data not shown). Also, removal of the ethyl group at the R1 position (INZ2-4) or modification at R3 on the indol moiety of INZ (INZ5) disabled the compound to induce p53 in cells (Fig 1B and C). These results indicate that a specific chemical structure with the intact triazino[5,6-b]indol-3-ylthio)butanoyl]-10H-phenothiazine is crucial for p53 activation in cells and suggest that this compound may bind to a specific target in cells. However, to our surprise, INZ or its close analogue INZ1 that induced p53 did not apparently affect the interaction between either MDMX and p53, or MDM2 and MDMX, or MDM2 and p53 in our in vitro fluorescence polarization and cell-based co-immunoprecipitation assays (data not shown).
Inauhzin inhibits cell growth in a p53-dependent fashion
To explore the effect of INZ treatment on the p53 pathway in human cancer cells and determine whether INZ reduces cell viability in a p53-dependent fashion, we first treated human lung cancer H460, A549, H1299, colon cancer HCT116, HT29, osteosarcoma U2OS and SJSA, breast cancer MCF7, ovarian cancer A2780, IGROV1 and SKOV3 and glioma U87 and U373 cells, as well as human embryonic fibroblast WI-38 and normal human fibroblast (NHF) cells with different doses of INZ for 18 h, and harvested them for IB. Indeed, INZ induced p53 level and activity in a dose-dependent fashion and at a dose as low as 0.5 µM as measured by detecting p53 and its targets p21 and Puma as well as cleaved PARP, which indicates apoptosis (Duriez & Shah, 1997), in p53-containing cancer cells including HCT116, H460 (Fig 1D), A549, MCF7, A2780, U87 (Fig S1 of Supporting Information), U2OS and SJSA cells (data not shown). In striking contrast, this effect was not observed in p53-null (H1299, HCT116−/−, SKOV3) or p53-mutated (HT29, IGROV1 and U373) cancer cells (Fig 1D and Fig S1 of Supporting Information). Intriguingly, it was much less potent in activation of p53 in WI-38 and NHF cells (Fig 1D and data not shown) in comparison of MI-63, a previously reported inhibitor of the MDM2–p53 interaction (Ding et al, 2006). In line with these results, INZ displayed much higher toxicity to p53-containing human cancer cells compared with WI-38 and NHF, or p53-mutant and null cancer cells. This is evident by IC50 values that in the former cell lines were 7–40 fold greater than that in the latter cell lines (Fig 1E). Consistently, silencing p53 with siRNA in A549 or H460 cells not only reduced p21, MDM2, PARP and Puma up-regulation induced by INZ, but also compromised the growth inhibition by INZ treatment (Fig S2A and B of Supporting Information and data not shown). In comparison, knockdown of p73 in H460 cells did not apparently affect p53 level, acetylation and p53-dependent cell growth suppression induced by INZ, although partially impaired the induction of p21 and MDM2 by INZ (Fig S2C–E of Supporting Information), suggesting that Inauhin can still activate p53 in p73-knocked down cells. Therefore, the ability of INZ to activate the p53 pathway is strongly correlated with its inhibition of cell survival. Taken together, these results demonstrate that INZ is a potent p53 activator and mediates p53-dependent cytotoxicity through its specific functional groups.
Inauhzin induces p53-dependent apoptosis
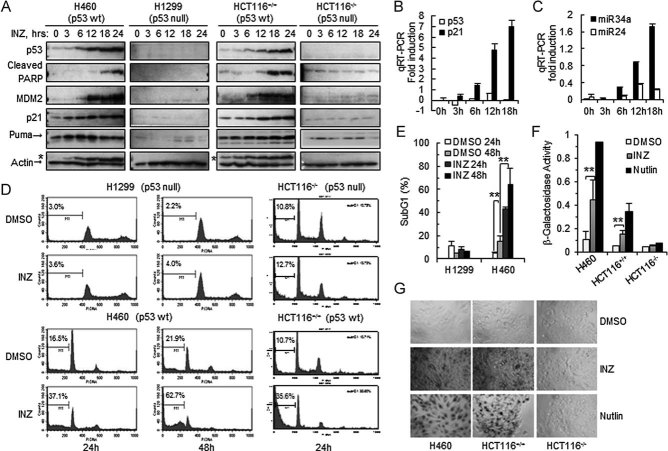
To further characterize the effect of INZ on p53 cellular functions, we performed a set of time course experiments using the same aforementioned approaches with 2 µM INZ, as this concentration was sufficient to significantly induce p53 level and activity (Fig 1D). We found that INZ induced p53 level in a time-dependent manner as early as 6 h post-treatment in both p53-containing H460 and HCT116 cells (Fig 2A). Correspondingly, three of p53 targets, MDM2, p21 and Puma, were also induced in a time-dependent manner in p53-containing H460 and HCT116, but not in p53-null H1299 and HCT116, cells (Fig 2A). Interestingly, Puma was induced earlier (3–6 h) and cleaved PARP was detected later on (∼12 h; Fig 2A). Apparently, cleaved PARP was p53-dependent as it was not detected in p53-null cells (Fig 2A). The induction of p53 targets was clearly at the transcriptional level as p21 mRNA and miR34a, but not p53 mRNA and miR24, were highly induced in H460 cells (Fig 2B and C).
Figure 2. INZ induces p53 level and activity as well as p53-dependent apoptosis.
- A. Cells were treated with 2 µM INZ for the indicated time and harvested for IB. * Indicates residual signals of p53.
- B-C. H460 cells were treated with 2 µM INZ and harvested for real-time PCR. Values represent mean ± SD (n = 3).
- D-E. Induction of apoptosis by 2 µM INZ analysed by FACS. The apoptotic cells, identified by sub-G1 DNA content, were presented in the M1 population. Quantification of apoptosis of H1299 and H460 cells was shown in (E). The results shown are representative of three-independent experiments. Values represent mean ± SD (n = 3), **p < 0.01.
- F-G. INZ induces p53-dependent senescence. Senescence-associated β-galactosidase staining was performed in cultured cells for 6 days in the presence of 2 µM INZ or 10 µM Nutlin-3. β-galactosidase activity was measured by the absorbance of 5,5′-dibromo-4,4′-dichloro-indigo at 650 nm generated by the β-galactosidase staining. Values represent mean ± SD (n = 3), **p < 0.01. Representative photomicrographs of the cells by β-galactosidase staining were shown in (G).
To investigate how INZ preferentially antagonize the proliferation of p53-containing cells, we examined whether the compound induces apoptosis in p53 WT, but not p53 null, cells. As shown in the Fig 2D and E, INZ at 2 µM induced drastic apoptosis in a time-dependent fashion in both p53-containing H460 and HCT116 cells by 30–40% increase of apoptotic cells, but not in p53-deficient H1299 and HCT116 cells. These results, in perfect line with the above results (Figs 1 and 2A and B), demonstrate that INZ not only induces p53 level, but also stimulates its transcriptional activity, consequently leading to p53-dependent apoptosis in a time-dependent fashion. Also, we determined if this compound promotes p53-dependent senescence as measured by the production of senescence-associated β-galactosidase. Notably, INZ induced senescence in H460 or HCT116p53+/+, but not in HCT116p53−/−, cells, though to a much less degree than did Nutlin-3 (Fig 2F and G; Efeyan et al, 2007). These results demonstrate that INZ inhibits cell proliferation by triggering both apoptosis and senescence in p53-containning cells, though it predominantly induces p53-dependent apoptosis.
Inauhzin stabilizes p53 without either directly inhibiting MDM2-mediated ubiquitylation or causing genotoxicity
The fact that INZ induces the protein, but not mRNA, level of p53 (Figs 1 and 2) suggests that this compound might regulate the stability of the p53 protein. To test this hypothesis, we assessed the half-life of endogenous p53 after the treatment of H460 or HCT116 cells with this compound. As shown in Fig 3A and B, 2 µM INZ extended the half-life of p53 from 30 min to more than 3 h (Fig 3A and B). This effect was specific to p53, as the half-life of p21 was not apparently influenced (Fig 3A) even though its level was remarkably elevated due to the activation of p53 (Figs 1D and 2A). Next, we determined if INZ stabilizes p53 by inhibiting its ubiquitylation in cells. Indeed, the ubiquitylation of both endogenous (Fig 3C) and exogenous (Fig 3D) p53 proteins was markedly inhibited by 2 µM INZ. However, the auto-ubiquitylation of MDM2 was not significantly affected by the treatment of INZ (Fig S3A of Supporting Information). Moreover, INZ did not appear to directly affect MDM2-mediated p53 ubiquitylation when it was titrated from 2 to 50 µM in an in vitro ubiquitylation assay using purified proteins (Fig S3B of Supporting Information). Taken together, these results demonstrate that INZ is able to prevent p53 from MDM2-mediated ubiquitylation and proteasomal degradation and also suggest that it may utilize a cellular mechanism that protects p53 without either directly inhibiting MDM2 activity towards p53 or interfering with MDMX/MDM2–p53 interaction (data not shown).
Figure 3. INZ stabilizes p53 and Inhibits its ubiquitylation.
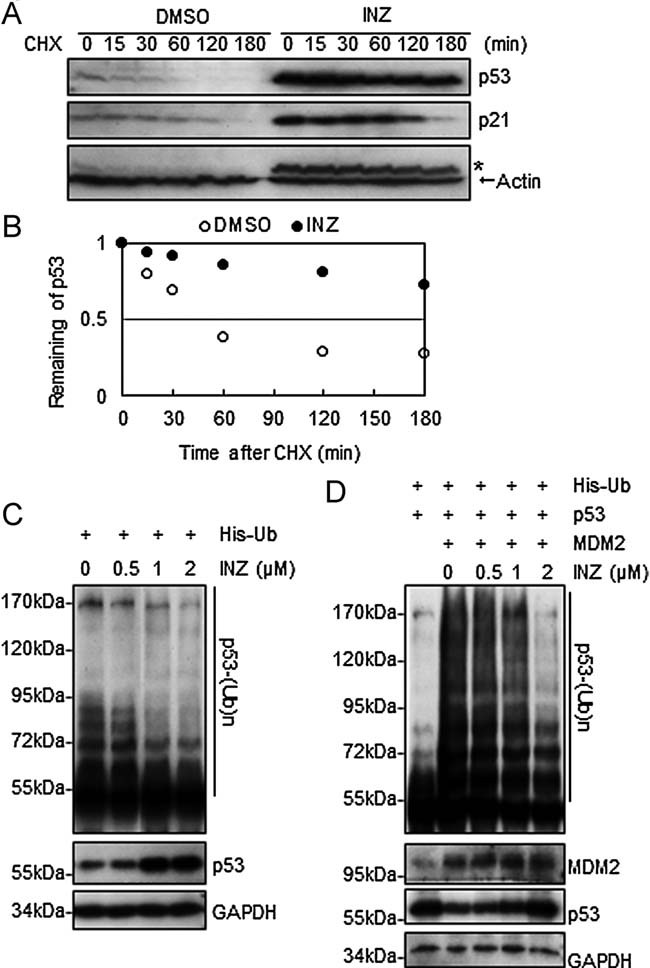
- A-B. H460 cells were treated with 2 µM INZ for 18 h followed by addition of 50 µg/ml cycloheximide (CHX) and harvested at indicated time points for IB. * Indicates residual signals of p53. The intensity of each band was quantified, and normalized with actin and plotted in (B).
- C. H460 cells transfected with His–Ub were treated with INZ for 18 h prior to addition of 10 µM MG132 and 20 µM ALLN for 8 h. Cell lysates were subjected to His pull-down by Nickel-NTA agarose and detected by IB with the anti-p53 (DO-1) antibody.
- D. HCT116−/− cells transfected with His–Ub, p53 and HA-MDM2 were treated with INZ for 4 h, followed by treatment with 10 µM MG132 for 8 h. Ubiquitylated p53 were purified by Nickel-NTA and detected by IB with the anti-p53 (DO-1) antibody. The expression levels of p53 and HA-MDM2 are shown in the lower panels. Also see Fig S3 of Supporting Information.
To elucidate possible cellular mechanisms underlying the protection of p53 by INZ from proteolysis in cells, we tested if this compound causes general genotoxicity to cells by conducting in vitro non-sequence-specific DNA-binding, in vivo immunofluorescence staining for H2AX Ser139 phosphorylation (γH2AX) and cellular p53 phosphorylation assays. We found that INZ is not genotoxic. First, it was a considerably poor DNA-binding agent in comparison with ActD, as the former hardly bound to DNA at 2 µM (Fig 4A), a dose that markedly induced p53 (Figs 1 and 2), while the latter bound to 50% of DNA molecules even at 0.3 µM (Fig 4A). Also, even though 2 µM INZ effectively induced p53 levels in cells compared to 10 µM Cisplatin (Cis), it did not appear to cause significant γH2AX focus formation, which is often used as a marker for DNA damage (Paull et al, 2000). As shown in Fig 4B and C and Fig S4 of Supporting Information, more than 80% of H460 cells treated with 10 µM Cis or 2 mM hydroxyurea (HU) were detected with more than 10 foci per nucleus, whereas, only less than 1.5% of H460 cells treated with 2 µM INZ contained such a high level of foci and ∼75% of INZ-treated cells were basically free of foci. Furthermore, the level of p53 phosphorylation at either serine 15 or serine 46 in INZ-treated H460 cells was not observed compared to the cells treated with Cis or Etoposide for 18 h (Fig 4D and E). Phosphorylation of p53 at serine 15 or serine 46 was previously shown to be responsive to severe DNA damage (Banin et al, 1998; Oda et al, 2000; Shieh et al, 1997). Finally, INZ did not activate AMPK (Fig 4F), which was also reported to activate p53 by phosphorylating serines 15 and 46 (Jones et al, 2005). All together, these results exclude the possibility that INZ might activate a kinase cascade that mediates p53 phosphorylation by causing DNA damage or activating AMPK.
Figure 4. INZ does not directly bind to DNA and does not cause significant DNA damage.
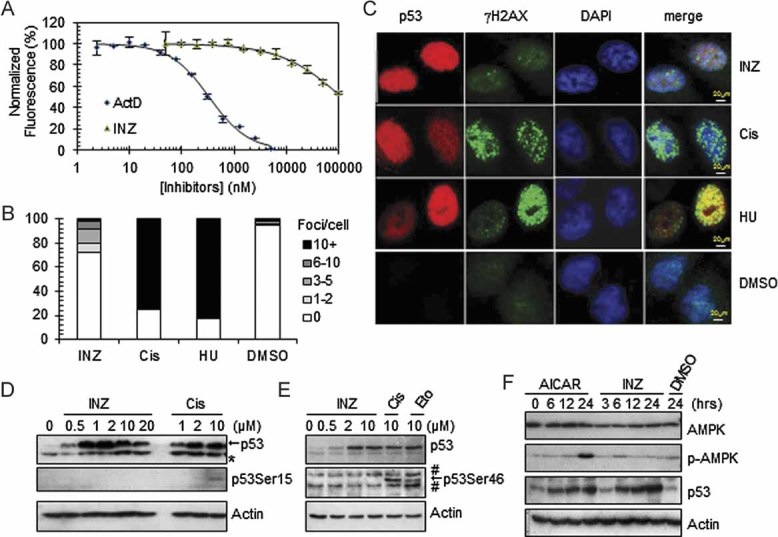
- A. Titration of INZ and ActD against DNA utilizing ethidium bromide as a fluorescence intercalator. Error bars represent standard deviation (n = 3).
- B-C. Immunofluorescent γH2AX foci in H460 cells treated with 2 µM INZ or 10 µM Cis for 18 h or 2 mM HU for 8 h. The percentage of nuclei with the indicated number of γH2AX foci was quantitated in (B). The number of γH2AX foci per nuclei in INZ-treated cells (1.3 ± 0.2 foci per cell, n = 394) is significantly less than that in Cis-treated cells (8.0 ± 1.2 foci per cell, n = 375; p = 0.0006). HU-treated cells (9.7 ± 0.6 foci per cell, n = 314); DMSO-treated cells (0.28 ± 0.1 foci per cell, n = 395). p-Values were calculated using two-tailed t-test. The quantification expressed as the mean number of foci per cell ± SD is shown in Fig S4 of Supporting Information. Bar, 20 µm.
- D-E. H460 cells were treated with INZ, Cis and Etoposide for 18 h. Cell lysates were immunoblotted for phosphorylated p53 at Ser15 (D) and Ser46 (E). Blots were exposed for longer than 15 min.
- F. H460 cells were induced with 1mM AICAR (AMP analogue, an AMPK activator) and 2 µM INZ for indicated times and subjected to IB with antibodies as indicated.
Inauhzin inhibits SIRT1 activity and induces acetylation of p53, but not tubulin
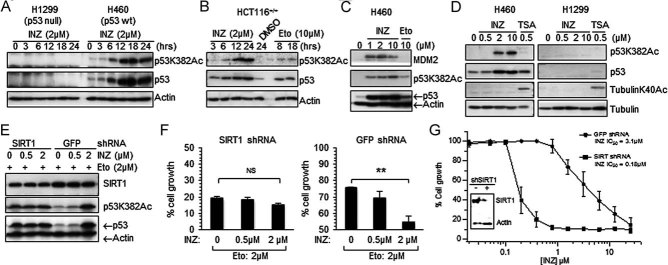
Previous studies have demonstrated that p53 is also modulated by reversible acetylation, which is inverse to ubiquitylation (Li et al, 2002) because the two post-translational modifications occur at similar lysine residues within p53. Hence, we tested whether INZ would influence p53 acetylation in cells. Indeed, at 2 µM it induced p53 acetylation at lysine 382 as detected by anti-acetylated K382 antibodies, which correlated well with the increment of p53 levels (Fig 5A) and more markedly than did Etoposide at 10 µM (Fig 5B and C). Interestingly, INZ induced acetylation of p53 in H460 cells, but not tubulin in constrast with trichostatin A (TSA), which induced acetylation of tubulin (Fig 5D) by inhibiting the activity of the HDAC family, such as HDAC1 and HDAC2 (Finnin et al, 1999).
Figure 5. INZ induces acetylation of p53, but not tubulin, in cells, which is affected by knockdown of SIRT1.
- A-D. Cells were treated with INZ, Etoposide or TSA as indicated. Total levels of p53 and acetylated p53 at lysine 382 were assessed by IB (70 µg of total proteins was used per lane; true to all panels in this figure).
- E. H460 cells were plated in 6-well plates 18 h prior to infection with SIRT1 shRNA or control GFP shRNA. To increase shRNA knockdown efficiency, cells were infected again 24 h later. At 24 h after second infection, cells were treated with Etoposide for 6 h followed by addition of INZ for 12 h.
- F-G. Cells infected with shGFP or shSIRT1 adenovirus in (E) were seeded at 3000 cells per well in 96-well culture plates and incubated overnight at 37C. Various concentrations of INZ (G) or INZ combined with 2 µM Etopside (F) were added into the plates. Cell growth inhibition was measured using WST cell growth assays. IC50 values are represented as mean ± standard deviation (n = 3). ** Indicates p < 0.01.
Because K382 is a target site for SIRT1 (Luo et al, 2001; Vaziri et al, 2001), we wondered whether knockdown of SIRT1 might affect INZ-induced p53 acetylation at K382. As shown in Fig 5E, knockdown of SIRT1 in H460 cells induced p53 acetylation and protein level in the presence of 2 µM Etoposide. However, additional treatment of the cells with 2 µM INZ failed to further induce p53 acetylation and level compared to the cells without SIRT1 knockdown. Consistently, knockdown of SIRT1 also impaired the ability of INZ to synergize the inhibition of cell growth by Etoposide (Fig 5F). By contrast, in the absence of Etoposide, INZ synergized the negative effect of SIRT1 knockdown on cell growth, as the IC50 value for INZ in cell growth analysis decreased by ∼17 fold when SIRT1 was partially depleted via SIRT1 shRNA (Fig 5G). Similar to INZ treatment (Figs 1 and 2 and 5A and B), knockdown of SIRT1 using SIRT1 specific siRNA induced the level and activity of p53 as well as p53 acetylation, leading to p53-dependent apoptosis and cell growth suppression in HCT116+/+ and H460 cells (Fig S5 of Supporting Information). These results indicate that INZ might induce p53 acetylation and suppress cell growth by inhibiting SIRT1 activity in cells.
To validate this possibility, we measured the effect of INZ on SIRT1 activity by conducing in vitro assays using acetylated p53 protein as a substrate and purified His–SIRT1 (Fig S6A and B of Supporting Information) as described in the Experimental Procedures Section of the Supporting Information. As shown in Fig 6A, INZ inhibited SIRT1 deacetylase activity in a dose-dependent fashion and effectively inhibited this activity at 3 µM. This inhibition was specific to INZ and its chemical analogue INZ1 (methyl substituted R1), which activated p53 (Fig 1B and C) and decreased SIRT1 activity in a dose-dependent fashion (Fig S7A of Supporting Information). By contrast, the analogues INZ5 (bromide substituted on R3) and INZ 15 or INZ 18 (lack of G1, data not shown) that failed to induce p53 did not significantly inhibit SIRT1 activity even at the highest concentration we tested (20 µM).
Figure 6. INZ inhibits SIRT1 activity and directly binds to SIRT1 in vitro.
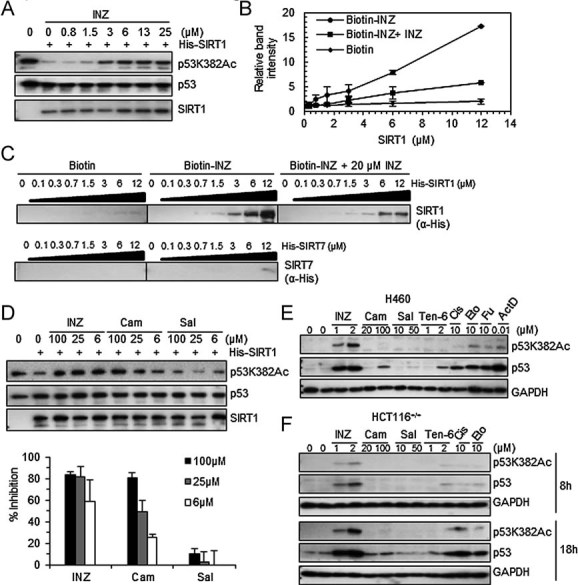
- A. INZ inhibits deacetylation of p53 at lysine 382 by SIRT1 in vitro in a dose-dependent fashion using acetylated p53 protein as a substrate as described in the Experimental Procedures Section of the Supporting Information.
- B-C. Purified SIRT1 was incubated at indicated concentrations with biotinylated INZ that was conjugated with avidin beads in the presence or the absence of 20 µM of non-biotinylated INZ. Purified SIRT7 was used as a negative control. The intensity of each band for bound SIRT1 as analysed using IB was quantified (B), and each sample was individually compared with the intensity of the samples without SIRT1. The results shown are representative of three-independent experiments. Values represent mean ± SD (n = 3).
- D. The inhibitory effects of INZ, Cambinol and Salermide on SIRT1 activity were measured by the increase of the levels of acetylated p53 at lysine 382 in vitro. The percentage of inhibition was calculated as described in the ‘Experimental Procedures’ Section of Supporting Information and shown in below. Values represent mean ± SD (n = 3).
- E-F. Effect of INZ, Cambinol, Salermide or Tenovin-6 on p53 acetylation and level in H460 (E) and HCT116+/+ (F) cells.
To obtain the evidence supporting the inhibition of SIRT1 by INZ through their direct interaction, we conjugated biotin to the R1 position of INZ since the analysis of the structure–activity relationships of INZ revealed that R1 could be substituted with a different chemical group. To our delight, this biotinylated INZ was as effective as INZ in the induction of p53 acetylation and level in both H460 and HCT116 cells (Fig S6C and D of Supporting Information) and in inhibition of SIRT1 activity in vitro using acetylated p53 protein as a substrate (data not shown). Using this newly synthesized biotin–INZ, we determined if SIRT1 could bind to INZ in vitro by performing a set of biotin–avidin pull down assays using SIRT7 (Fig S6A of Supporting Information) as a control. SIRT7 was recently reported to deacetylate p53 as well (Lavu et al, 2008). As shown in Fig 6C, SIRT1, but not SIRT7, specifically bound to biotin–INZ in a dose-dependent manner. This binding was markedly reduced by 20 µM INZ, further validating the specificity of the INZ–SIRT1 binding (Fig 6B and C). Additionally, biotin–INZ formed complexes with SIRT1 in vitro as detected by native polyacrylamide gel electrophoresis (PAGE) analyses (Fig S6E of Supporting Information). These observations suggest that INZ inhibits SIRT1 activity by directly binding to this deacetylase.
In consistence with the results showing that INZ induces acetylation of p53K382 (Figs 5 and 6) and Histone H3K9 (Fig S7C of Supporting Information), but not tubulin K40 (Fig 5D), INZ selectively inhibited the activity of SIRT1, but not SIRT2, SIRT3 or HDAC8, with the IC50 of 0.7–2 µM using Fluor-de-Lys fluorimetric assays (Fig S7B of Supporting Information). As K382 of p53 and K9 of Histone H3 have been indicated the acetylated target sites for HDAC1 (DiTacchio et al, 2011; Luo et al, 2000), we tested the inhibitory effect of INZ on HDAC1. Flag-HDAC1 was prepared from H1299 cells transfected with Flag-HDAC1 by immunoaffinity purification and the deacetylase assay was performed similarly using acetylated p53 protein as a substrate. In contrast to the complete rescue of p53 acetylation by 1 µM of TSA, a drug known selectively inhibits HDAC, but not the Sirtuins, INZ had little effect at 5 µM, but mild effect at 25 µM, on HDAC1 activity (Fig S7D of Supporting Information). These results, together with the results of Figs 5 and 6, indicate that INZ appears to be more specific to SIRT1 than to other members of the HDAC family.
To further delineate biochemical mechanisms underlying the inhibition of SIRT1 activity by INZ, we conducted a set of competition assays with limited titration of the compound, acetylated p53 peptide substrates and NAD+, a co-factor of SIRT1 (Tanny et al, 1999). As shown in Fig S7E of Supporting Information, INZ did not compete with the substrate, as both of Vmax and Km values were not changed significantly by increasing amounts of INZ. However, in the case of NAD+, INZ reduced both Vmax and Km values in a dose-dependent manner (Fig S7F of Supporting Information), suggesting that this compound might utilize an uncompetitive mechanism to inhibit SIRT1 activity. All together, these results demonstrate that INZ is able to inhibit SIRT1 activity in vitro and in cells, consequently leading to p53 aceylation and activation, and also suggest that it might employ an uncompetitive mechanism influencing the binding of NAD+ to SIRT1, which requires further examination using biophysical and biochemical approaches in the near future.
Inauhzin is more effective than cambinol or salermide in inhibition of SIRT1 activity and activation of p53
We then compared INZ with some published SIRT1 inhibitors (Heltweg et al, 2006; Lain et al, 2008; Lara et al, 2009) by performing in vitro p53-deacetylation and cellulous p53-activation assays. INZ was shown to be more effective in inhibiting SIRT1 activity compared to cambinol or Salermide in in vitro assays using acetylated p53 proteins as a substrate, as neither of the latters could inhibit SIRT1 activity at 6 µM, at which INZ markedly recovered p53 acetylation (Fig 6D). Consistently, INZ was also more effective than these two compounds in the induction of p53 acetylation and level in H460 (Fig 6E and Fig S8A of Supporting Information) and HCT116 cells (Fig 6F and Fig S8B of Supporting Information) as well as in the inhibition of cancer cell growth (Fig S8C of Supporting Information). These results demonstrate that INZ is a better bioactive inhibitor of SIRT1 and a more effective activator of p53 than these two known SIRT1 inhibitors. Even though Tenovin-6 was able to induce p53, it was less effective than INZ in the induction of p53 acetylation and level in the colon and lung cancer cells as tested here (Fig 6E and F and Fig S8A and B of Supporting Information). Also, Tenovin-6 was much more toxic than INZ to human primary embryonic fibroblasts (WI-38) and NHF-1 (Fig S8C of Supporting Information). Together, these results indicate that INZ is tumour selective and more effective in inhibiting SIRT1 activity and activating p53.
Inauhzin suppresses growth of human xenograft tumours harbouring p53
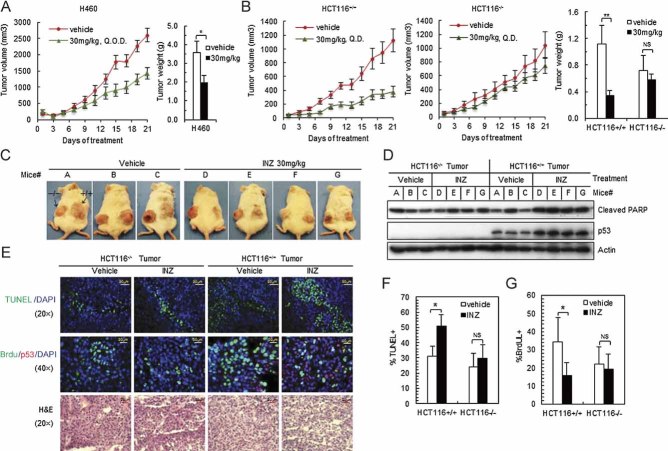
To investigate the biological significance of the activation of p53 by INZ, we carried out a set of animal experiments to evaluate the effect of this compound on the growth of human xenograft tumours. First, we tested if INZ would affect the growth of xenograft tumours derived from H460 cells in severe combined immunodeficiency (SCID) mice since this compound markedly induced p53 level and activity at 2 µM as well as p53-dependent apoptosis in this cell line (Figs 1 and 2). Once H460 xenograft tumours grew into the size of 100 mm2, we started to administer the mice with 30 mg/kg INZ via intraperitoneal (i.p.) injection once every other day (Q.O.D.) for 3 weeks. As shown in Fig 7A, the tumours grew significantly more slowly in the INZ treated animals than in those animals treated with respective vehicle (5% dimethyl sulphoxide; DMSO; p < 0.05). INZ significantly reduced the average tumour weight at the end of the experiment by nearly 40% (p < 0.05, Fig 7A and Fig S9C of Supporting Information). Through this experimental period, both groups of animals had been healthy except bearing tumours and without apparent changes in their behaviour, food appetite and body weight. At the end of the experiment, 1, 2, 4, 8 and 10 h after the last dose, we obtained sera and harvested tumours at 4 and 8 h. Quantitative high-performance liquid chromatography (HPLC)-MS/MS (API 4000, Applied Biosystems) analysis of the sera revealed that the average level of INZ peaked at 1.5 µg/ml (equivalent to 3 µM concentration) 1 h after the i.p. administration (Fig S9A of Supporting Information). The result was consistent with our pharmacokinetics results (data not shown). INZ levels in the tumors reached to 653.8 ng/g at 4 h and then decreased by 8 h (Fig S9B of Supporting Information), indicating INZ was able to penetrate tumours and persist within tumour tissues after i.p. administration. Moreover, INZ-treated H460 tumours displayed elevated p53 compared to the vehicle-treated tumours by IB (data not shown). These results suggest that INZ has good tumour tissue penetration and is able to inhibit tumour growth by inducing p53.
Figure 7. INZ induces p53 and p53-dependent apoptosis in vivo and suppresses the growth of human xenograft tumours.
- A. Mice bearing H460 xenografts were treated with INZ or 5% of DMSO (vehicle) by i.p. at the dose as indicated at every other day (Q.O.D.) for 21 days. Mean tumour volumes ± SEM are shown in curves (left), and tumour weight is shown in columns (right; n = 5 mice per group; *p < 0.05).
- B-C. Mice with HCT116p53−/− and HCT116p53+/+ tumours were treated with INZ or 5% of DMSO at doses as indicated by i.p. once every day (Q.D.) for 21 days. Mean tumour volumes ± SEM are shown in curves (left), and tumour weight is shown in columns (right; n = 7 mice per group; *p < 0.05 and **p < 0.01). Photos for representative mice bearing HCT116p53−/− and HCT116p53+/+ tumours were shown in (C).
- D. IB for proteins extracted from HCT116p53−/− and HCT116p53+/+ tumour samples in (C).
- E. INZ induces apoptosis and inhibits proliferation in xenograft tumours. Representative H&E, BrdU and TUNEL-stained xenograft tumour sections from INZ or vehicle treated mice as presented in (B–D). Bars for 50 µm (TUNEL) and 20 µm (BrdU), respectively.
- F-G. Quantification of BrdU (G) and TUNEL (F) stainings (n = 4 mice per group analysed). Mean values ± SEM are indicated, *p < 0.05. p-Values were calculated using two-tailed t-test.
To further assess the tumour suppression activity of INZ and determine if the p53 pathway contributes to this tumour suppression function of this compound in vivo, we implanted p53-containing and p53-null HCT116 cells into the same SCID mouse (one at each side of its back) to generate p53+/+ and p53−/− tumours, as shown in the representative Fig 7C, to minimize possible variations between the two cell lines caused by individual animals. Also, we modified the strategy of drug administration since the animals in H460 xenograft experiments did not show any apparent abnormality. Once palpable tumours were detected, pairs of mice were randomized to receive either 30mg/kg (n = 7) once per day (Q.D.) or vehicle (5% DMSO). As a result, INZ was more effective in retarding the tumour growth in HCT116+/+ tumours, as it more significantly reduced tumour growth and weight by ∼70% at the end of the experiment (Fig 7B). Furthermore, this inhibition was p53-dependent, as INZ had moderate effect on the growth of HCT116−/− tumours (Fig 7B and Fig S9C of Supporting Information). INZ-treated HCT116+/+ tumours were significantly smaller than their respective controls of vehicle treatment (p < 0.01), whereas, there were marginal differences between INZ and vehicle treatments in p53-null HCT116 xenografts (p > 0.1). Correspondingly, p53 level and activity as indicated with induction of cleaved PARP were highly induced in INZ-treated p53-harbouring, but not in p53-null, HCT116 tumours (Fig 7D). This induction in the p53-harbouring tumours was also well correlated with a significant increase in apoptosis within the tumour as measured by TdT-mediated dUTP nick end labelling (TUNEL) assays and a marked decrease in proliferation as measured by 5-bromodeoxyuridine (BrdU) staining (Fig 7E–G). However, there was no significant difference observed in apoptosis and proliferation between INZ and vehicle treatments in p53-null HCT116 xenografts. Remarkably, INZ also potently induced apoptosis specifically in tumour cells (Fig 7E and F) without measurable cell death in the high proliferative of normal tissues (Small intestine, spleen and stomach; Fig S9D of Supporting Information). Collectively, these results demonstrate that INZ effectively induces apoptosis and suppresses tumour growth in p53-harbouring tumours.
DISCUSSION
Our study as presented here identifies INZ as a novel small molecule that possesses an ability to induce p53 level and activity, consequently leading to p53-dependent apoptosis. As a result, this compound inhibits the growth of xenograft tumours from p53-containing lung and colon cancer cell lines, but exhibits minimum effect on tumours from p53-null HCT116 cells. By using a rationale-based strategy and a reverse target-identification approach (identifying the target(s) of a compound after unveiling its cellular phenotype or biological activity), we excavated a likely mechanism that attributes to the activation of p53 by this compound, that is inhibition of SIRT1 activity (Fig 8). Our initial biochemical analyses indicate that INZ does not appear to compete with a substrate for the active site of SIRT1, but might affect the binding of NAD+ to SIRT1 via an uncompetitive mechanism, though this mode of action needs to be further investigated.
Figure 8. A model for INZ action in cancer cells.
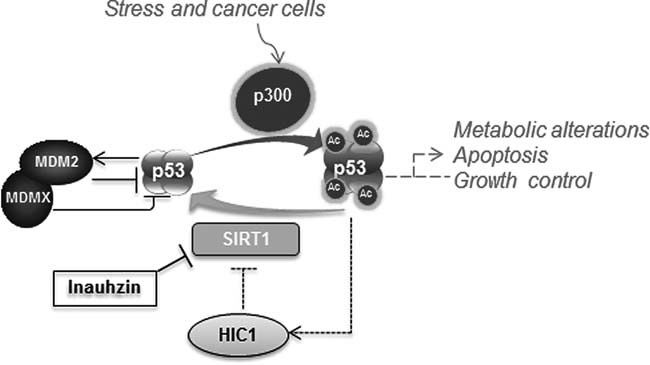
Upon stress, p53 is significantly acetylated by p300 and thus prevented from ubiquitylation and degradation mediated by MDM2/MDMX. SIRT1 deacetylates p53, not only inhibiting its activity, but also rendering p53 into an ideal substrate for MDM2/MDMX-mediated ubiquitylation and degradation. INZ can induce p53 acetylation and level, hence reactivating p53 by inhibiting SIRT1 deacetylase activity, as SIRT1 is often highly expressed in cancers or cancer cells due to the lack of expression of its repressor HIC-1 via promoter hypermethylation as indicated in dotted lines; Otherwise, in normal cells where its promoter is not hypermethylated, HIC-1 can be induced by p53 to repress SIRT1 expression at the mRNA level in response to stress.
It is intriguing that INZ does not effectively induce p53 level and activity in human embryonic fibroblast WI-38 cells and human fibroblast NHF cells (Fig 1D and data not shown). Likewise, it is also much less toxic to these normal cells even though they contain WT p53 (Fig 1E and Fig S8 of Supporting Information). This is distinct from MDM2 inhibitors, such as Nultin or MI-63, both of which can activate p53 in normal fibroblast cells (Shangary et al, 2008). Although the SIRT inhibitor, Tenovin-1, was also reported to resist in the normal human dermal fibroblasts (Lain et al, 2008), the comparison of its analogue Tenovin-6 with INZ revealed that the latter is much less toxic than the former to primary human fibroblast cells (Fig S8C of Supporting Information). Although it remains to be investigated why INZ does not activate p53 in normal fibroblast cells, it is possible that in the absence of stress, SIRT1 might not be needed to inactivate p53 as p53 is not acetylated by p300/CBP in normally growing cells. Consistent with this assumption, it was recently shown that SIRT1 has no significant role in the growth of murine embryonic stem (ES) cells under normal conditions, but SIRT1 could control mitochondrial localization of p53 by deacetylating it in response to oxidative stress in ES cells (Han et al, 2008). Another possibility is that SIRT1 might be more active in some cancer cells than in normal cells. Indeed, SIRT1 is highly expressed in several human cancers including lung, colon and prostate cancers due to the inactivation of its repressor HIC-1 (Fleuriel et al, 2009; Fukasawa et al, 2006; Nakae et al, 2006; Tseng et al, 2009). HIC-1 suppresses the expression of SIRT1 at the transcriptional level in response to p53 activation (Chen et al, 2005; Wales et al, 1995), but is often turned off in cancers due to promoter hyper-methylation (Fleuriel et al, 2009; Fukasawa et al, 2006; Hayashi et al, 2001; Fig 8). Thus knockdown of SIRT1 by siRNA or inhibition of SIRT1 activity by INZ conveys a more significant effect on p53 activation in cancer cells than in normal cells (Figs S5 and S8 of Supporting Information). Because of this, INZ is much less toxic to normal cells, and this feature would be conducive to clinical therapy, as it would minimize its side effect on cancer-bearing patients.
Over the past decade or so, several small molecule inhibitors of the Sirtuin family have been reported (Alcain & Villalba, 2009). Some of them were quite potent in test tubes (Trapp et al, 2007) and effective in yeast or plants (Grozinger et al, 2001). However, none of them has been shown to be bioactive in suppression of tumour growth in animal models until recently when Tenovins were identified to inhibit SIRT2 as well as SIRT1 with a relatively weak activity (Lain et al, 2008). Tenovins induce p53 acetylation and activity resulting in suppression of xenograft melonoma and Burkitt's lymphoma growth in vivo at 50–90 mg/kg via IP. By contrast, INZ is a specific and more potent inhibitor of SIRT1 (Figs 5 and 6 and Figs S7 and S8 of Supporting Information) and also considerably effective in exerting p53-dependent suppression of xenograft lung and colon cancer growth in vivo at 30 mg/kg via i.p. (Fig 7). More importantly, INZ is more selective between cancer cells and normal cells compared to Tenovin-6 (Fig S8C of Supporting Information). This discrepancy might be due to the following possibilities: (1) Tenvoins affect multiple members of the Sirtuin family; (2) INZ and Tenovins perhaps inhibit SIRT1 through different mechanisms or bind to different forms of SIRT1 in different cellular locations (Byles et al, 2010; Lynch et al, 2010; Nasrin et al, 2009), and for example, INZ might bind to phosphorylated SIRT1 which is more active in cancer cells (data not shown); (3) Tenovin and INZ might be transported through cellular membranes by distinct transporters, whose expression levels could be different between normal and cancer cells. Some of the possibilities will be worth to be further examined. Also, INZ was more effective than two other known SIRT1 inhibitors, Cambinol or Salermide, in inhibition of SIRT1 activity in vitro and activation of p53 in cells (Fig 6D–F and Fig S8 of Supporting Information). Although another SIRT1-specific inhibitor, EX527, which effectively inhibited SIRT1-mediated p53 acetylation in vitro, had little influence on p53 acetylation and level in MCF7 cells (Peck et al, 2010), it did affect the SIRT1–p53 pathway in rodent tissues (Velasquez et al, 2011). These seemingly contradictory results suggest that EX527 might not be permeable to certain cancer cell lines, such as MCF7 cells. By contrast, INZ was able to activate p53 in all of the p53-containing cancer cells we tested, including MCF7 cells (Figs 1 and 2 and Fig S1 of Supporting Information). Therefore, our initial comparison of INZ with the existing small molecule SIRT1 inhibitors indicates that INZ distinguishes itself with following features: (1) more effective in inhibiting SIRT1 activity in vitro; (2) more potent in p53 activation in cells; (3) less toxic to normal cells and tissues; (4) more bioactive and bioavailable to all of the cancer cell lines tested. Based on these special characters, INZ appears to be a good candidate for further developing into an anti-cancer drug.
Although there might be the existence of other potential protein targets for INZ, our results clearly show that this compound at lower doses specifically triggers p53-dependent apoptosis and suppression of cell proliferation in both cultured cells and xenograft tumours. Another p53 family member, p73, was previously shown to be a target for SIRT1 (Dai et al, 2007). Indeed, knockdown of p73 partially impaired the induction of p21 and MDM2 levels by INZ (Fig S2C of Supporting Information), which could partially explain why p21 induction by this compound occurred earlier than p53 induction in Fig 2A. However, depletion of p73 by siRNA did not apparently affect the induction of the level, acetylation and apoptotic activity of p53 by INZ (Fig S2C–E of Supporting Information), indicating that this compound indeed suppresses cell growth by mainly activating p53 and inducing p53-dependent apoptosis (Figs 1–8), which is in line with the previous reports showing the close link of p53 acetylation with p53-dependent apoptosis (Tang et al, 2006). Therefore, our study uncovers INZ, which is structurally distinct from any of the published Sirtuin inhibitors, as the first SIRT1 inhibitor that can induce p53 acetylation, level and activity without causing genotoxicity and disrupting MDM2/MDMX–p53 interaction in cancer cells, leading to p53-dependent apoptosis and suppression of tumour growth (Fig 8). This unexpected finding not only validates the negative regulation of p53 by SIRT1 in cancer cells, but also divulges a new class of target-specific small molecules as another highly promising contender for future therapy of p53-bearing human lung, colon and prostate cancers that highly express SIRT1 (Jung-Hynes & Ahmad, 2009; Nosho et al, 2009), although it has been debating if SIRT1 plays a role in cancer development and whether SIRT1 would be an appropriate target for cancer therapy (Bosch-Presegue & Vaquero, 2011; Fang & Nicholl, 2011; Herranz & Serrano, 2010; van Leeuwen & Lain, 2009). Since SIRT1 is also involved in aging and metabolic disorders (Guarente, 2000), identification of INZ as an inhibitor of SIRT1 would offer a useful tool for studying molecular events or mechanisms underlying these diseases and a potential candidate for their therapeutic development. Finally, it would be important and interesting to explore if INZ could synergize the anti-cancer effect of the known small molecules that target the p53 pathway or of the existing chemotherapy or radiotherapy in the near future.
MATERIALS AND METHODS
Compounds
The compounds for the cell-based screen, INZ and its analogues were purchased from Asinex, ChemDiv and ChemBridge. INZ and INZ 1–5 were re-validated by LC/MS on an Agilent 1200 LC/MS system (Agilent Technology) at the Chemical Genomics Core Facility on the campus. INZ used for the animal experiments was synthesized, purified and identity verified by ChemBridge Inc. The minimum purity of all compounds is higher than 90%. MI-63 was generously provided by Shaomeng Wang (University of Michigan). ActD, Cis, Etoposide and TSA were purchased from Sigma. 5-Aminoimidazole-4-carboxamide-1-β-d-ribofuranoside (AICAR) was purchased from Toronto Research Chemicals Inc., North York, Ontario, Canada. Cambinol, Salermide and Tenovin-6 were from Cayman Chemical Company. Tenovin-6 was also provided by Sonia Lain (University of Dundee) as a gift. Biotinylated INZ was synthesized and characterized by NMR and LC-MS (Supporting Information).
Cell viability assay
To assess cell growth, the cell counting kit (Dojindo Molecular Technologies Inc., Gaithersburg, Maryland) was used according to manufacturer's instructions. Cell suspensions were seeded at 5000 cells per well in 96-well culture plates and incubated overnight at 37°C. Compounds were added into the plates and incubated at 37°C for 72 h. Cell growth inhibition was determined by adding WST-8 at a final concentration of 10% to each well, and the absorbance of the samples was measured at 450 nm using a Microplate Reader (Molecular Device, SpecrtraMax M5e).
In vivo ubiquitylation assay
For detection of ubiquitylation of endogenous p53, H460 cells in the 60 mm plates were transfected with (His)6–ubiquitin (His–Ub; 3 µg). At 24 h after transfection, cells were treated with various concentrations of INZ for 18 h, and then 10 µM MG132 and 20 µM ALLN for 8 h. For detection of ubiquitylation of exogenous p53, HCT116−/−cells were transfected with His–Ub (3 µg), p53 (0.2 µg) and HA-MDM2 (2 µg) expression plasmids as indicated in Fig 3D and Fig S3A of Supporting Information. At 36 h after transfection, cells were treated with INZ for 4 h followed by addition of 10 µM MG132 for 8 h. Cells were harvested and split into two aliquots, one for IB and the other for His pull-down by Nickel-NTA agarose (Thermo Scientific) as described previously (Dai et al, 2006; Sun et al, 2007) and analysed by IB.
Immunofluorescence for detection of H2AX Ser139 phosphorylation (γH2AX) foci
H460 cells at 50–70% confluence were treated with 2 µM of INZ or 10 µM Cis for 18 h or 2 mM HU for 8 h. Cells were fixed in 4% formaldehyde/PBS for 10 min, permeabilized and blocked with 0.3% Triton-100, 8% BSA/PBS. The primary antibodies used were polyclonal Phospho-Histone H2A.X (Ser139) antibodies in 1:250 dilution (20E3, Cell Signaling) and monoclonal p53 antibodies (DO-1, Santa Cruz Biotechnology) according to the manufactural instruction. Alex488 secondary antibodies were used to detect protein signals (Invitrogen). Images were taken with a Zeiss Axiovert 200M fluorescent microscope and measured using AxioVision 4.7.2.0 software.
Deacetylation activity assays using full-length acetylated p53 proteins as a substrate
H1299 cells were transfected with Flag-p53 plasmid and then infected with p300 adenovirus (Zeng et al, 2003) for 24 h. The cells were treated with 0.4 µM TSA for 18 h and harvested for purification of p53 proteins by using anti-Flag M2 agarose (Sigma). Bound proteins were eluted in TBS buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl) containing 0.2 mg of synthetic Flag peptides/ml, and then dialysed in deacetylation buffer (50 mM Tris–HCl, pH 8.8, 50 mM NaCl, 4 mM MgCl2, 0.2 mM PMSF, 1 mM DTT, 5% Glycerol, 0.1%NP-40).
Deacetylation reaction containing purified Flag-p53 proteins and titrated INZ was pre-incubated at room temperature for 10 min and initiated by adding 1.0 unit of SIRT1 enzyme (Enzo Life Sciences) and 50 µM NAD+. Reactions were incubated at 30°C for 1 h and stopped by addition of SDS loading buffer. Samples were analysed by IB and the acetylated Flag-p53 was detected with anti-p53K382Ac antibodies and total Flag-p53 was detected with anti-p53 DO-1 antibodies (North et al, 2005).
The inhibition experiments of INZ, Cambinol and Salermide on SIRT1 activity were carried out using the same conditions as above. For each sample, the level (band intensity) of p53K382Ac was quantified and normalized against total p53 levels. Inhibitory activity was calculated as the mean value of negative controls minus the average sample value divided by the mean value of negative controls minus the mean value of positive controls, multiplied by 100. Positive controls (100% inhibition) contained the acetylated p53 protein substrate only (first lane), and negative controls (0% inhibition) contained the substrate and SIRT1 enzymes (second lane). See Fig 6D.
Binding of biotinylated INZ to SIRT1
Recombinant SIRT1 and SIRT7 were expressed in Escherichia coli (E. coli) BL21-CodonPlus (DE3)-RIPL and purified through Ni-NTA agarose beads. Purified proteins was stained with Coomassie blue staining and quantified with BSA as a standard. The activity of purified SIRT1 was measured by the decrease in the levels of Ac-Lys382-p53 with acetylated-p53 protein as a substrate. For detection of binding of biotinylated INZ to SIRT1, the indicated concentrations of purified His–SIRT1 was incubated with biotinylated INZ that was conjugated to NeutrAvidin Agarose (Thermo Scientific) at 4°C overnight by gently agitating in the presence or the absence of 20 µM of free and non-biotinylated INZ. The beads were washed three times with 0.5% w/v NP-40, 0.2% w/v Tween 20/Tris buffered saline. Equal volumes of 2× SDS sample buffer were then added to each bead sample. The samples were boiled for 5 min and subjected to SDS–PAGE and immunoblotted with anti-His antibodies.
The paper explained
PROBLEM
The p53 tumour suppressor is one of the most important proteins that protect humans from the development of cancers. Although this gene is highly mutated in late stages of cancers, approximately 50% of all types of human cancers still contain WT p53. However, p53 in these types of cancers is often deactivated through a concerted action by its abnormally elevated suppressors, such as MDM2, MDMX or SIRT1. SIRT1 is highly expressed due to loss of its repressor HIC-1 via promoter hypermethylation in cancer cells, keeps p53 in a deacetylated status and facilitates its ubiquitylation and degradation by MDM2/MDMX. Thus, reactivation of p53 by targeting these p53 suppressors in p53-containing cancers has become an attractive approach for the development of anti-cancer therapy. Regardless of the intensive endeavour over the past decade, so far none of the known small molecules that target this p53 pathway has yet been developed into a clinically effective therapy. Thus, identifying more effective small molecules that specifically target this pathway in cancer has still remained challenging and exciting.
RESULTS
In our initial attempt to screen small molecules that may block MDM2/MDMX–p53 binding, we surprisingly identified a novel small molecule called INZ that effectively activates p53 by inhibiting SIRT1 activity without genotoxicity. Remarkably, INZ suppressed the growth of xenograft tumours derived from p53-containing human lung and colon cancer cells in a p53-dependent fashion. More remarkably, this small molecule was less toxic to normal cells and tissues. Hence, our study as described here unravels INZ as a novel class of small molecules that can activate p53 by inhibiting SIRT1 and repress tumour growth in xenograft tumour models.
IMPACT
Identification of INZ as a new class of small molecules that can activate p53 and induce p53-dependent apoptosis and senescence without causing genotoxicity as well as suppress tumour growth with little toxicity to normal cells and tissues offers another golden opportunity in the field of translational cancer research for the development of target-specific anti-cancer therapy either as an individual drug or as a component in combined therapy.
Mouse xenograft studies
Five-weeks-old female SCID mice were purchased from In Vivo Therapeutics (IVT) Core, Indiana University Simon Cancer Center (IUSCC) and housed in a BSL2 environment. Mice were subcutaneously inoculated with 5 × 106 H460 or 3 × 106 HCT116 cells. Tumour growth was monitored every other day with electronic digital calipers (Fisher Scientific) in two dimensions. Tumour volume was calculated with the formula: tumour volume (mm3) = (length × width2)/2(Figg & McLeod, 2004). When the mean tumour volume reached approximately 100 mm3 after 7–9 days, animals were dosed by i.p. injection with vehicle (5% DMSO) or INZ. Inhibition of tumour growth was calculated on the last day of treatment. To detect p53 activation in vivo, tumours were harvested and disrupted in RIPA buffer containing a protease inhibitor mixture (Sigma). Tumour lysates were analysed by IB. Cell proliferation in tumours was assessed by BrdU labeling followed by Immunostaining. 200 mg/kg body weight of BrdU (Sigma) was administrated to mice via i.p. injection 2 h before mice are sacrificed. Apoptosis was examined by TUNEL staining, using the Fluorescein In situ cell death detection kit (Roche) according to manufacturer's instructions. All animal experiments comply with protocols approved by The IUSM Institutional Animal Care and Use Committee (IACUC).
For more detailed Materials and Methods see the Supporting Information.
Acknowledgments
We thank Wei Gu, Hal Broxmeyer, Daniela Elena, X. Charlie Dong, Shaomeng Wang, Jiandong Chen and Sonia Lain for generously offering reagents including cell lines, plasmids, antibodies, MI-63 and Tenovin-6. We are grateful for the technical supports from the Chemical Genomics Core Facility, David R. Jones at IUSCC Clinical Pharmacology Analytical Core and Karen Pollok at IUSCC-In Vivo Therapeutic Core. Also, we gratefully acknowledge Sergio C. Chai for kinetics and inhibition analysis, and Christopher Burlak and Susan Downey for tissue sectioning and immunofluorescence. This work was partially supported by a CTSI pilot grant to H. L., who was also supported by NIH-NCI grants CA127724, CA095441 and CA129828. S. M and Q. Z. Y were supported by NIH grants.
Supporting information is available at EMBO Molecular Medicine online.
The authors declare that they have no conflict of interest.
Author contributions
QZ, SXZ and HL designed the experiments; QZ and HL analysed the data and composed the manuscript with participation of SXZ; HL drafted the manuscript; QZ conducted most of in vitro assays with assistance of SXZ; QZ and SXZ conducted cell-based assays; QZ, SXZ, YuZ and YiZ carried out animal experiments; QY and DD designed and conducted the synthesis of Biotin–INZ; SOM performed computational screening.
Supplementary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Alcain FJ, Villalba JM. Sirtuin inhibitors. Expert Opin Ther Pat. 2009;19:283–294. doi: 10.1517/13543770902755111. [DOI] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. Mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegue L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer. 2011;2:648–662. doi: 10.1177/1947601911417862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, Dai Y. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int J Biol Sci. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, Lu H. Regulation of the MDM2–p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J Biol Chem. 2006;281:24304–24313. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai JM, Wang ZY, Sun DC, Lin RX, Wang SQ. SIRT1 interacts with p73 and suppresses p73-dependent transcriptional activity. J Cell Physiol. 2007;210:161–166. doi: 10.1002/jcp.20831. [DOI] [PubMed] [Google Scholar]
- Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, Gao W, Qin D, Stuckey J, Krajewski K, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2–p53 interaction. J Med Chem. 2006;49:3432–3435. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duriez PJ, Shah GM. Cleavage of poly(ADP-ribose) polymerase: a sensitive parameter to study cell death. Biochem Cell Biol. 1997;75:337–349. [PubMed] [Google Scholar]
- Efeyan A, Ortega-Molina A, Velasco-Miguel S, Herranz D, Vassilev LT, Serrano M. Induction of p53-dependent senescence by the MDM2 antagonist nutlin-3a in mouse cells of fibroblast origin. Cancer Res. 2007;67:7350–7357. doi: 10.1158/0008-5472.CAN-07-0200. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Lozano G. p53 and MDM2: Antagonists or partners in crime. Cancer Cell. 2009;15:161–162. doi: 10.1016/j.ccr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Fang Y, Nicholl MB. Sirtuin 1 in malignant transformation: Friend or foe. Cancer Lett. 2011;306:10–14. doi: 10.1016/j.canlet.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Figg WD, McLeod HL. Handbook of Anticancer Pharmacokinetics and Pharmacodynamics. Totowa, NJ: Humana Press; 2004. [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- Fleuriel C, Touka M, Boulay G, Guerardel C, Rood BR, Leprince D. HIC1 (hypermethylated in cancer 1) epigenetic silencing in tumors. Int J Biochem Cell Biol. 2009;41:26–33. doi: 10.1016/j.biocel.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M, Kimura M, Morita S, Matsubara K, Yamanaka S, Endo C, Sakurada A, Sato M, Kondo T, Horii A, et al. Microarray analysis of promoter methylation in lung cancers. J Hum Genet. 2006;51:368–374. doi: 10.1007/s10038-005-0355-4. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001;276:38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, Parant J, Lozano G, Yuan ZM. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem. 2002;277:19251–19254. doi: 10.1074/jbc.C200150200. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Tokuchi Y, Hashimoto T, Hayashi S, Nishida K, Ishikawa Y, Nakagawa K, Tsuchiya S, Okumura S, Tsuchiya E. Reduced HIC-1 gene expression in non-small cell lung cancer and its clinical significance. Anticancer Res. 2001;21:535–540. [PubMed] [Google Scholar]
- Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53–HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Jung-Hynes B, Ahmad N. Role of p53 in the anti-proliferative effects of Sirt1 inhibition in prostate cancer cells. Cell Cycle. 2009;8:1478–1483. doi: 10.4161/cc.8.10.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobet E, Zeng X, Zhu Y, Keller D, Lu H. MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc Natl Acad Sci USA. 2000;97:12547–12552. doi: 10.1073/pnas.97.23.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, McCarthy A, Appleyard V, Murray KE, Baker L, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13:454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara E, Mai A, Calvanese V, Altucci L, Lopez-Nieva P, Martinez-Chantar ML, Varela-Rey M, Rotili D, Nebbioso A, Ropero S, et al. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene. 2009;28:781–791. doi: 10.1038/onc.2008.436. [DOI] [PubMed] [Google Scholar]
- Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins – novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Shah ZH, Allison SJ, Ahmed SU, Ford J, Warnock LJ, Li H, Serrano M, Milner J. SIRT1 undergoes alternative splicing in a novel auto-regulatory loop with p53. PloS ONE. 2010;5:e13502. doi: 10.1371/journal.pone.0013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Olson AJ. Using AutoDock for ligand–receptor docking. Curr Protoc Bioinform. 2008;8:14. doi: 10.1002/0471250953.bi0814s24. [DOI] [PubMed] [Google Scholar]
- Nakae J, Cao Y, Daitoku H, Fukamizu A, Ogawa W, Yano Y, Hayashi Y. The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J Clin Invest. 2006;116:2473–2483. doi: 10.1172/JCI25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PloS ONE. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Schwer B, Ahuja N, Marshall B, Verdin E. Preparation of enzymatically active recombinant class III protein deacetylases. Methods. 2005;36:338–345. doi: 10.1016/j.ymeth.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Nosho K, Shima K, Irahara N, Kure S, Firestein R, Baba Y, Toyoda S, Chen L, Hazra A, Giovannucci EL, et al. SIRT1 histone deacetylase expression is associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2009;22:922–932. doi: 10.1038/modpathol.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2:1–8. [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD, Lam EW. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- Popowicz GM, Czarna A, Rothweiler U, Szwagierczak A, Krajewski M, Weber L, Holak TA. Molecular basis for the inhibition of p53 by Mdmx. Cell Cycle. 2007;6:2386–2392. doi: 10.4161/cc.6.19.4740. [DOI] [PubMed] [Google Scholar]
- Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, van der Eb AJ, Jochemsen AG. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J Biol Chem. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Trapp J, Meier R, Hongwiset D, Kassack MU, Sippl W, Jung M. Structure–activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins) ChemMedChem. 2007;2:1419–1431. doi: 10.1002/cmdc.200700003. [DOI] [PubMed] [Google Scholar]
- Tseng RC, Lee CC, Hsu HS, Tzao C, Wang YC. Distinct HIC1–SIRT1–p53 loop deregulation in lung squamous carcinoma and adenocarcinoma patients. Neoplasia. 2009;11:763–770. doi: 10.1593/neo.09470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen I, Lain S. Sirtuins and p53. Adv Cancer Res. 2009;102:171–195. doi: 10.1016/S0065-230X(09)02005-3. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Velasquez DA, Martinez G, Romero A, Vazquez MJ, Boit KD, Dopeso-Reyes IG, Lopez M, Vidal A, Nogueiras R, Dieguez C. The central Sirtuin 1/p53 pathway is essential for the orexigenic action of ghrelin. Diabetes. 2011;60:1177–1185. doi: 10.2337/db10-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales MM, Biel MA, el Deiry W, Nelkin BD, Issa JP, Cavenee WK, Kuerbitz SJ, Baylin SB. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med. 1995;1:570–577. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Zeng SX, Jin Y, Kuninger DT, Rotwein P, Lu H. The acetylase activity of p300 is dispensable for MDM2 stabilization. J Biol Chem. 2003;278:7453–7458. doi: 10.1074/jbc.M209030200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.