SUMMARY
The redox-regulated chaperone Hsp33 protects organisms against oxidative stress that leads to protein unfolding. Activation of Hsp33 is triggered by the oxidative unfolding of its own redox-sensor domain, making Hsp33 a member of a recently discovered class of chaperones that require partial unfolding for full chaperone activity. Here we address the long-standing question of how chaperones recognize client proteins. We show that Hsp33 uses its own intrinsically disordered regions to discriminate between unfolded and partially structured folding intermediates. Binding to secondary structure elements in client proteins stabilizes Hsp33’s intrinsically disordered regions, and this stabilization appears to mediate Hsp33’s high affinity for structured folding intermediates. Return to nonstress conditions reduces Hsp33’s disulfide bonds, which then significantly destabilizes the bound client proteins and in doing so converts them into less-structured, folding-competent client proteins of ATP-dependent foldases. We propose a model in which energy-independent chaperones use internal order-to-disorder transitions to control substrate binding and release.
INTRODUCTION
Molecular chaperones areinvolved in maintaining a functional proteome (Tyedmers et al., 2010). Divided into various unrelated yet highly conserved protein families, chaperones have in common the ability to bind unfolded polypeptides and prevent their nonspecific aggregation. Many chaperones, including the DnaK/DnaJ/ GrpE system, are classified as “foldases” as they promote protein folding in an ATP-dependent process. The other category of chaperones is “holdases.” They include stress-specific chaperones that are needed to protect proteins against aggregation under distinct stress situations (Haslbeck et al., 2005; Jakob et al., 1999). Usually ATP independent, holdases bind tightly to unfolding proteins until nonstress conditions resume. Then, client proteins are released for refolding in a process that is often mediated by foldases (Haslbeck et al., 2005; Hoffmann et al., 2004).
The highly conserved, redox-regulated Hsp33 is a stress-specific holdase, which protects bacteria against severe oxidative stress conditions, including bleach treatment (Winter et al., 2005, 2008). Under nonstress conditions, Hsp33 is a compactly folded zinc-binding protein with negligible affinity for unfolded proteins. However, when exposed to oxidative stress conditions that lead to widespread protein unfolding, Hsp33 undergoes massive conformational rearrangements. These changes occur via Hsp33’s C-terminal redox-switch domain, which consists of an ~50 amino acid (aa) flexible linker region (aa 178–231) and an adjacent, redox-sensitive zinc center. Triggered by oxidative disulfide bond formation and zinc release, the redox-switch domain of Hsp33 unfolds (Graf et al., 2004; Ilbert et al., 2007). That activation of Hsp33 requires its unfolding makes Hsp33 a member of a new class of chaperones that are active when intrinsically disordered (Tompa and Csermely, 2004). Previous studies revealed that the specific activation of Hsp33 compensates for the inactivation of ATP-dependent folding chaperones, which are unable to function efficiently due to the oxidative stress-mediated drop in intracellular ATP levels (Winter et al., 2005). Release of client proteins from Hsp33 requires restoration of reducing nonstress conditions and the presence of a functional DnaK system (Hoffmann et al., 2004). Analysis of the in vivo substrate-binding specificity of Hsp33 and DnaK revealed an extensive overlap in client proteins, a necessary prerequisite for the synergistic action of the two classes of chaperones (Winter et al., 2005).
Activation of Hsp33 requires its native unfolding, making Hsp33 a particularly intriguing example of a protein that apparently needs to lose its structure to gain function. Several other chaperones, including the acid-activated HdeA (Tapley et al., 2009) and the heat-activated small heat shock proteins (Jaya et al., 2009), have been shown to use localized protein unfolding for activation. In addition, many other chaperones have been shown to constitutively contain regions of intrinsic disorder (Tompa and Csermely, 2004). Due to a lack of structural information about chaperone-substrate complexes and intrinsically disordered proteins in general, little is known about the precise roles played by these regions. Here we used a combination of peptide-binding analysis, measurements of protein stability by rates of hydrogen/deuterium (H/D) exchange and mass spectrometry, and limited proteolysis studies to demonstrate that chaperones, like Hsp33, use their intrinsically disordered regions to specifically recognize early unfolding intermediates and trigger unfolding processes necessary to return proteins onto a productive folding pathway.
RESULTS
Identification of Hsp33’s Peptide-Binding Specificity
We were intrigued by the question of how Hsp33 can effectively bind hundreds of different client proteins while studiously avoiding binding to its own intrinsically disordered C-terminal redox-switch domain, even though this domain is unfolded and present at very high local concentrations. We thus decided to determine the substrate-binding specificity of Hsp33 by screening a peptide array comprised of 3,914 peptides, encompassing the complete sequences of 18 different proteins (Table S1 available online). A large subset of these peptides was derived from previously identified Hsp33 and DnaK client proteins (Rüdiger et al., 1997; Winter et al., 2008). We used 12-mer peptides spanning the entire sequence of the individual proteins. Each peptide overlapped the adjacent peptide by 10 residues.
The peptides were synthesized in situ on microfluidic chips and immobilized via a long (equivalent to 30 aa) C-terminal poly-ethylene glycol linker (Pellois et al., 2002). As shown in Figure 1A, in contrast to reduced, inactive Hsp33red, which did not reveal significant binding, active Hsp33ox bound strongly to select sets of overlapping peptides (i.e., repetitive binding). We normalized and scored the fluorescence intensities from 0 to 1 according to a flow scheme depicted in Figure S1A (see Experimental Procedures for details). Peptides that bound to at least three directly adjacent peptides with individual scores of ≥0.8 were defined as “good binders,” whereas peptides with at least three repetitive low scores of ≤0.2 were defined as “nonbinders.”
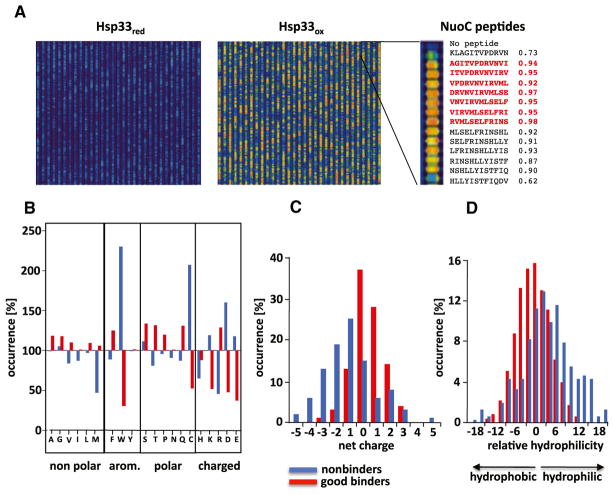
Figure 1. Peptide-Binding Specificity of Active Hsp33ox.
(A) A peptide array was incubated with inactive Hsp33 (Hsp33red) or active Hsp33 (Hsp33ox), and Hsp33 was visualized with Cy5-fluorescently labeled anti-Hsp33 antibodies. False-color imaging of the signal intensities ranges from blue (no binding) to red (maximal binding). The order of overlapping peptides in the array, including normalized binding scores, is shown for select NuoCD peptides. Sequences of peptides that comprise the good binder PepNuoC are shown in bold red.
(B–D) Relative amino acid occurrence (B), net charge (C), and hydrophilicity (D) in good binders (red) and nonbinders (blue) were calculated and normalized to the occurrence of the same amino acid or feature distribution in the entire library. A value of 100% indicates that occurrence of a specific amino acid is identical to occurrence in the total peptide library.
To assess the peptide-binding specificity of Hsp33ox, we determined the relative occurrence of amino acids in good binders versus nonbinders compared to the relative occurrence of all 20 amino acids in the total peptide library. We found only a few amino acids that were slightly more abundant in good binders (Figure 1B, red) and less abundant in nonbinders (Figure 1B, blue) as compared to the total peptide library. In contrast, however, we found that the two negatively charged residues Asp and Glu, as well as Trp, Cys, and Lys, were strongly disfavored in good binders and highly enriched in peptides that failed to bind to Hsp33 (Figure 1B). Analysis of the amino acid sequence in either good binders or nonbinders did not reveal any specific sequence motif or position specificity for any of these residues (Figure S1B). However, Hsp33-peptide interactions appeared to largely depend on net charge and hydrophobicity of the peptides, with nonbinding peptides being significantly more acidic and less hydrophobic than good binders (Figures 1C, 1D, and S1B). To predict Hsp33-binding sites in proteins, we developed an algorithm that takes amino acid distribution, hydrophobicity, net charge, and secondary structure into account (see Extended Experimental Procedures). This algorithm showed high accuracy in predicting good binders (83%) and nonbinders (91%) of Hsp33.
Validation of Hsp33’s Peptide-Binding Specificity in Solution
To validate the peptide array binding results in solution assays, we synthesized PepNuoC, a 25 aa peptide exhibiting one of the highest scores in our peptide array over seven repeats (Figure 1A, right panel). To assess whether PepNuoC binds to Hsp33ox in solution, we determined its competitive activity by measuring how increasing amounts of PepNuoC influence the ability of active Hsp33ox to suppress the aggregation of chemically unfolded citrate synthase (CS), a known substrate of Hsp33 (Figure 2A). We found that the ability of activated Hsp33ox to prevent CS aggregation diminished with increasing amounts of PepNuoC, implying that this peptide functions as Hsp33 substrate also in solution. To determine the precise affinity of Hsp33ox for PepNuoC, we conducted surface plasmon resonance (SPR) experiments in which we immobilized PepNuoC on a chip (Figure 2B). Whereas Hsp33red failed to bind to the peptide even at the highest concentration used (i.e., 750 nM), Hsp33ox showed specific binding with a dissociation constant (KD) of 35 ± 0.6 nM per active Hsp33 dimer (Figures 2B and S2A and Table S2A). Note that this KD is very similar to the KD values (30–300 nM) that were previously determined using the SUPREX technique to quantify binding of Hsp33ox to thermally unfolded CS (Xu et al., 2010). Taken together, these results strongly suggest that PepNuoC and full-length substrates share the same binding site in Hsp33.
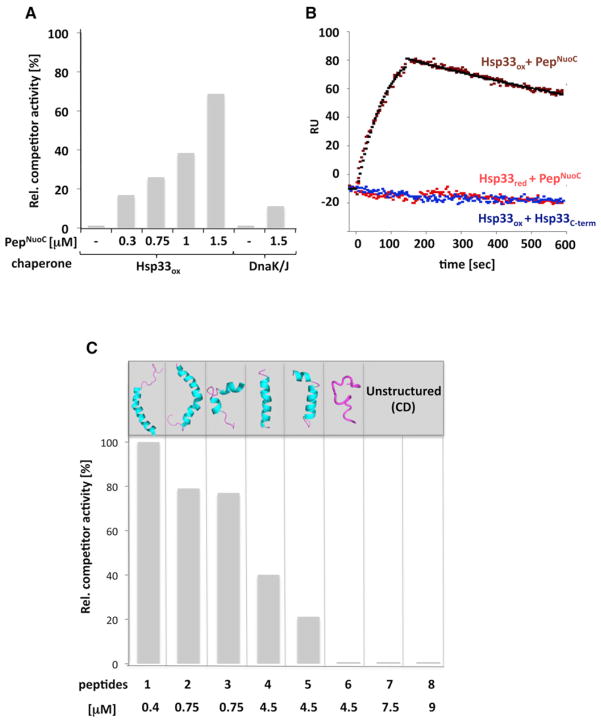
Figure 2. Validation of Peptide Binding in Solution and KD Measurements.
(A) Competitor activity of PepNuoC was assessed by determining its ability to compete with 0.075 μM chemically unfolded CS for binding to 0.15 μM Hsp33ox or 0.375 μM DnaK/0.075 μM DnaJ. Complexes between chaperones and competitors were allowed to form for 4 min before chemically denatured CS was added. Light-scattering measurements of CS at 30°C were conducted as read-out for competitor activity. The light-scattering signal of CS in the presence of fully active chaperones (i.e., absence of competitor) was set to 0% competitor activity, whereas the light-scattering signal of CS in the absence of functional chaperone (i.e., full competition) was set to 100%. Presence of PepNuoC had no significant influence on the aggregation behavior of CS in the absence of chaperones.
(B) SPR sensorgrams to monitor interactions between Hsp33ox or Hsp33red and immobilized PepNuoC or oxidized Hsp33C-term. The data were fitted (black symbols) using BIAevaluation software.
(C) Competitor activity of predicted Hsp33 good binder peptides and nonbinder peptides was assessed by determining their ability to compete with 0.075 μM chemically unfolded CS for binding to 0.15 μM Hsp33ox. Light-scattering measurements were conducted as described above. The upper panel illustrates the NMR structures of the respective peptides according to PDB (Table S2B). Far-UV CD spectroscopy revealed an α helix for Pep1 and random coils for Pep6–8 (see Figure S2D).
Several other high-affinity peptides of Hsp33ox identified by the array turned out to be insoluble upon synthesis. We thus decided to use a complementary approach to validate our peptide array results. We reasoned that our prediction algorithm, if accurate, should identify good binders and nonbinders of Hsp33 from a list of commercially available, soluble peptides. We scored ~670 commercially available peptides for Hsp33 binding and screened the peptides against a database of peptides for which nuclear magnetic resonance (NMR) structures have been solved. By using this approach, we would be able to test our binding predictor and gain access to atomic resolution structures of potentially binding and nonbinding peptides. Whereas the NMR structures of many of our predicted good binders had been solved, none of our predicted nonbinders had structures available. We obtained six predicted good binders with known structures as well as two nonbinders (Table S2B) and tested their competitive activity in our chaperone assay (Figure 2C). All but one of the predicted good binders (i.e., Pep6, see below) prevented Hsp33ox from suppressing CS aggregation in a concentration-dependent manner, indicating that they interact with active Hsp33 (Figures 2C and S2C). The peptide with the apparently highest affinity to Hsp33 was Pep1 (i.e., neuropeptide Y), which fully competed with full-length CS for binding to Hsp33 at a Pep1 to CS ratio of 5:1. In contrast, none of our predicted nonbinder peptides (Pep7, Pep8) competed with CS for binding to Hsp33 even when present in 100-fold excess to CS (Figure 2C). These results strongly suggest that the peptide-binding specificity as determined in our peptide array and used as dataset for our binding predictor reflects well the binding specificity of active Hsp33 in solution.
Hsp33 Avoids Self-Binding by Disfavoring Natively Unfolded Polypeptides
Peptide-binding analysis revealed that Hsp33’s substrate recognition might rely more on the chemical and structural features of the amino acids and peptides than on specific amino acid sequence motifs. It was furthermore intriguing to observe that overrepresentation of the same amino acids that are abundant in Hsp33’s nonbinders characteristically defines natively disordered segments (Prilusky et al., 2005). In fact, all of the amino acids that Hsp33ox excludes from binding, i.e., Asp, Glu, Cys, and Lys, are highly enriched in natively unfolded proteins, including Hsp33’s own C-terminal redox-switch domain (Figure S1C). These results suggested that Hsp33 avoids self-binding by disfavoring binding to natively disordered regions.
Localization analysis of our nonbinders and good binders in the available crystal structures of the tested proteins fully agreed with these conclusions and revealed that 59% of nonbinders are indeed found in loops and nonstructured regions. In contrast, the majority (76%) of Hsp33’s good binders are located in the structured regions of the proteins (Figure 3A), suggesting that Hsp33 might require secondary structure elements for binding. Although it is experimentally difficult to assess whether secondary structures persist within the dense spots of individual 12-mer peptides on the array, secondary structure elements are known to occur in peptides as short as 12 aa (Voelz et al., 2009). Moreover, our peptide-binding studies shown in Figure 2C were entirely consistent with this model. All five of our Hsp33-binding peptides (Pep1–5) contained significant amounts of secondary structure as illustrated by their NMR structures (Figure 2C) and confirmed for Pep1 by far-UV circular dichroism (CD) spectroscopy (Figure S2D). In contrast, far-UV CD spectra of our non-binding peptides (Pep6–8) revealed that they are predominantly random coil in aqueous solution (Figure S2D; Table S2C). This included Pep6 (i.e., obestatin), the only predicted good binder of Hsp33 that failed to compete with CS for binding to active Hsp33ox. Protein Data Bank (PDB) searches revealed that Pep6 is α helical when present in micelles yet adopts random-coil structure in water (Figures 2C and S2D). It thus appears that our predictor identified this peptide as a potentially good Hsp33 binder based on the fact that this peptide has secondary structure-forming potential. The peptide apparently failed to bind to Hsp33, however, as it was unfolded in our buffer system.
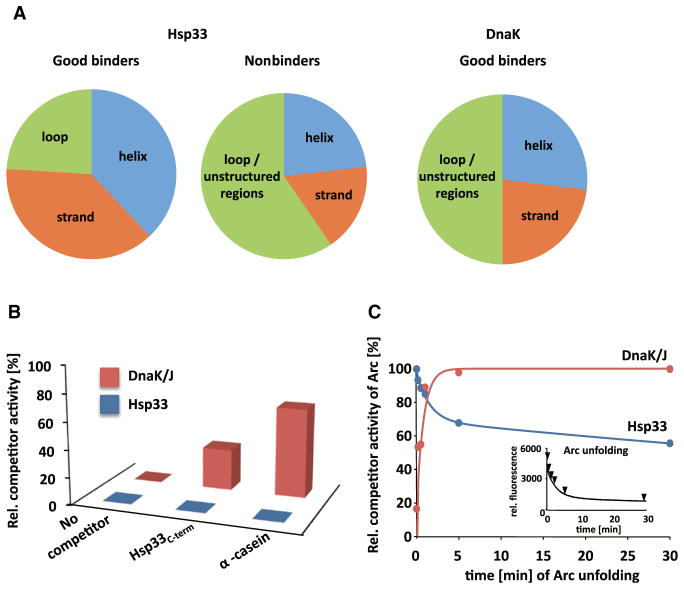
Figure 3. Hsp33 and DnaK Show Distinct Peptide-Binding Specificity.
(A) Comparison of secondary structure features in good binders and nonbinders of Hsp33ox with good binders of DnaK. Analysis of DnaK’s non-binders was not possible due to the semi-quantitative nature of the cellulose peptide array used to collect this data (Rüdiger et al., 1997).
(B) Influence of 0.325 μM Hsp33C-term or α-casein on the ability of 0.13 μM active Hsp33ox or 0.65 μM DnaK /0.13 μM DnaJ to suppress the aggregation of 0.065 μM chemically denatured luciferase at 30°C. Competitor activity was determined as described above.
(C) Time course of Arc unfolding and competitor activity. Arc unfolding was initiated by diluting Arc (0.75 μM) into HEPES buffer (pH 7.5) at 30°C. At distinct time points after start of the unfolding process, 0.15 μM active Hsp33ox or 0.187 μM DnaK/0.075 μM DnaJ was added to allow complex formation. After 10 s, 0.075 μM chemically unfolded CS was added. Light scattering of CS was monitored and competitor activity of Arc was determined as described above. Inset: Unfolding of 0.75 μM Arc was monitored by following the fluorescence of its single tryptophan residue located at the monomer-monomer interface upon dilution into buffer. Arrows indicate the time points at which Arc was analyzed for competitor activity.
See also Figure S3.
To experimentally address the question of whether Hsp33ox interacts with natively disordered proteins, we performed chaperone competition experiments in the presence of increasing concentrations of its own oxidized C-terminal fragment, Hsp33C-term (aa 218–287), which is unfolded when oxidized (Graf et al., 2004), or α-casein, a well-established natively unfolded protein (Gaspar et al., 2008). We found that neither polypeptide competed with chemically unfolded luciferase (Figure 3B) or CS (Figure S3A) for Hsp33ox binding. SPR binding experiments with the immobilized Hsp33C-term as a substrate confirmed these results (Figure 2B). These results suggest that Hsp33 avoids self-binding by disfavoring binding to natively disordered regions and favoring binding to secondary structure elements.
DnaK and Hsp33 Have Overlapping Client Proteins but Distinct Folding-State Specificities
Our results fit well with the physiological niche that Hsp33 occupies in vivo, which is to prevent aggregation of mature proteins that slowly unfold as cells encounter stress conditions (Winter et al., 2005, 2008). In contrast, Hsp33’s synergistic interaction partner DnaK interacts with unfolded polypeptide chains as they emerge from the ribosome (Deuerling et al., 1999). Analysis of DnaK’s previously conducted peptide array (Rüdiger et al., 1997) with regard to the predicted secondary structure features of DnaK’s high-affinity peptides agreed with this conclusion and showed that many high-affinity binding peptides of DnaK are predicted to be localized in unstructured regions of the substrate proteins (Figure 3A). Moreover, localization analysis of the high-affinity binding peptides of Hsp33 and DnaK on the primary sequences of the seven common substrate proteins revealed that many binding sites only partially overlapped, suggesting that the chaperones often recognize regions adjacent in the primary structures of the substrate proteins (Figure S3B).
To directly compare the in vitro substrate-binding specificity of the two chaperones, we conducted competition experiments with DnaK in the presence of either Hsp33C-term or α-casein, the two natively unfolded proteins that failed to interact with Hsp33ox (Figure 3B), or PepNuoC (Figure 2A), which we identified as a good binder of Hsp33ox. These experiments were conducted in the presence of the cochaperone DnaJ, which is required to mediate stable complex formation between DnaK and substrate proteins (Lu and Cyr, 1998). Both natively unfolded proteins decreased the influence of the DnaK/DnaJ system on the aggregation of the client protein luciferase, indicating that the unfolded poly-peptides compete for DnaK’s binding site (Figure 3B). In contrast, however, the presence of a 20-fold molar excess of PepNuoC only marginally affected the chaperone function of the DnaK system, which agrees with SPR measurements that showed that the affinity of DnaK to PepNuoC is almost 10-fold lower than the affinity of Hsp33ox for this peptide (Figure S2A and Table S2A). These results confirmed the preference of DnaK for unstructured polypeptides (Deuerling et al., 1999), including the intrinsically disordered region of activated Hsp33ox.
To obtain independent confirmation that activated Hsp33ox and the DnaK system have the same client proteins but recognize distinct structural features, we investigated their influence on the 55 aa Arc repressor, a client protein of both chaperones (Figure S3C). Arc has been previously shown to rapidly dissociate and slowly unfold upon dilution into salt-free buffer without forming aggregates (Figure 3C, inset) (Bowie and Sauer, 1989). We predicted, based on our previous results, that activated Hsp33ox should interact with more structured folding intermediates of Arc that appear early in the unfolding process of Arc, whereas the DnaK/DnaJ system should recognize folding intermediates at later points during Arc’s unfolding process. We thus added activated Hsp33ox or the DnaK/DnaJ system at defined time points after start of Arc’s unfolding process and allowed complexes to form before we added unfolded CS to analyze Arc’s competitive activity. As shown in Figure 3C, the results were in excellent agreement with our prediction. We found that the ability of Arc to compete with CS for binding to Hsp33ox decreased with progressing unfolding of Arc, whereas competition for binding to the DnaK/DnaJ system increased with increased unfolding. These results strongly support our conclusion that the two chaperone systems share their client proteins but exert significantly different folding-state specificity.
Intrinsically Disordered Regions in Chaperones: Flexible Binding Sites for Structured Folding Intermediates?
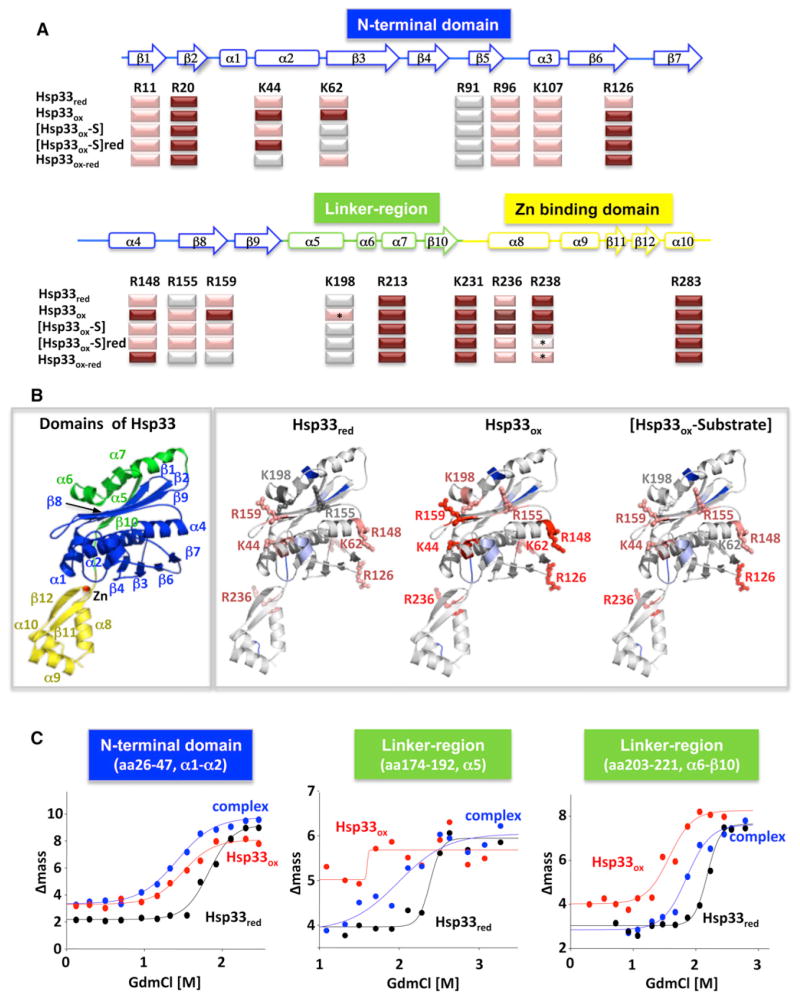
Our results raised a number of intriguing conceptual questions, particularly with regard to the mechanism by which substrate transfer between Hsp33 and the DnaK system is coordinated and the potential role that Hsp33’s intrinsically disordered region might play in this process. We thus decided to directly monitor the structural changes that Hsp33 undergoes upon substrate binding, paying particular attention to Hsp33’s flexible linker region and zinc-binding domain (i.e., C-terminal redox-switch domain). In one set of experiments, we used a limited proteolysis assay with a mass spectrometric readout to characterize the exposed and buried regions in Hsp33 by means of their accessibility to trypsin. We first used different ratios of trypsin to Hsp33 and digestion times to compare the proteolytic sensitivity of inactive Hsp33red and active Hsp33ox in the absence of substrate proteins. In each case, the peptides were analyzed by liquid chromatography mass spectrometry (LC-MS) and mapped onto the modeled structure of reduced Escherichia coli Hsp33 (Cremers et al., 2010). Under all conditions tested, we found differential protection patterns in Hsp33red and Hsp33ox, consistent with large structural rearrangements upon Hsp33’s oxidative activation. Residues that were more accessible in Hsp33ox as compared to Hsp33red were K198, which is located in the central α5 helix of Hsp33’s metastable linker region, and R236, located in the zinc-binding domain of Hsp33 (Figures 4A, 4B, S4A, and S4B). These results confirmed earlier CD and fluorescence measurements, which provided the initial indication that Hsp33’s linker region and zinc-binding domain unfold in response to disulfide bond formation and zinc release (Graf et al., 2004; Ilbert et al., 2007). We also observed increased accessibility of R159, located in β8, the nearby R155, and K44, a residue of the central α2 helix, upon activation of Hsp33 (Figures 4A, 4B, S4A, and S4B). These results suggest that structural changes in α5 disrupt interactions with β8, one of the four β strands shown to be involved in linker binding in inactive Hsp33red and thought to contribute to substrate binding in active Hsp33ox (Cremers et al., 2010). Oxidative activation of Hsp33 also affected accessibility for three residues (i.e., K62, R126, and R148) that are located adjacent to Hsp33’s highly conserved monomer-monomer interface (Graumann et al., 2001), reflecting the structural changes that accompany dimerization of oxidized Hsp33 molecules.
Figure 4. Hsp33’s Intrinsically Unfolded Linker Region Is involved in Substrate Binding.
(A) Inactive Hsp33red, active Hsp33ox, and Hsp33ox in complex with thermally unfolded luciferase [Hsp33ox-S] before or after reduction with DTT ([Hsp33ox-S]red) were digested with trypsin for 5, 10, or 15 min, and cleavage products were analyzed by LC-MS. The most accessible sites (cleaved within 5 min) are indicated in dark red, and sites that were not cleaved within 15 min are indicated in gray. Peptides with very low abundance are indicated with a star. Color-coded secondary structure elements of Hsp33 are shown for reference.
(B) Differences in the proteolytic patterns of Hsp33red, Hsp33ox, and Hsp33ox-substrate were mapped onto the modeled E. coli Hsp33 structure. Relative accessibility of the individual sites is indicated by color intensity, with the most accessible residues shown in dark red and the least accessible shown in gray. Unaltered trypsin cleavage sites are indicated in light blue. The color-coded domain structure of Hsp33 with labeled secondary structure elements is shown for reference.
(C) Representative SUPREX curves of two fragments from the flexible linker region (aa 174–192, aa 203–221, after 5 min H/D-exchange) and one peptide from the stable N-terminal region (aa 26–47, after 2 min H/D exchange) obtained from Hsp33red (black), Hsp33ox (red), or the Hsp33ox-CS complex (blue). Solid lines are best fits of the data to a four-parameter sigmoidal equation, used to obtain C1/2 values (Table S3).
To determine how substrate binding affects the conformation of Hsp33ox, we analyzed the proteolytic accessibility of Hsp33 in complex with thermally unfolded luciferase (Figures 4A and 4B) or CS (Figure S4A). We reasoned that regions in Hsp33ox that are either directly or indirectly involved in substrate interaction should become more protected upon substrate binding. The regions that we found to be affected by binding to either substrate involved the flexible linker of Hsp33 (K198) as well as the adjacent β8 strand and central α2 helix (Figures 4A and 4B). Accessibility of these sites (i.e., R159, K44) decreased upon substrate binding, implying that interaction of Hsp33ox with substrate proteins either directly obscures the cleavage sites and/or sterically blocks trypsin from gaining access to the sites. Other residues in Hsp33ox whose accessibility was decreased by client protein binding were identified to be close to Hsp33’s dimerization interface, consistent with functional studies that showed that substrate binding stabilizes the Hsp33 dimer (Hoffmann et al., 2004). No other region in Hsp33ox was found to have substantially altered proteolytic behavior upon substrate binding at this or less stringent trypsin concentrations. This included the zinc-binding domain, whose proteolytic accessibility was unaltered by the presence of substrate proteins. This result agreed with previous studies that showed that absence of the zinc-binding domain affects Hsp33’s redox regulation but not its ability to bind client proteins (Ilbert et al., 2007). These results suggest that client protein binding involves the flexible linker of Hsp33 as well as potentially a loop region located directly underneath the central α5 helix of Hsp33’s linker.
Limited proteolysis is a good tool to assess global conformational changes in proteins but is restricted by cleavage site locations. We therefore decided to employ the SUPREX technique to directly compare the relative changes in thermodynamic stability of Hsp33 upon oxidative activation and substrate binding (Xu et al., 2010). In SUPREX, the H/D exchange properties of globally protected amide protons in a protein are probed as a function of chemical denaturant concentration to acquire information about a protein’s conformational stability (Xu et al., 2010). SUPREX experiments have previously shown that Hsp33ox is globally destabilized by 23.8 kJ/mol per dimer upon oxidative activation and that substrate binding stabilizes Hsp33ox by 7.1–10.0 ± 0.9 kJ/mol per dimer, depending on the substrate protein (Tang et al., 2007). In order to map which regions of the Hsp33 structure undergo the most significant conformational changes upon oxidation and upon substrate binding, we utilized the SUPREX methodology and combined it with a pepsin digestion protocol (Xu et al., 2010). The overall coverage of Hsp33 was 60% (Figure S4B; Table S3). A MALDI-MS analysis was used to determine the extent of H/D exchange (i.e., Δmass) for each peptide. Plots of Δmass versus [GdmCl] (i.e., SUPREX curves) were generated for each peptide, which revealed transition midpoints (i.e., C1/2 values) and initial H/D exchange levels, used to determine sequence-specific stability changes in Hsp33 (Table S3). In excellent agreement with our limited proteolysis results, we found that the most dramatic thermodynamic changes upon oxidative activation and substrate binding take place in two regions of Hsp33, aa 174–192 (i.e., α5) and aa 203–221 (i.e., α6–β10), which span nearly all of Hsp33’s linker region (Figure 4C). Our thermodynamic analyses with C1/2 value changes observed for peptides derived from this region indicate that the linker region of Hsp33 is destabilized upon oxidative activation, as indicated by the 0.5 to 0.8 M C1/2 value shifts that were observed for the Hsp33 (aa 174–192) and Hsp33 (aa 203–221) peptides (Figure 4C, compare black and red data; Table S3). Notably, this region of Hsp33ox’s structure regained some of its stability (i.e., C1/2 values were shifted to a higher denaturant concentration) upon binding thermally unfolding CS (Figure 4C, compare red and blue data; Table S3). These results provide additional evidence that the natively unfolded linker region of Hsp33 is directly involved in substrate binding. The stabilization that we observed in Hsp33’s linker region is most likely through a combination of direct substrate binding and substrate-mediated refolding and contributes to the overall affinity of Hsp33 for its client proteins.
Reduction of Hsp33-Substrate Complexes Destabilizes Bound Substrate Proteins
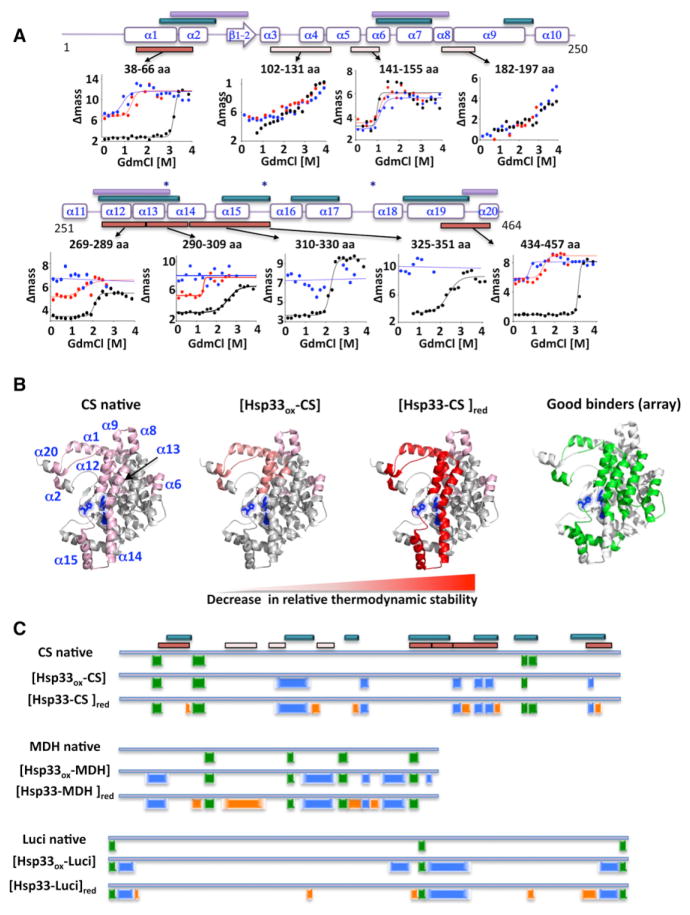
Release of client proteins from Hsp33ox requires the restoration of reducing nonstress conditions and the presence of the DnaK system (Hoffmann et al., 2004). Our studies now raise the question of how the DnaK system is able to trigger substrate release from Hsp33, given the only partially overlapping peptide-binding specificity of the two chaperone systems. To address this question, we undertook structural studies on the bound substrate proteins primarily using SUPREX analysis of CS, both alone and in complex with Hsp33 before and after DTT treatment. This analysis provided region-specific stability information of approximately 50% of the CS molecule (Figures 5A and S5A). A comparison of the transition midpoint values (C1/2) obtained on these peptides from native CS and thermally unfolded CS in complex with Hsp33ox revealed that the relative thermodynamic stabilities of several regions (helices α3, α4, α5/6, and α8/9) of the CS structure were unchanged (Figures 5A and 5B and Table S4). However, we identified three regions of CS involving helices α1–α2, α12–α15, and α19–α20 that appear to be strongly destabilized in the complex (Figures 5A and 5B). Importantly, these regions coincide well with the dimerization interface of CS; helices α12 and α13 (aa 269–298) are part of a largely hydrophobic, four-stranded antiparallel α helix sandwich, which constitutes the main component of the monomer-monomer interface of CS, whereas helices α2 (aa 57–70) and α20 (aa 453–460) contribute to dimerization by deeply burying themselves into the other monomer (Figure S5A). Two additional CS peptides located near the monomer-monomer interface containing helix α14/15 (aa 310–330, aa 325–351) were detected in native CS but not detected in the MALDI readout of the complex. Absence of these peptides was not due to signal suppression effects in the MALDI readout (Figure S5B) but presumably due to Hsp33 directly protecting these CS sites against proteolysis (Figures 5A and 5B). These results are remarkably consistent with our peptide array studies, which showed high-affinity binding to peptides spanning helices α1, α12, α13, and α15 (Figure 5B). They strongly suggested that Hsp33ox interacts with parts of CS’s hydrophobic dimerization interface (Figure S5A).
Figure 5. Reduction of Hsp33-Substrate Complexes Destabilizes Bound Substrate Proteins.
(A) Representative SUPREX curves of CS fragments obtained from native CS (black) or thermally unfolded CS in complex with Hsp33ox before (red) or after (blue) complex reduction with 5 mM DTT. Solid lines represent best fits of the data. Secondary structure elements of CS are shown for reference. Peptide regions that show a significant change in relative thermodynamic stability are indicated by red bars, whereas regions that do not significantly change are indicated by pink bars (see also Table S4). Regions of CS defined as good binders by peptide array are shown by dark cyan bars, whereas regions of CS involved in monomer-monomer interactions are indicated by purple bars. Stars depict active site residues.
(B) Comparison of the relative thermodynamic stability profile of CS peptides obtained from SUPREX analysis in native CS, in thermally unfolded CS in complex with Hsp33ox ([Hsp33ox-CS]), or upon DTT reduction ([Hsp33-CS]red). All identified peptides were mapped onto the CS structure (PDB: 2CTS), and changes in relative thermodynamic stability are depicted as color changes from pink to red as stability decreases. Right panel: Localization of Hsp33’s good binders in CS structure as defined by peptide array is depicted in green.
(C) Comparison of limited proteolysis profiles of CS, MDH, or luciferase either in native form or upon thermal unfolding in the presence of Hsp33ox before or after reduction with DTT. Peptides detected in native form and upon complex formation are depicted in green, peptides found in complex both before and after reduction are shown in blue, and peptides that are only detected after complex reduction are indicated in orange. Regions of CS defined as good binders by peptide array are indicated by dark cyan bars. Peptides of CS identified in SUPREX analysis are indicated by red and pink bars, respectively (see Figure 5A for reference).
To assess whether reduction of the Hsp33-substrate complex affects the conformation of CS, we treated the complex with DTT for 30 min ([Hsp33-S]red). This incubation is sufficient to reduce Hsp33’s disulfide bonds (Hoffmann et al., 2004) and causes moderate conformational rearrangements in Hsp33, particularly in its zinc-binding domain (Figure 4A). Importantly, complex formation is maintained unless the DnaK system is added (Hoffmann et al., 2004). Upon reduction of the Hsp33ox-CS complex, we found that peptides covering the four helices α12–α15 (aa 269–351) of CS all lacked transition regions and showed significant loss in the number of globally and/or subglobally protected amide protons (Figure 5A and Table S4). Such a loss of protected amides is most likely a result of a significant destabilization and/or structural changes of the monomer-monomer interface upon reduction of the Hsp33-substrate complex. Limited proteolysis analysis of CS, MDH, or luciferase in complex with Hsp33ox yielded qualitatively very similar results (Figure 5C). In each case, return to non-stress conditions uncovered proteolytic sites in the substrate proteins that were inaccessible in the native substrates or upon complex formation with Hsp33ox. Notably, thiol trapping experiments of the thermally unfolded substrate proteins in complex with Hsp33ox before and after complex reduction excluded significant thiol oxidation within the substrate proteins, implying that the conformational changes that we observed in the client proteins are induced by Hsp33 reduction and not by the reduction of the bound proteins. These results strongly suggest that reduction-mediated refolding of Hsp33’s C-terminal redox-sensor domain triggers conformational rearrangements in the bound proteins that are consistent with increased substrate unfolding and the loss of stabilizing Hsp33-substrate interactions. These appear to be the necessary prerequisites for the successful transfer of folding intermediates to the DnaK system.
DISCUSSION
Intrinsically Disordered Regions Confer “Folding-State” Specificity to Substrate Binding
Hsp33 is a member of a new group of unrelated ATP-independent chaperones, which become rapidly activated when cells encounter particularly problematic stress conditions that cause both widespread protein unfolding and inactivation of essential housekeeping chaperones (Jakob et al., 1999; Tapley et al., 2009). One common feature that these stress-specific chaperones share is their ability to quickly convert large parts of their structure into intrinsically disordered protein segments in response to activating stress conditions. Only upon reaching this state of native disorder, either stress-induced or by rational design (Cremers et al., 2010), do these chaperones gain their full substrate-binding potential. Based on the widely accepted notion that molecular chaperones utilize distinct hydrophobic binding sites (Tyedmers et al., 2010), it was thought that these chaperones contain specialized hydrophobic surfaces that serve double duty: as stabilizing interfaces for metastable protein segments (i.e., Hsp33) or monomer-monomer interactions (i.e., HdeA) under nonstress conditions and as high-affinity substrate-binding sites for unfolding proteins under stress conditions (Ilbert et al., 2007; Tapley et al., 2009). This model, however, raised several conceptual questions, including why these chaperones would prefer binding of unfolding substrate proteins over interacting with their own intrinsically disordered regions. By studying the redox-regulated chaperone Hsp33, we have uncovered one solution to this apparent quandary. We found that activated Hsp33 uses parts of its own intrinsically disordered segment, an ~50 aa linker region, as interaction sites for binding early protein-unfolding intermediates. By using two unrelated methods, limited proteolysis and SUPREX analysis, both combined with mass spectrometry, we independently confirmed significant conformational rearrangements and stabilization of this intrinsically disordered region upon interaction with two different substrate proteins. Involving these intrinsically disordered segments in substrate binding provides large flexibility in substrate recognition without requiring sequence specificity. These results concur with previous studies on the small heat shock protein PsHsp18-1, which showed that different unfolded substrate proteins crosslink most strongly with amino acids of the flexible N terminus (Jaya et al., 2009).
Our binding studies revealed that Hsp33 preferentially binds early unfolding intermediates with residual secondary structure. This result is consistent with the binding specificity of many other intrinsically disordered proteins whose binding partners (shown to be native proteins, DNA, or membranes) are thought to act as stabilizing scaffolds that promote the refolding of the disordered regions (Tompa and Csermely, 2004). Based on the conformation-selection theory proposed for intrinsically disordered proteins, it is entirely possible that secondary structure elements, which are abundantly present in early unfolding intermediates, capture native-like states of Hsp33’s linker region, thus increasing linker stability. These results strongly suggest that Hsp33’s high specificity for structured early unfolding intermediates (i.e., folding-state specificity) is guided by their ability to act as folding scaffolds for the intrinsically disordered linker of Hsp33. The DnaK system, however, whose primary role is to support the folding of nascent polypeptide chains, has a discrete peptide-binding pocket that is well-suited to bind short extended peptides largely devoid of secondary structure elements (Chen et al., 2006). These results serve to illustrate how the peptide-binding specificities of distinct classes of chaperones have adapted to serve specific physiological needs.
Reversible Order-to-Disorder Transition Controls Substrate Binding and Release
Holdases like Hsp33 or sHsps rely on foldases to support the refolding of their bound substrate proteins upon return to non-stress conditions. This raises the question as to how substrate transfer is coordinated between these chaperone systems, given the apparent difference in binding specificity. One model, the “entropy transfer model,” which was proposed by Csermely and Tompa, states that refolding of intrinsically disordered regions upon substrate binding might provide the entropic energy necessary to unfold the bound substrate proteins, thus returning them onto a more productive folding pathway (Tompa and Csermely, 2004). However, because unfolding intermediates do not occupy discrete conformational states and are highly prone to form insoluble aggregates, structural or thermodynamic differences between unfolding proteins in the chaperone-free and chaperone-bound forms are very difficult to determine. Fortunately, the unique nature of Hsp33’s substrate-binding and release mechanism provided us with the exciting opportunity to directly compare two distinct, soluble substrate protein populations. One population is freshly unfolded in the presence of Hsp33ox and is protected against further protein unfolding and aggregation but is unable to be released to and/or interact with the DnaK system. The second population, generated upon return of Hsp33-substrate complexes to a reducing environment, is also protected against spontaneous dissociation but is rapidly released and refolded when the DnaK system is present. Thermodynamic analysis and limited proteolysis studies revealed substantial destabilization in the second population of substrates. These results suggest that the same thermodynamic linkage that exists between the redox status of Hsp33’s cysteines and the stability of Hsp33’s linker region and that is responsible for the unfolding of Hsp33’s linker region upon disulfide bond formation (Ilbert et al., 2007) acts during the inactivation process of Hsp33 and is utilized to shift the equilibrium of the bound substrate protein toward a more unfolded, destabilized conformation (Figure 6). Increased destabilization in the substrate proteins concomitant with increased stability in the linker region will inevitably decrease Hsp33’s affinity for the substrate proteins, marking them for release once DnaK is present. A similar mechanism might also apply to the acid-activated chaperone HdeA, in which return to neutral pH triggers the disorder-order transition of the chaperone, sufficient to cause the slow release of substrate proteins in a folding-competent conformation (Tapley et al., 2010). Our results provide mechanistic insights into how, in the absence of ATP, interactions between natively unfolded regions and early protein-unfolding intermediates are used to translate environmental changes into conformational rearrangements in the substrate proteins that are conducive to productive refolding.
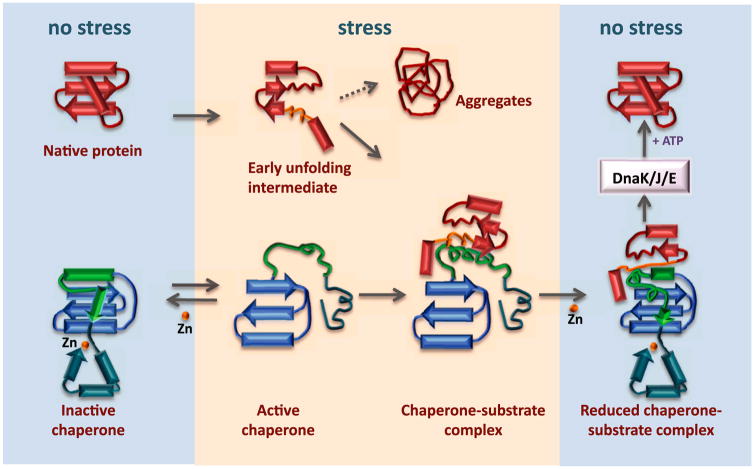
Figure 6. Schematic Model of Hsp33 as an Example of Stress-Specific Intrinsically Disordered Chaperones.
Hsp33 senses unfolding conditions by converting stably folded regions into intrinsically disordered segments (green and cyan), which participate directly in the highly flexible binding of a wide variety of early unfolding intermediates. Client protein binding confers stability to intrinsically disordered regions of the chaperone. Return to nonstress conditions further stabilizes the chaperone (cyan regions), leading to destabilization/unfolding of the bound client protein, which appears to be necessary for substrate release and refolding by foldases. To simplify the model, the dimerization equilibrium of active Hsp33 was omitted.
EXPERIMENTAL PROCEDURES
Protein Purification
Purification of wild-type Hsp33, Hsp33C-term, DnaK, and DnaJ and preparation of inactive reduced and active HOCl-oxidized Hsp33 were conducted as described (Graf et al., 2004; Winter et al., 2008). PepNuoC was synthesized by Genscript and included an added tyrosine residue for peptide concentration determination. Peptides described in Table S2B were obtained from American Peptide Company or Genscript. They were either diluted in DMSO (Pep2–5) or in buffer (Pep1, Pep6–8). Peptide concentrations were determined using the appropriate extinction coefficient or, in the absence of Tyr or Trp residues, were estimated based on the amount of peptide provided by the company. Arc repressor protein was generously provided by Dr. Lewis Kay.
Screening of Peptide Array for Hsp33 Binding
The peptide library comprised of 3,914 peptides of 12 aa was synthesized by LC Sciences using PepArray technology (Pellois et al., 2002). Hsp33 binding was visualized with Cy5-labeled anti-Hsp33 antibodies. Details about data analysis and the development of the Hsp33-binding predictor are described in the Extended Experimental Procedures.
Binding Affinity Measurements with SPR
Kinetic constants for peptide binding were determined by SPR using the ProteOn XPR36 Protein Interaction Array System (Bio-Rad) in KH2PO4 (pH 7.4) with 0.003% surfactant P20 at 25°C. PepNuoC or Hsp33C-term were immobilized by amine coupling to the GLC sensor chip. Hsp33red or Hsp33ox were applied at six different concentrations (0–1.5 μM) using the simultaneous application mode. The binding sensorgrams were analyzed with BIAevaluation (Biacore).
Chaperone Competition Assays
The influence of Hsp33 or DnaK/DnaJ on the aggregation of chemically denatured CS (Sigma) or luciferase (Promega) was determined as described (Graf et al., 2004), using chaperone and competitor concentrations given in the figure legends. For competition studies, chaperones were preincubated with the competitors for 4 min (peptides), 2 min (Hsp33C-term, α-casein), or 10 s (un-folding Arc) at 30°C before CS or luciferase was added. Light scattering was monitored in 40 mM HEPES (pH 7.5) at λex/λem of 360 nm using a Hitachi F4500 fluorescence spectrophotometer equipped with a temperature-controlled cuvette holder and stirrer. Competition assays with PepNuoC were conducted in 0.003% Tween P20 to increase solubility of PepNuoC. Tryptophan fluorescence of 0.75 μM Arc was followed in 40 mM HEPES (pH 7.5) to monitor Arc unfolding. λex 295 nm, λem 332 nm, and ex/em slits of 10/5 nm were used.
Formation of Hsp33-Substrate Complexes
Complexes between Hsp33 and thermally unfolded CS, malate L-dehydrogenase (MDH) (Roche), or luciferase were formed as described (Xu et al., 2010). The final Hsp33 concentration was 3–5 μM for limited proteolysis or 114–124 μM for SUPREX.
Limited Proteolysis Coupled with LC-MS
Tryptic digests were carried out at 1:10 ratios of trypsin to protein in 40 mM K2HPO4 (pH 7.4), 25°C. Aliquots were taken at defined time points (0–15 min), and the digest was stopped with 10% TFA. Samples were applied to a reverse phase C18 column (Zorbax 300SB-C18, 1 × 50 mm, 3.5 μ) coupled to a Q-TOF (Agilent) dual ESI LC-MS for peptide identification. Peptides were eluted using a 2%–80% gradient of acetonitrile at a flow rate of 0.3 μl/min at 30°C in 15 min. The BioConfirm software (Agilent) was used for peptide identification.
SUPREX Analysis
Detailed protocols of our SUPREX analysis are provided in the Extended Experimental Procedures. In short, a series of H/D exchange buffers containing 20 mM Na2DPO4, pD 7.4 and different GdmCl concentrations were prepared. H/D exchange reactions were started by diluting proteins 1:10 into SUPREX buffers. At specified exchange times, reactions were quenched with HCl (pH 0.9), followed by a 30 s pepsin digest. One microliter of the digested samples was added to 9 μl matrix solution and spotted onto a MALDI target for MS analysis. The uptake of deuterons (i.e., Δmass) was calculated for each singly charged peptide by subtracting the undeuterated peptide mass from the deuterated peptide mass determined in the MALDI-TOF analysis. Ten MALDI spectra were collected at each denaturant concentration, and Δmass values were averaged. SUPREX curves (plots of Δmass versus [GdmCl]) were generated for each peptide and fitted to a four-parameter sigmoidal equation using a nonlinear regression routine in SigmaPlot to obtain the C1/2 value, which represents the denaturant concentration at the transition midpoint.
Supplementary Material
Acknowledgments
We thank Dr. Rüdiger for providing us with the raw data of the DnaK peptide array, Drs. Yosseff and Schreiber for the use of ProteOn and the analysis software, and Dr. Kay for purified Arc repressor protein. We thank Dr. Mayer for running the DnaK binding predictor on the CS sequence. We are grateful to Drs. Bardwell, Xu, and Walter for critically commenting on the manuscript. This project was supported by National Institutes of Health grants GM065318 to U.J. and GM084174 to M.C.F. and by the European Molecular Biology Organization and Human Frontiers postdoctoral fellowship to D.R.
Footnotes
Supplemental Information includes Extended Experimental Procedures, five figures, and four tables and can be found with this article online at doi:10.1016/j.cell.2012.01.045.
References
- Bowie JU, Sauer RT. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989;28:7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- Chen Z, Kurt N, Rajagopalan S, Cavagnero S. Secondary structure mapping of DnaK-bound protein fragments: chain helicity and local helix unwinding at the binding site. Biochemistry. 2006;45:12325–12333. doi: 10.1021/bi0612263. [DOI] [PubMed] [Google Scholar]
- Cremers CM, Reichmann D, Hausmann J, Ilbert M, Jakob U. Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. J Biol Chem. 2010;285:11243–11251. doi: 10.1074/jbc.M109.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- Gaspar AM, Appavou MS, Busch S, Unruh T, Doster W. Dynamics of well-folded and natively disordered proteins in solution: a time-of-flight neutron scattering study. Eur Biophys J. 2008;37:573–582. doi: 10.1007/s00249-008-0266-3. [DOI] [PubMed] [Google Scholar]
- Graf PC, Martinez-Yamout M, VanHaerents S, Lilie H, Dyson HJ, Jakob U. Activation of the redox-regulated chaperone Hsp33 by domain unfolding. J Biol Chem. 2004;279:20529–20538. doi: 10.1074/jbc.M401764200. [DOI] [PubMed] [Google Scholar]
- Graumann J, Lilie H, Tang X, Tucker KA, Hoffmann JH, Vijayalakshmi J, Saper M, Bardwell JC, Jakob U. Activation of the redox-regulated molecular chaperone Hsp33—a two-step mechanism. Structure. 2001;9:377–387. doi: 10.1016/s0969-2126(01)00599-8. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U. Identification of a redox-regulated chaperone network. EMBO J. 2004;23:160–168. doi: 10.1038/sj.emboj.7600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci USA. 2009;106:15604–15609. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- Pellois JP, Zhou X, Srivannavit O, Zhou T, Gulari E, Gao X. Individually addressable parallel peptide synthesis on microchips. Nat Biotechnol. 2002;20:922–926. doi: 10.1038/nbt723. [DOI] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Hopper ED, Tong Y, Sadowsky JD, Peterson KJ, Gellman SH, Fitzgerald MC. H/D exchange- and mass spectrometry-based strategy for the thermodynamic analysis of protein-ligand binding. Anal Chem. 2007;79:5869–5877. doi: 10.1021/ac0700777. [DOI] [PubMed] [Google Scholar]
- Tapley TL, Körner JL, Barge MT, Hupfeld J, Schauerte JA, Gafni A, Jakob U, Bardwell JC. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc Natl Acad Sci USA. 2009;106:5557–5562. doi: 10.1073/pnas.0811811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley TL, Franzmann TM, Chakraborty S, Jakob U, Bardwell JC. Protein refolding by pH-triggered chaperone binding and release. Proc Natl Acad Sci USA. 2010;107:1071–1076. doi: 10.1073/pnas.0911610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- Voelz VA, Shell MS, Dill KA. Predicting peptide structures in native proteins from physical simulations of fragments. PLoS Comput Biol. 2009;5:e1000281. doi: 10.1371/journal.pcbi.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Schmitt S, Tang L, Jakob U, Fitzgerald MC. Thermodynamic analysis of a molecular chaperone binding to unfolded protein substrates. Biochemistry. 2010;49:1346–1353. doi: 10.1021/bi902010t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.