Energy is a basic requirement for growth of all cells. It has been estimated that during aerobic growth of bacteria on simple carbon sources, approximately 50 grams of ATP are consumed for each gram dry weight of cells produced (1). Energy for ATP synthesis is derived from electron transport through the chemiosmotic scheme proposed by Mitchell (2). During electron transport, protons are expelled from the cell, creating a difference in proton concentration (ΔpH) and electrical charge or membrane potential (Δψ) across the cytoplasmic membrane. These potential energies, collectively termed proton motive force (Δp), are used for a variety of energy-consuming processes, including synthesis of ATP. During aerobic growth, oxygen serves as the terminal electron acceptor of the electron transport chain with approximately 1 mol of oxygen atoms consumed for every 2 mol of ATP produced.
Oxygen is essential for efficient generation of cellular energy in aerobically grown cells, but it is also a cause of toxicity due to production of reactive superoxide and hydroxyl radicals. Thus, it is not surprising that motile cells have developed strategies for seeking environments with optimal concentrations of oxygen. This phenomenon, called aerotaxis, was documented over a century ago by Engelmann, who observed accumulation of bacteria around air bubbles (3). Beginning in the 1960s (4), the molecular basis of aerotaxis and chemotaxis has been actively investigated. A central issue has been the mechanism by which cells detect oxygen gradients and transduce signals that direct migration during aerotaxis (5). A major step toward answering this question recently has been provided by the identification of a novel sensor Aer (6, 7), and a new role ascribed to the previously characterized serine chemoreceptor, Tsr, described by Rebbapragada et al. (6) in the current issue of the Proceedings.
Aer, the sensor/signal transducer named for aerotaxis and energy responses, was identified independently by Rebbapragada et al. (6) and Bibikov et al. (7) as the product of an ORF discovered in the Escherichia coli genome sequencing project (8). The encoded 506-aa protein exhibits the basic features that had been predicted for the elusive aerotaxis sensor/transducer. Specifically, the sequence contains a hydrophobic region that could anchor the protein to the cytoplasmic membrane, an oxygen-sensing domain homologous to domains of other oxygen sensors and a signal-transducing domain homologous to those of chemoreceptors (Fig. 1). Existence of the latter domain was expected based on the observation that aerotaxis requires the phosphotransfer signaling components CheW, CheA, and CheY that comprise the chemotaxis signal transduction pathway (9). It is perhaps notable that Aer, so readily identifiable from its sequence, had not been previously identified by genetic screens. The advances afforded by identification of this new sensor/transducer provide another example of the depth of information that can be gleaned from genome sequencing.
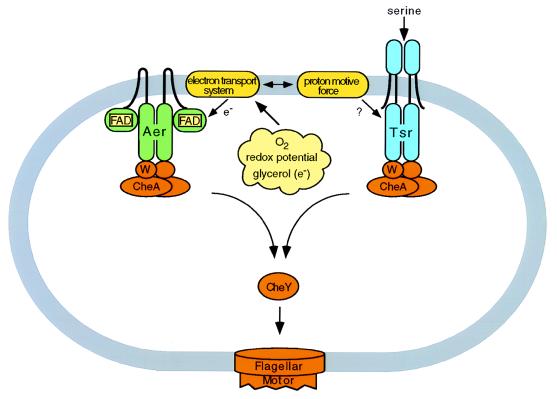
Figure 1.
Schematic diagram of the aerotaxis mechanism proposed by Rebbapragada et al. (6). The recently identified sensor Aer (depicted here as a dimer by analogy to the oligomeric structure of chemoreceptors) and the serine chemoreceptor Tsr mediate taxis responses to oxygen, redox effectors, and glycerol, substances that modulate electron transport and proton motive force. Homologous signaling domains relay information to the cytoplasmic phosphotransfer signaling components CheW, CheA, and CheY.
Identification of the aer gene provided a foundation for examining its role in aerotaxis. In a variety of behavioral assays, cells with disrupted aer genes were shown to exhibit decreased aerotaxis, while maintaining normal chemotaxis toward sugars and amino acids (6, 7). Aerotaxis could be restored by introduction of a plasmid-encoded aer gene, further supporting the identification of Aer as an aerotaxis transducer. However, the lack of complete ablation of the aerotaxis response in aer-deficient cells led Rebbapragada et al. to postulate the existence of an additional transducer for aerotaxis. Earlier studies, examining the complementation of mutants with aberrant aerotaxis, had suggested a role for the serine chemoreceptor/transducer Tsr (10). Although a tsr mutant showed normal aerotactic behavior, aerotaxis was completely abolished in an aer tsr double mutant and could be restored by either plasmid-encoded aer or tsr. From these observations Rebbapragada et al. conclude that Aer and Tsr function as independent sensor/transducers for aerotaxis.
The Aer and Tsr sensor/transducers allow cells to migrate with respect to oxygen gradients in search of environments where the concentration of oxygen is neither too high nor too low. But what chemical or physical entity is being sensed by these proteins? From data accumulated over a number of years, Taylor’s group has begun to hone in on the signal. Aerotaxis has been shown to require a functional electron transport chain (11). Additionally, cells respond to other compounds, that like oxygen, affect the electron transport chain, including redox effectors such as quinone analogs (electron carriers) and rapidly metabolized carbon sources such as glycerol (electron donors) (12, 13). Responses to all of these compounds were abolished in the aer tsr double mutant (6). Thus, Aer and Tsr appear to be sensors of the cellular energy state, with increased or decreased energy levels causing positive or negative taxis, respectively. The tight coupling between electron transport and the proton motive force preclude resolution of which specifically provides the signal for energy-dependent behavior.
The N-terminal 120-residue sensor domain of Aer is homologous to domains of NifL (14) and other proteins known to sense oxygen. Both NifL, which regulates transcription of nitrogen-fixation genes in a redox-dependent manner (15), and Aer (6, 7) have been shown to contain noncovalently associated FAD. It seems likely that Aer uses FAD to monitor redox potential, possibly through exchange of electrons with components of the electron transport chain.
Recent findings suggest that the energy-sensing mechanism used by Aer may be universal. Aer contains a PAS domain (16), previously proposed to be involved in protein–protein interactions in proteins associated with photoreception and circadian clocks in species ranging from bacteria to mammals (17). Within the PAS domain, Zhulin et al. (16) have identified highly conserved regions of approximately 30 amino acids that have been designated S-boxes for their putative role as sensory motifs. S-boxes have been identified within a large family of proteins with representatives from all kingdoms. The common functional feature shared by these proteins is the sensing of some form of energy-related stimuli such as oxygen, light, or redox. This finding suggests a mechanism for signaling by PAS domain proteins in which energy-related stimuli sensed through the S-box motifs may be further transmitted through protein–protein interactions of the PAS domains.
The intramolecular signaling mechanism of Aer remains to be defined. The C-terminal domain of Aer is very similar to the signaling domains of the chemotaxis sensor/transducer proteins that interact with proteins of the phosphotransfer signal transduction pathway that carries information to the flagellar motors. Like the chemoreceptors, Aer also contains an N-terminal sensing domain and transmembrane regions. But the chemoreceptors have periplasmic sensor domains and cytoplasmic signaling domains, whereas both domains of Aer are predicted to be located in the cytoplasm. In the chemoreceptors, the importance of the transmembrane regions in transmitting signals between the periplasmic ligand-binding domains and the cytoplasmic signaling domains has been clearly established (18–20). The topological arrangement of Aer raises some interesting questions. Given the similarity in the signaling domains of Aer and the chemoreceptors it is logical to expect that similar conformational changes are involved in communication with the downstream signaling components, but are the transmembrane regions of Aer involved in this process? Or alternatively, as previously proposed for archaebacterial transducers (21, 22), could direct domain–domain interactions within the cytoplasm produce similar conformational changes as those resulting from transmembrane signaling in the chemoreceptors?
An even larger mystery is posed by the newly ascribed energy-sensing role of the chemoreceptor Tsr. This role adds yet another category of stimuli to the responses mediated by Tsr, a list that already includes serine, external pH, weak acid repellents (cytoplasmic pH), temperature, hydrophobic amino acids, and indole (23). Tsr contains no redox-sensitive cofactor, thus it is possible that proton motive force rather than direct interaction with the electron transport chain is used by Tsr to monitor cellular energy levels. As Tsr contains no specialized energy-sensing domain it seems that energy sensing has been acquired through a minor adaptation of some region present in other chemoreceptors, which despite a high degree of sequence similarity, apparently do not have this capability.
Like the discovery of most new genes, the identification of Aer opens new directions for research. Further investigations of Aer undoubtedly will provide insight into details of aerotaxis and the maintenance of optimal energy levels in bacteria. The existence of similar sensor motifs in Aer and other proteins raises the possibility that the molecular mechanism used by Aer may represent a common strategy used by cells in all organisms for monitoring their intracellular energy state.
References
- 1.Bauchop T, Elsden S R. J Gen Microbiol. 1960;23:457–469. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P. Nature (London) 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 3.Engelmann T W. Pflüegers Arch Gesamte Physiol Menschen Tiere. 1881;25:285–292. [Google Scholar]
- 4.Adler J. J Bacteriol. 1966;92:121–129. doi: 10.1128/jb.92.1.121-129.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor B L. Trends Biochem Sci. 1983;8:438–441. [Google Scholar]
- 6.Rebbapragada A, Johnson M S, Harding G P, Zuccarelli A J, Fletcher H M, Zhulin I B, Taylor B L. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibikov S I, Biran R, Rudd K E, Parkinson J S. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels D L, Plunkett G, Burland V, Blattner F R. Science. 1992;257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 9.Roswell E H, Smith J M, Wolfe A, Taylor B L. J Bacteriol. 1995;177:6011–6014. doi: 10.1128/jb.177.20.6011-6014.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang C V, Niwano M, Ryu J-I, Taylor B L. J Bacteriol. 1986;166:275–280. doi: 10.1128/jb.166.1.275-280.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazlo D J, Taylor B L. J Bacteriol. 1981;145:990–1001. doi: 10.1128/jb.145.2.990-1001.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bespalov V A, Zhulin I B, Taylor B L. Proc Natl Acad Sci USA. 1996;93:10084–10089. doi: 10.1073/pnas.93.19.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhulin I B, Roswell E H, Johnson M S, Taylor B L. J Bacteriol. 1997;179:3196–3201. doi: 10.1128/jb.179.10.3196-3201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco G, Drummond M, Woodley P, Kennedy C. Mol Microbiol. 1993;9:869–879. doi: 10.1111/j.1365-2958.1993.tb01745.x. [DOI] [PubMed] [Google Scholar]
- 15.Hill S, Austin S, Eydmann T, Jones T, Dixon R. Proc Natl Acad Sci USA. 1996;93:2143–2148. doi: 10.1073/pnas.93.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhulin I B, Taylor B L, Dixon R. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 17.Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 18.Falke J J, Koshland D E., Jr Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- 19.Lynch B A, Koshland D E., Jr Proc Natl Acad Sci USA. 1991;88:10402–10406. doi: 10.1073/pnas.88.23.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee G F, Dutton D P, Hazelbauer G L. Proc Natl Acad Sci USA. 1995;92:5416–5420. doi: 10.1073/pnas.92.12.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao V J, Spudich J L. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krah M, Marwan W, Oesterhelt D. FEBS Lett. 1994;353:301–304. doi: 10.1016/0014-5793(94)01068-4. [DOI] [PubMed] [Google Scholar]
- 23.Macnab R M. In: Escherichia coli and Salmonella typhimurium: Molecular and Cellular Biology. Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Washington, D.C.: Am. Soc. Microbiol.; 1987. pp. 732–759. [Google Scholar]