Abstract
Analysis of the local immunological microenvironment in colorectal cancer lesions yielded prognostic markers. Harnessing these insights for clinical application however requires the use of sophisticated technology and algorithms, especially the robust and reproducible quantification of immune cells. These technologies are available and will allow individualized treatment decisions beyond the current standard.
Keywords: chemotherapy, colorectal cancer, immunomap, lymphocytes, prognosis, T cells, whole slide imaging
The role of the immune system in the course of cancer—and especially colorectal cancer—has been debated for decades. Recent data support a prognostic role of the local tumor microenvironment to the point, where an application in clinical settings should be discussed. The technical and immunological aspects that govern such an endavour are discussed here and show the development in this field.
Historically, the lack of appropriate tools for analysis of immune cell infiltrates precluded precise investigations in the past. Therefore, the presence of immune cells in cancer lesions was viewed as a general inflammatory response promoting cancer growth or a specific immune response with respect to the immunosurveillance theory or it was simply dismissed as not relevant. In fact, pathologists typically do not evaluate the immune cell presence, but it were pathologists who also noted the association between the presence of large infiltrates and a better prognosis in colorectal cancer patients.1 With more sophisticated methodology at hand, analyses became more focused on the immunological setup within the cancer microenvironment and factors weighing in on the interaction between immune cells and cancer cells were better characterized.2-4 The use of immunohistochemistry to delineate the effects of different immune cell populations and their spatial distribution within and around cancer lesions became more sophisticated and generally identified T cells as a driving force behind a better prognosis in colorectal cancer patients.5-9 The distinct interplay between immune cell subpopulations and their diverging roles allowed a better differentiation: dendritic cells, macrophages (with anti-tumor properties), Th1 T cells and especially cytotoxic T cells and natural killer cells are seen as protective factors for the host, while cancer-associated macrophages (formerly termed M2), myeloid-derived suppressor cells, neutrophils, Th2 and Th17 T cells, and FOXP3-positive regulatory T (Treg) cells are seen as cancer-promoting.10,11 The precise function and effect on cancer cells and other immune cells of the above mentioned cells in different cancer entities, their composition and the identification of novel phenotypic subpopulations is still ongoing and highlights the complexity of the microenvironment.
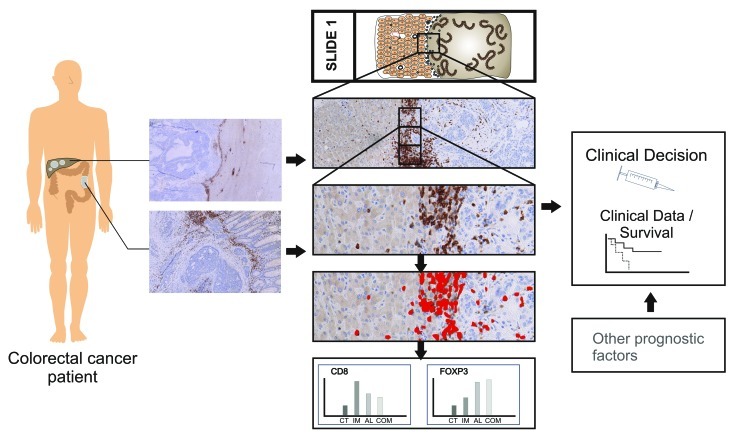
Following the early observations more sophisticated and systematic analyses on large cohorts of patients were conducted.12 These elegant studies then could convincingly identify the prognostic role of T cell infiltrates in the center and at the invasive margin of the primary tumor.13 Further analysis of specific stages of colorectal cancer brought more insight into the effects of different immune cell populations.14-16 The role of regulatory T cells within the colorectal cancer microenvironment remains controversial. These cells are mainly identified by FOXP3 expression and their presence is either attributed to a favorable prognosis or a worse prognosis. Further delineation of regulatory T cell and regulatory immune cell subpopulations and better ways to identify these will most likely yield clarifying insights. This matter however highlights a fundamental problem in the analysis of cells on histology sections, especially immune cells. Quantification of immune cells with robust and reproducible results is problematic for human observers. This problem of quantification is long known and leads to difficulties in reproducibility and robustness. Human observers are especially good at discerning extremes: high densities vs. low densities. But gradients beyond “black and white” are very problematic, an issue that is prominently present in HER2/neu quantification. Even for direct counting of low cell numbers, the reproducibility for the same observer is low.17 It is impossible for a human observer to reliably count cells in conglomerates and as such a semi-quantitative estimation is the common solution. However, all approaches with one ore more human observers are extremely time consuming and can only analyze a minute fraction of the actual tumor tissue. A solution to this is the use of computational image analyses, where the quantification is based directly on morphological and spectral information of detected cells.18 Using thin sections, one can ascertain that no overlapping cells in conglomerates are present. Conglomerates are analyzed based on their area and conformation and a statistical dataset then allows the reproducible deduction of the immune cells present within. Robustness and reproducibility are however only one side of the benefits of an automated quantification algorithm. Coupling this methodology to whole slide scanning, the question of immune cell heterogeneity can be adressed systematically. Using an artifical grid as overlay, regions of 1 mm2 area can be quantified and the whole lattice then can be visualized (Fig. 1). The observed heterogeneity for primary colorectal cancer showed enormous variability and in the end leads to a technical question: can we reliably measure the cell numbers based on a single selected area of approximately 1 mm2? A typical core from a tissue microarray has a surface area of 0.3 mm2. The answer for individual patients is: no, the analyzed area has to be much larger for robustness.19 The average number of CD3+ T cells converges if more then five fields of 1 mm2 are analyzed for primary colorectal cancer. So it is necessary to analyse a tissue surface area large enough to achieve reliability for a single patient. This immune cell heterogeneity within the tumor tissue is evident, even when corrected for e.g., necrotic areas.19 It is important to see, that for each tumor entity and for each marker analyzed (e.g., CD3, FOXP3, CD8, CD45RO, etc.), this minimum surface tissue area has to be calculated for robustness. Combining whole slide imaging with a powerful image analysis algorithm allows the robustness and reproducibility needed for personalized diagnostics. Coupling these technologies to a dedicated tissue preparation workflow is going to deliver a basis for treatment decisions (Fig. 2). Colorectal cancer prognosis is influenced by the presence of T cells in the stroma, within the invasive front, and in the parenchyma, in an intraepithelial localization. So what is the best region for quantification: the invasive margin or the center of the tumor, or the stroma? Heterogeneity is present in all regions and one of the obvious aspects is the accumulation of immune cells in distinct regions or compartments. The stroma, either peritumoral or within the tumor lesion, harbors the vast majority of immune cells, only small percentages of T cells are in close contact with the tumor epithelium.20 In contrast, macrophage populations are in direct contact with a high percentage of infiltrating T cells.3 Furthermore, the presence or absence of broad peritumoral stroma does not automatically dictate the quantity of immune cells present in the tumor or around the tumor.20
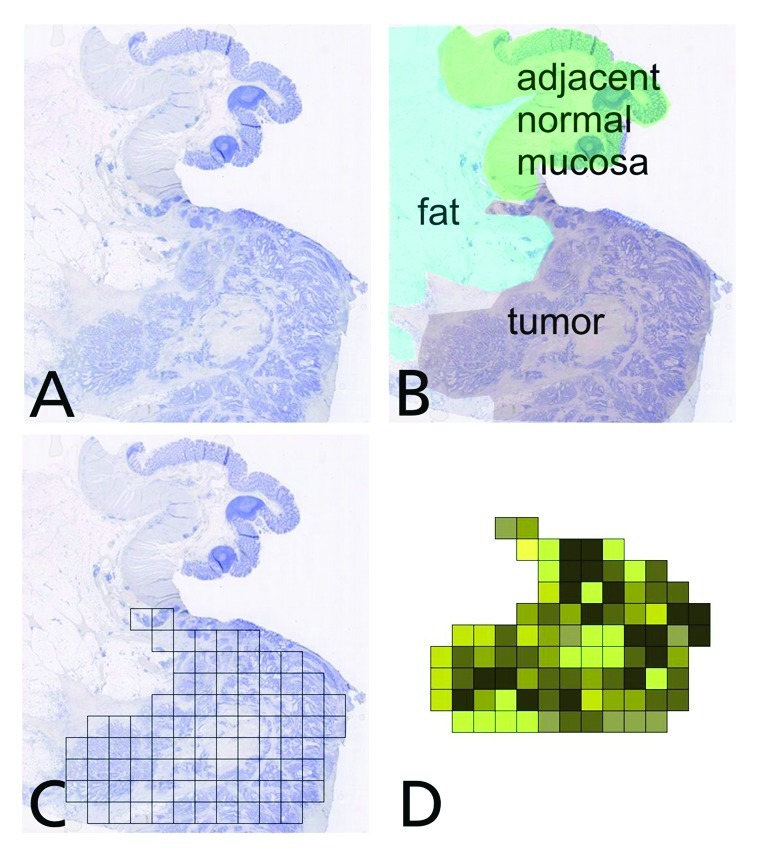
Figure 1. Workflow example for the generation of a whole slide “immunomap”, visualizing the immune cell density and heterogeneity across the tissue section. (A) Overview of the tissue section (B) Different regions of interest for analysis (C) Grid construction on the tumor lesion (grid size is typcially 1 mm2 per tile) (D) Coloring of grid tiles according to immune cell quantities revealing the heterogeneity.
Figure 2. Individualized analysis of the immune cell profiles for the stratification of patients.
Colorectal cancer liver metastases have not been analyzed often with a focus on immunology.21,22 For the medical oncologist a patient with advanced cancer and liver metastases however is a daily challenge. While surgical approaches and chemotherapy regimens have improved, the prognosis for patients with irresectable metastatic disease is still around 24 mo. The tantalizing data from the large systematic analyses showed that there are indeed at least two immunological patient subgroups for each UICC stage: patients with a high infiltrate density and patients with a low infiltrate density. The latter fare much worse and their prognosis is sometimes even worse then the prognosis for patients with a more advanced UICC stage. So the next question was: how is the local immune infiltration in colorectal cancer liver metastases and is it related to the clinical outcome? One surprising observation was that one could not predict the infiltrate density at the liver metastasis from looking at the primary tumor.20 While sometimes there was concordance in the infiltrate density between primary tumor and metastasis, more often there was discordance. Furthermore it was remarkable that there is a clear border at the invasive margin of liver metastases, showing a strong variability in T cell densities between samples from different patients. The T cell infiltrate at the invasive margin of the liver lesion also did not correspond to the infiltrate density within the metastasis. So all combinations were observed: strong infiltrate at the invasive margin and almost no infiltration within the metastasis, less infiltrate at the invasive margin but strong infiltration of the metastatic lesion, almost no infiltrate at the invasive margin and almost no infiltrate within the metastasis and so on. In metastatic lesions, the microenvironment of the inner tissue is more often governed by necrosis and cell debris, making it difficult to compare these inner regions between different patients. Surprisingly, the invasive margin was found to be informative for prognosis of these patients and is informative in the prediction of chemotherapy response. In a selected cohort of patients, from which samples with the invasive margin of liver metastases could be analyzed, the score based on the infiltrate density of CD3, CD8 or Granzyme B positive immune cells allowed to predict treatment response in these patients.23 While in previous studies, only the completely resected primary tumor was analyzed and therefore the “escaped” tumor cells could not be analyzed,24 the patients for this new analysis either had a diagnostic excision (during resection of the primary tumor or laparoscopy) or in a few cases had a biopsy containing a large stretch of the invasive margin of the liver lesion. So the analysis of the tissue is the analysis of the actual tissue that is then treated with (first line) chemotherapy, allowing for the first time to analyze large stretches of invasive margin and correlate this with the clinical course. In essence, the TIL densities at the invasive margin of liver metastases allowed the prediction of response to chemotherapy with a sensitivity of 79% and specificity of 100%. The association of high density values with longer progression free survival under chemotherapy was also statistically significant. This raises of course several important questions: what is the effect of these cells during or after chemotherapy? Two hypotheses can be proposed: either chemotherapy leads to immunomodulation, i.e., suppression of inhibitory immune cell function, allowing the peritumoral T cells to infiltrate and attack cancer cells. This is a hypothesis that is in line with the cancer immunosurveillance theory developed by R. Schreiber and coworkers.25 Or, as an alternative or additional explanation, chemotherapy induces an immunogenic cell death that leads to renewed activation of the effector immune cells, the recognition of novel epitopes or a general unspecific activation of the immune system and thereby provides new targets to immune cells.10,26,27
Overall, these findings extend the impact of the local immune response on the clinical course from the primary tumor also to metastatic lesions. While untreated reference samples cannot be expected to be available in larger numbers due to ethical reasons, efforts to compile a set of these rare specimens are ongoing. This then can clarify whether the immune infiltrate density is prognostic and predictive in the strictest sense. It also remains to be seen whether this observation from large stretches of invasive margin can be transferred to unselected biopsies from patients with liver metastases from colorectal cancer. The clinical need is there and an improvement beyond the almost 50% chemotherapy failure rate is desireable. A trial will adress this question soon. But also another question rises from these observations: what dictates the presence of these lymphocytes within the tissue and especially at the invasive margin? And can this be exploited? From the primary tumor we have ample data that identifies the cytokines and chemokines involved in this process.28-30 Key factors are CXCL10 and CXCL9, which may attract (memory) T cells as well as CX3CL1 (also known as fractalkine), which may attract Th1 cells to the tumor site. It is surprsing to see, that these cytokine regulation is weakened in colorectal cancer liver metastases and even more surprising to see that natural killer cells are generally scarce in colorecal cancer, despite the presence of activating and recruiting cytokines and chemokines.30 The analysis of the metastatic situation is ongoing and new therapeutic options are of pressing need for the clinician. From a clinical perspective, the measurement of a distinct protein in the peripheral blood that reflects the immunologic situation within the tumor lesion is highly desireable. So far, there has been no good candidate for this application and the major source for relevant parameters for patient stratification is the analysis of the local tumor bed.
In the future, treatment decisions based on immune cell profiling require automated and robust approaches to open the door for personalized medicine in oncology. Multiplex immunohistochemistry on single slides will allow the efficient analysis of samples and speed up the process of quantification. High detail levels in the analysis of immune cells and their cytokines and chemokines are broadening the understanding of the processes in the microenvironment. The interplay between immune cells, antigenicity of the tumor cells and immunosuppressive mechanisms at the local site has the potential to open new therapeutic avenues. Especially in the light of spatial relations and quantitative biology there is plenty of translational research that can be performed.
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/18460
References
- 1.Pihl E, Nairn RC, Milne BJ, Cuthbertson AM, Hughes ES, Rollo A. Lymphoid hyperplasia: a major prognostic feature in 519 cases of colorectal carcinoma. Am J Pathol. 1980;100:469–80. [PMC free article] [PubMed] [Google Scholar]
- 2.McDougall CJ, Ngoi SS, Goldman IS, Godwin T, Felix J, DeCosse JJ, et al. Reduced expression of HLA class I and II antigens in colon cancer. Cancer Res. 1990;50:8023–7. [PubMed] [Google Scholar]
- 3.Ohtani H, Naito Y, Saito K, Nagura H. Expression of costimulatory molecules B7-1 and B7-2 by macrophages along invasive margin of colon cancer: a possible antitumor immunity? Lab Invest. 1997;77:231–41. [PubMed] [Google Scholar]
- 4.Ohtani H. Pathophysiologic significance of host reactions in human cancer tissue: desmoplasia and tumor immunity. Tohoku J Exp Med. 1999;187:193–202. doi: 10.1620/tjem.187.193. [DOI] [PubMed] [Google Scholar]
- 5.Ropponen KM, Eskelinen MJ, Lipponen PK, Alhava E, Kosma VM. Prognostic value of tumour-infiltrating lymphocytes (TILs) in colorectal cancer. J Pathol. 1997;182:318–24. doi: 10.1002/(SICI)1096-9896(199707)182:3<318::AID-PATH862>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen HJ, Hansen U, Christensen IJ, Reimert CM, Brunner N, Moesgaard F. Independent prognostic value of eosinophil and mast cell infiltration in colorectal cancer tissue. J Pathol. 1999;189:487–95. doi: 10.1002/(SICI)1096-9896(199912)189:4<487::AID-PATH484>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 7.Menon AG, Janssen-van Rhijn CM, Morreau H, Putter H, Tollenaar RAEM, van de Velde CJH, et al. Immune system and prognosis in colorectal cancer: a detailed immunohistochemical analysis. Lab Invest. 2004;84:493–501. doi: 10.1038/labinvest.3700055. [DOI] [PubMed] [Google Scholar]
- 8.Prall F, Duhrkop T, Weirich V, Ostwald C, Lenz P, Nizze H, et al. Prognostic role of CD8+ tumor-infiltrating lymphocytes in stage III colorectal cancer with and without microsatellite instability. Hum Pathol. 2004;35:808–16. doi: 10.1016/j.humpath.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 9.Diederichsen ACP. Hjelmborg JvB, Christensen PB, Zeuthen J, Fenger C. Prognostic value of the CD4+/CD8+ ratio of tumour infiltrating lymphocytes in colorectal cancer and HLA-DR expression on tumour cells. Cancer Immunol Immunother. 2003;52:423–8. doi: 10.1007/s00262-003-0388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zitvogel L, Kepp O, Kroemer G. Immune parameters affecting the efficacy of chemotherapeutic regimens. Nat Rev Clin Oncol. 2011;8:151–60. doi: 10.1038/nrclinonc.2010.223. [DOI] [PubMed] [Google Scholar]
- 11.Pagès F, Galon J, Dieu-Nosjean MC, Tartour E, Sautes-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29:1093–102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 12.Pagès F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–66. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 13.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 14.Pagès F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol. 2009;27:5944–51. doi: 10.1200/JCO.2008.19.6147. [DOI] [PubMed] [Google Scholar]
- 15.Salama P, Phillips M, Grieu F, Morris M, Zeps N, Joseph D, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27:168–9. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 16.Sinicrope FA, Rego RL, Ansell SM, Knutson KL, Foster NR, Sargent DJ. Intraepithelial effector (CD3+)/regulatory (FoxP3+) T-cell ratio predicts a clinical outcome of human colon carcinoma. Gastroenterology. 2009;137:1270–9. doi: 10.1053/j.gastro.2009.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs TJ, Wild PJ, Moch H, Buhmann JM. Computational Pathology Analysis of Tissue Microarrays Predicts Survival of Renal Clear Cell Carcinoma Patients. Med Image Comput Comput Assist Interv 200811:1-8 [DOI] [PubMed] [Google Scholar]
- 18.Halama N, Zoernig I, Spille A, Westphal K, Schirmacher P, Jaeger D, et al. Estimation of Immune Cell Densities in Immune Cell Conglomerates: An Approach for High-Throughput Quantification. PLoS ONE. 2009;4:e7847. doi: 10.1371/journal.pone.0007847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halama N, Zoernig I, Spille A, Michel S, Kloor M, Grauling-Halama S, et al. Quantification of prognostic immune cell markers in colorectal cancer using whole slide imaging tumor maps. Anal Quant Cytol Histol. 2010;32:333–40. [PubMed] [Google Scholar]
- 20.Halama N, Michel S, Kloor M, Zoernig I, Pommerencke T, von Knebel Doeberitz M, et al. The localization and density of immune cells in primary tumors of human metastatic colorectal cancer shows an association with response to chemotherapy. Cancer Immun. 2009;9:1. [PMC free article] [PubMed] [Google Scholar]
- 21.Katz SC, Pillarisetty V, Bamboat ZM, Shia J, Hedvat C, Gonen M, et al. T Cell Infiltrate Predicts Long-Term Survival Following Resection of Colorectal Cancer Liver Metastases. Ann Surg Oncol. 2009;16:2524–30. doi: 10.1245/s10434-009-0585-3. [DOI] [PubMed] [Google Scholar]
- 22.Okano K, Maeba T, Moroguchi A, Ishimura K, Karasawa Y, Izuishi K, et al. Lymphocytic infiltration surrounding liver metastases from colorectal cancer. J Surg Oncol. 2003;82:28–33. doi: 10.1002/jso.10188. [DOI] [PubMed] [Google Scholar]
- 23.Halama N, Michel S, Kloor M, Zoernig I, Benner A, Spille A, et al. Localization and Density of Immune Cells in the Invasive Margin of Human Colorectal Cancer Liver Metastases Are Prognostic for Response to Chemotherapy. Cancer Res. 2011;71:5670–7. doi: 10.1158/0008-5472.CAN-11-0268. [DOI] [PubMed] [Google Scholar]
- 24.Morris M, Platell C, Iacopetta B. Tumor-infiltrating lymphocytes and perforation in colon cancer predict positive response to 5-fluorouracil chemotherapy. Clin Cancer Res. 2008;14:1413–7. doi: 10.1158/1078-0432.CCR-07-1994. [DOI] [PubMed] [Google Scholar]
- 25.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 26.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6:715–27. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 27.Zitvogel L, Apetoh L, Ghiringhelli F, Andre F, Tesniere A, Kroemer G. The anticancer immune response: indispensable for therapeutic success? J Clin Invest. 2008;118:1991–2001. doi: 10.1172/JCI35180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mlecnik B, Tosolini M, Charoentong P, Kirilovsky A, Bindea G, Berger A, et al. Biomolecular network reconstruction identifies T cell homing factors associated with survival in colorectal cancer. Gastroenterology. 2010;138:1429–40. doi: 10.1053/j.gastro.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 29.Bindea G, Mlecnik B, Fridman WH, Pages F, Galon J. Natural immunity to cancer in humans. Curr Opin Immunol. 2010;22:215–22. doi: 10.1016/j.coi.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Halama N, Braun M, Kahlert C, Spille A, Quack C, Rahbari N, et al. Natural Killer Cells are Scarce in Colorectal Carcinoma Tissue Despite High Levels of Chemokines and Cytokines. Clin Cancer Res. 2011;17:678–89. doi: 10.1158/1078-0432.CCR-10-2173. [DOI] [PubMed] [Google Scholar]