Abstract
Exosomes are small vesicles secreted in relative abundance by cancer cells, which may prove useful as disease markers. However, exosomes also exhibit potent functions; modulating the behavior of immune- and other cells. Bridging our understanding of their molecular phenotype and functional mechanisms will provide key insight into their importance in cancer.
The field of exosome research has seen renewed interest in recent years, particularly in the realm of disease biomarkers. Such small vesicles are composed of a sub-proteome of the originating cell, enriched in many immunologically-relevant antigens, and also harbor an enriched set of certain mRNA and micro-RNA’s. Hence substantive information about the parent cell status can be gained through relatively non-invasive methods, by analyzing exosomes present in urine1 or in plasma.
While the concept of using exosomes as a “liquid biopsy” continues to evolve at some pace, it is important not to loose sight of some fundamental questions surrounding these vesicles, principally; what are their biological functions? And what are the molecular mechanism(s) underlying these?
Some of our earliest experiments in the realm of exosome-function(s) provided us with some striking results, suggesting these vesicles have some considerable impact on immune effector cells. For example the potent proliferative responses of lymphocytes to a variety of mitogens including PHA, MLR CD3/CD28 cross-linking (our unpublished observations) or IL-22, could be essentially abolished in the presence of cancer cell derived exosomes. Exosomes from solid tumors like pleural malignant mesothelioma, prostate cancer and others exhibited this anti-proliferative property. While a precise mechanistic explanation for this observation has not been fully elucidated, this exosome-driven inhibitory effect was completely lost when fractionating cells into pure CD8+ T cells, but was fully retained within pure CD4+ T cells. One possible explanation therefore was the exosome-mediated activation of a CD4+ suppressor cell population, capable of influencing effector T cells of either subset. We revealed an elevation in the proportion of regulatory T cells (CD4+CD25hiFoxp3+) following exosome treatment, with a corresponding potent elevation in their suppressor function as a result. Cancer exosomes can therefore recruit regulatory T cells, and limit the proliferative expansion of effector T cells.2 How exosomes achieve regulatory T cell activation is not yet definitively understood, but is a phenomenon independent of MHC class-II expression, requiring instead TGFβ which is expressed by some cancer cell line derived exosomes,2,3 and expressed by exosomes isolated from tumor effusions.4
Ultracentrifugation of exosomes on continuous sucrose gradients reveal that TGFβ and commonly used exosomal markers like TSG101, co-isolate at classical exosome densities of around 1.1–1.2g/ml.3 We found that TGFβ is tethered to the exosomal surface through association with the heparan sulfate proteoglycan; betaglycan (TGFβ-Receptor-III). This is predominantly (98%) latent-TGFβ which nevertheless remains fully available to recipient cells for activating SMAD dependent and independent signaling.3 Blocking exosomal-TGFβ can abrogate their anti-proliferative influence over IL-2 stimulated lymphocytes, and some evidence suggests when comparing exosomal-TGFβ with soluble-TGFβ at matched doses, the former exhibits significantly higher functional potency.2,3
Regulatory T cells however are certainly not the only target of cancer-exosomes. Cytotoxic cells including NK cells and CD8+ T cells are also subject to functional modulation by such exosomes. One targeting mechanism is through expression of membrane associated NKG2D ligands by cancer exosomes,5 which deliver exosomes to the surface of such effector cells and trigger a selective downregulation of NKG2D. Thus this critically important innate activation molecule is suppressed, with a concomitant decrease in IFNγ production and cytotoxic function as a result.4
Several other groups have also documented important mechanisms whereby cancer exosomes may interfere with correct immune function(s), through expression of ligands like galectin-9 or CD95L (as reviewed in ref. 6). Surprisingly, however, direct physical interaction between exosomes and immune cells may not always be needed in order for exosomes to exert a modulating influence. Proteomic analyses of exosomes from diverse cancer sources, reveal that they seem replete with enzymes.7 This poses an intriguing possibility whereby exosomes may modulate the environment through the action of such enzymes; through diminished substrates or accumulating product(s). One example of this includes observations of the ATP-hydrolytic activity of exosomes, that may be driven sequentially by surface bound CD39 and CD73 and likely also other exosomal constituents, to form adenosine.8 Like TGFβ expression, this is indeed a property which is present on exosomes isolated directly from tumor effusions, as well as from several cultured cell lines. Elevated adenosine within cancerous microenvironments can attenuate several effector T cell functions by signaling through the adenosine A2A-receptor. Hence exosomes have the capacity to modulate the peritumor solute environment and adversely impact cellular immune functions through the actions of these catalytically active enzymes.8
The immuno-biology of cancer exosomes continues to dominate the literature, but increasingly we are also beginning to appreciate broader effects of exosomes, whereby non-immune cells may also be profoundly influenced. Critically important processes in cancer including angiogenesis and recruitment of reactive stroma3 appear to be at least in part under the control of exosomes. For example, differentiation of fibroblasts to myofibroblasts is a major cellular alteration resulting in wholesale changes in the cytoskeleton, pericellular coats, extracellular matrix turnover and growth factor production. Exosomally expressed TGFβ1 is a potent inducer of this differentiation, which leads to a myofibroblastic phenotype that is surprisingly distinct from that phenotype driven by soluble TGFβ1.
Future proteomics, RNA-arrays and metabolomics data sets will undoubtedly provide a wealth of information about the composition of exosomes, but such a focus may risk losing sight of the basic questions posed early in this commentary; namely what do exosomes do? and how do they do it? Greater insight into exosome-driven mechanisms of immune-modulation and other contributory pathological processes will be important for defining truly disease-relevant biomarkers, and for highlighting novel pathways or processes rife for future therapeutic interventions.
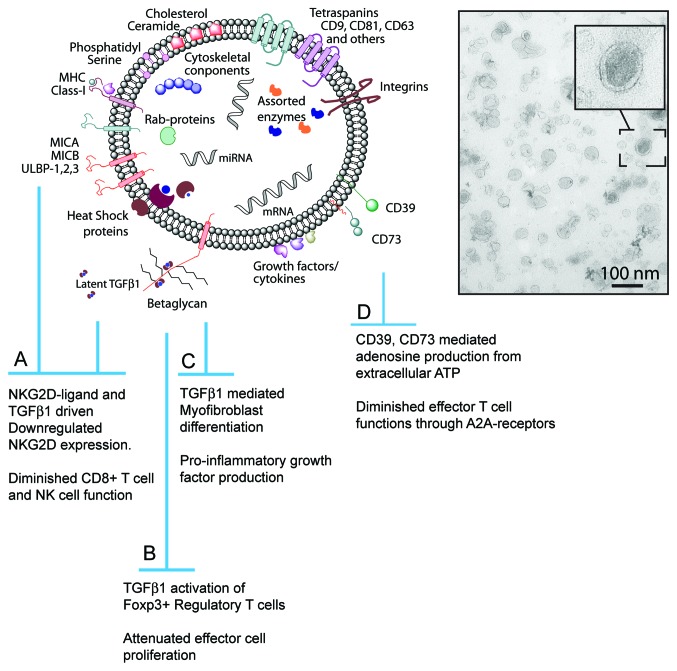
See Figure 1.
Figure 1.
Cancer derived exosomes, some key features and functions. Exosomes are nano-meter sized vesicular structures, typically under 100nm diameter. They comprise an assortment of membrane associated and intra-luminal proteins, including tetraspanins, heat shock proteins, MHC molecules and others. The lipid bi-layer membrane is cholesterol and ceramide rich. The lumen contains protected mRNA and miRNA and likely other non-coding RNA’s. Ligands of NKG2D together with membrane-associated TGFβ1 can downregulate cytotoxic cell NKG2D expression and attenuate NKG2D-dependent target cell recognition, cytokine production and cytolysis (A). TGFβ1 also plays an important role in activating regulatory T cell phenotype and function (B). Non-immune cells like fibroblasts are also influenced by exosomal-TGFβ1, differentiating into a pro-inflammatory myofibroblastic cell (C). Enzymes within, or on the exosome surface may alter environmental substrates, forming products like adenosine that attenuate T cell functions (D).
Disclosure of Potential Conflicts of Interest
The author has no such conflicts to declare.
Acknowledgments
The author is grateful to Cancer Research Wales, Velindre NHS Trust and the June Hancock Mesothelioma Research Fund for financially supporting these studies.
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/17826
References
- 1.Mitchell PJ, Welton J, Staffurth J, Court J, Mason MD, Tabi Z, et al. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med. 2009;7:4. doi: 10.1186/1479-5876-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–66. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 3.Webber J, Steadman R, Tabi Z, Mason MD, Clayton A. Cancer exosomes activate fibroblast to myofibroblast differentiation in a TGFβ dependent manner. Cancer Res. 2010;70:9621–30. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 4.Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol. 2008;180:7249–58. doi: 10.4049/jimmunol.180.11.7249. [DOI] [DOI] [PubMed] [Google Scholar]
- 5.Clayton A, Tabi Z. Exosomes and the MICA-NKG2D system in cancer. Blood Cells Mol Dis. 2005;34:206–13. doi: 10.1016/j.bcmd.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 7.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–38. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clayton A, Al-Taei S, Webber J, Mason MD, Tabi Z. Cancer exosomes express CD39 and CD73, which suppress T cells through adenosine production. J Immunol. 2011;187:676–83. doi: 10.4049/jimmunol.1003884. [DOI] [PubMed] [Google Scholar]