Abstract
Designer T cells expressing transgenic T cell receptors (TCR) with anti-tumor specificity offer new treatment options for cancer patients. We developed a three phase procedure to identify T cells of high avidity based on the fact that T cells recognizing peptides presented by allogeneic MHC efficiently kill tumor cells. Autologous dendritic cells (DC) are co-transfected with ivt-RNA encoding an allogeneic MHC molecule and a selected antigen to allow them to express allogeneic MHC-peptide complexes that activate allo-restricted peptide-specific T cells. This approach provides great flexibility for obtaining high-avidity T cells as potential sources of TCR for adoptive T cell therapy.
Keywords: adoptive T cell therapy, dendritic cells, human cytotoxic T lymphocytes, T cell receptor (TCR), TCR gene therapy
Introduction
The rapidly expanding field of adoptive T cell transfer for immunotherapy of cancer has revealed the important need to have sources of T cells that can effectively recognize and eliminate different types of malignant cells. The power of adoptive cell therapy to induce complete tumor remission lasting over many years has been clearly demonstrated in patients undergoing allogeneic hematopoietic stem cell transplantation (HSCT). In this setting, genetic differences between the donor and the patient enable strong T cell responses directed against minor histocompatibility determinants to eradicate malignant leukemia clones of the patient.1-3 Complete remissions often occur only after several months up to one year following donor leukocyte infusion (DLI), due to the need for specific T cells to be activated and expanded to adequate numbers in vivo. This time lag places limits on the efficacy of DLI in the treatment of acute leukemia after HSCT, since the growth of residual malignant cells often outpaces the expansion capacity of specific immune cells.4,5 Furthermore, this therapy option is not available for many patients who cannot undergo HSCT.
For immunotherapy of solid tumors, the adoptive transfer of tumor-infiltrating lymphocytes (TIL) has provided substantial clinical benefit for numerous patients, particularly those with melanoma.6-8 However the failure to obtain adequate numbers of TIL with good function hinders the treatment of many patients. Many TIL recognize self-MHC molecules that present peptides derived from self-proteins.6 These T cells are usually of low avidity because negative selection of high-avidity T cells recognizing self-proteins occurs in the thymus to prevent autoimmunity7 however high-avidity T cells are needed to effectively eradicate tumor cells. Furthermore, the isolation and large-scale expansion of tumor-associated antigen (TAA)-specific T cells for each patient is expensive and time consuming. Thus, a new therapeutic strategy aims to rapidly imbue activated patient T cells with tumor specificity and high functional avidity by using transgenic expression of selected TCR sequences (TCR gene therapy).
Clinical studies have utilized several patient-derived transgenic TCR (tg-TCR) in phase I trials of advanced melanoma.8 When compared with adoptive transfer of heterogenous TIL, these initial trials suggested that clinical efficacy may require use of mini-repertoires of lymphocytes expressing different tg-TCR. Unfortunately, tg-TCR therapy was often accompanied by substantial toxicity due to cross-recognition of normal tissues, particularly when a TCR of higher avidity was used.8 Animal studies have also revealed that adoptive transfer of high-avidity tg-TCR specific for p53 leads to recognition of stem cells, causing hematological cell loss and death of recipient animals.9 Therefore, it is of critical importance to select tg-TCR for clinical application that will provide the best gain in clinical efficacy with the lowest toxicity for self-tissues. This requires careful selection of the TAA that should serve as targets for tumor recognition, as well as careful selection of the corresponding therapeutic tg-TCR. While a number of TAA candidates have been identified over the past decades that may prove to be suitable target molecules, including tissue-restricted proteins and cancer-testis antigens,10 there is still a critical need to have access to more TCR that can be utilized for TCR gene therapy. In the end, rigorous selection of both the target TAA and the corresponding tg-TCR sequence will be needed to achieve optimal discrimination between tumor cells and normal tissues. However, after these parameters have been established, a tg-TCR can be widely used for large numbers of patients whose tumors express the corresponding tg-TCR ligand.
The problem of acquiring only low-avidity T cells as sources of tg-TCR, due to negative selection, can be bypassed because this selection process is limited to self-MHC molecules that are expressed in the thymus.11 Therefore, it is possible to isolate T cells of high avidity that recognize peptides derived from any TAA if they are presented by allogeneic MHC molecules. Previous work showed that transgenic lymphocytes that express allo-restricted peptide-specific tg-TCR can effectively eliminate tumor cells in pre-clinical models.12 The most common approach to obtain allo-restricted T cells uses peptide-pulsed T2 cells as antigen-presenting cells (APC). However, many T cell clones generated in this manner fail to kill tumor cells. As an alternative, activated B cells have been used following coating with allogeneic peptide-MHC (pMHC) monomers.13 This method takes advantage of the professional APC capacity of activated B cells, but it requires the development of many different pMHC-monomers if it is to be applied for numerous TAA. Allo-restricted peptide-specific T cells can also be obtained using peptide-loaded APC or tumor cells to stimulate T cells derived from HLA partial-mismatched individuals if suitable donor pairs can be identified. Here a major drawback is the rarity of individuals that differ by only one HLA allotype of choice, restricting the ability to use some HLA allotypes for peptide presentation. Therefore all of these approaches have major limitations for generating allo-restricted TAA-specific T cells with high functional avidity.
To overcome these obstacles we developed a procedure to isolate allo-restricted peptide-specific T cells as sources of high avidity tg-TCR which uses in vitro transcribed RNA (ivt-RNA)-transfected dendritic cells (DC) as APC. This method utilizes the important capacity of mature DC (mDC) to prime naïve lymphocytes.14 In the experimental procedure, T cells are co-cultured with autologous mDC that have been loaded with ivt-RNA which encodes a selected TAA, combined with ivt-RNA encoding a selected HLA allele that is not carried by the donor. Both proteins are expressed by the mDC and thereby new allo-pMHC ligands are presented at the cell surface where they function to activate allo-restricted peptide-specific responses in autologous T cells.
Results
T cells are generated using a three phase experimental procedure
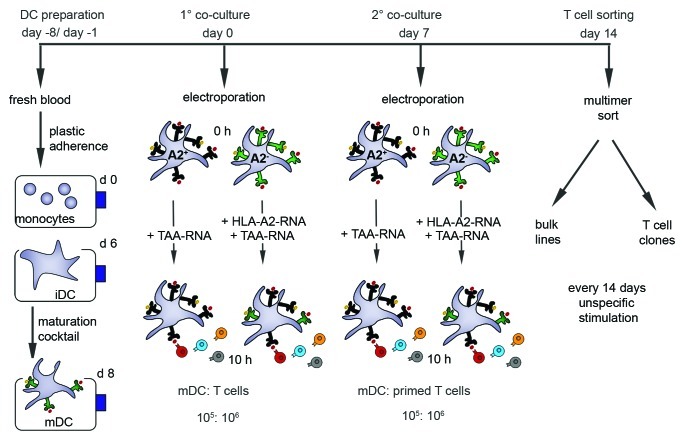
The generation of allo-restricted peptide-specific T cells using mDC pulsed with ivt-RNA encompasses an experimental procedure that includes three distinct phases (Fig. 1). The first phase involves the generation of mDC from peripheral blood monocytes. This phase is spread over a time period of eight days, beginning on day -8 with respect to initiation of the primary (1°) mDC-T cell co-cultures that are established on day 0. A similar procedure must also be performed to prepare fresh mDC for use in the secondary (2°) co-cultures of mDC and 1° T cells. Therefore, a new set-up is performed in an identical manner, whereby the monocytes are isolated from fresh peripheral blood samples on day -1, in order to have mDC available for harvest and use when the 2° co-cultures are established on day 7.
Figure 1. Time schedule of a de novo stimulation protocol for naïve CD8-enriched T cells with ivt-RNA-pulsed DC. The generation of DC of HLA-A2+ and HLA-A2- donors is started eight days before priming (day -8). Monocytes are isolated from PBMC by plastic adherence on day 0 and cultured for 6 d with GM-CSF and IL-4 to produce immature DC (iDC) which are then incubated with a maturation cocktail. After 48 h, mature DC (mDC) are harvested and used for the T cell priming (day 0). Mature DC of HLA-A2+ donors are electroporated with TAA-ivt-RNA and mDC of HLA-A2- donors are transfected with ivt-RNA encoding the allo-MHC molecule HLA-A2, in combination with ivt-RNA encoding the TAA. RNA concentrations depend on the MHC molecules and the TAA that are used. Ten hours after electroporation, priming (1°) is initiated by co-culture of autologous CD8-enriched T cells with ivt-RNA-loaded mDC in a ratio of 10:1. The second stimulation (2°) is performed seven days later (day 7) using freshly prepared ivt-RNA-pulsed mDC. On day 14, the TAA-specific HLA-A2-restricted T cell cultures are stained with multimer and sorted via fluorescence activated cell sorting. The sorted T cells are cloned in limiting dilution cultures or expanded as bulk lines. Thereafter T cells are non-specifically stimulated every 14 d.
The second phase of the experimental procedure involves the co-culture of CD8-enriched T cells with the ivt-RNA-pulsed mDC. The 1° co-cultures are established on day 0 and use mDC that are freshly loaded with ivt-RNA via electroporation. After ivt-RNA-loading, the mDC are cultured for 10 h in DC medium supplemented with 100 ng/ml GM-CSF and 20 ng/ml IL-4. After 10 h in DC medium, the mDC are harvested, counted and added to the pre-seeded T cells. The co-culture is performed in T cell medium. CD8+ T cells are generated from freshly drawn peripheral blood samples obtained on day 0. After Ficoll density centrifugation the lymphocytes undergo CD8-enrichment through depletion of contaminating lymphocyte populations with the use of a commercial kit. After washing and counting, mDC and CD8+ T cells are co-cultured at a ratio of 1:10. We aim to stimulate 2x107 T cells in the 1° co-cultures. This number can vary dependent upon the numbers of viable mDC that are available after loading of ivt-RNA. The 1° co-cultures are continued for seven days, at which time the T cells are harvested, counted and replated with freshly prepared ivt-RNA-pulsed mDC, established from freshly isolated monocytes on day -1. The primed T cells are recovered on day 7 from the 1° co-cultures, washed, counted and resuspended in fresh culture medium. A ratio of 1:10 for mDC to T cells is also used in the 2° co-cultures. The numbers of primed T cells that can be replated in 2° co-cultures is dependent upon the numbers of viable mDC that are available for restimulation.
The third phase of the experimental procedure involves an enrichment step based on MHC-multimer sorting to obtain T cells that express TCR which recognize a pMHC ligand that is newly expressed on the mDC following transfection with both ivt-RNA. The T cells recovered from the 2° co-cultures are incubated with PE-labeled pMHC-multimer and APC-labeled antibody specific for CD8. The double-positive cells are gated and sorted using either MoFlo or FACS Aria instruments. The multimer+ T cells are then immediately cloned in limiting dilution cultures. The remaining T cells are cultured in bulk to establish corresponding T cell lines. Both the T cell clones and the T cell lines are restimulated non-specifically every 14 d with anti-CD3 antibody and feeder cells.
Allogeneic MHC molecules are provided to mDC by ivt-RNA
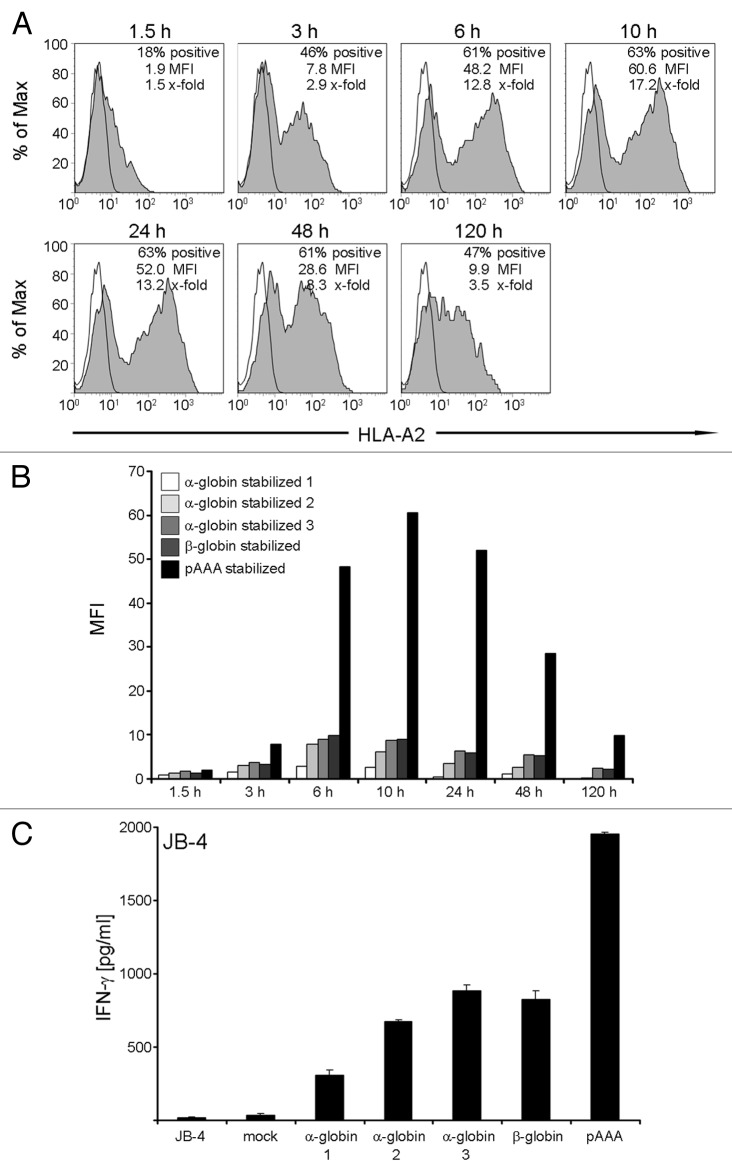
In order to stimulate allogeneic T cells, some of which are peptide-specific, the mDC must express allogeneic MHC molecules at the cell surface after transfer of ivt-RNA encoding an HLA allotype. Since mDC display high levels of endogenous MHC class I, the surface expression of transgenic allogeneic MHC must be monitored using HLA allotype-specific monoclonal antibodies to analyze cells by flow cytometry. For example, expression of transgenic HLA-A2 molecules can be studied on mDC prepared from HLA-A2- donors using a primary HLA-A2-specific BB7.2 monoclonal antibody, followed by a secondary PE-conjugated antibody. As an example HLA-A2 surface expression was already detected on the mDC surface at 1.5 h following electroporation of 25 μg ivt-RNA encoding the HLA-A*0201 allele (Fig. 2A). In this case, HLA-A2 expression reached a peak at 10 h, measured as mean fluorescent intensity (MFI), and then decreased over time. Nevertheless, around 50% of the mDC were still positive for HLA-A2 at 120 h. The greatest percentage of HLA-A2+ mDC (63%) was also seen at 10 h.
Figure 2. Kinetics of HLA-A2 expression and stimulatory capacity of ivt-RNA-transfected mDC from an HLA-A2- donor. (A) HLA-A2- mDC are electroporated with 25 µg HLA-A2 ivt-RNA prepared by the standard method. Surface staining of HLA-A2 is performed at different time points (1.5 h, 3 h, 6 h, 10 h, 24 h, 48 h and 120 h) after transfection and analyzed by flow cytometry. Stained samples are represented by filled gray curves and corresponding controls by empty curves. Percent positive cells, mean fluorescence intensity (MFI) and x-fold expression are shown in the upper right corner of each histogram. The x-fold expression is calculated by dividing the MFI of the positive sample with the MFI of the control. (B) For comparison, mDC of an HLA-A2- donor are transfected with 25 µg of 5 different species of HLA-A2 ivt-RNA. The quality of the standard poly-A tail (pAAA)-stabilized HLA-A2 ivt-RNA is compared with α-globin-stabilized (1, 2, 3) and β-globin-stabilized HLA-A2 ivt-RNA. The expression at different time points is depicted in a bar histogram as MFI. (C) mDC transfected with the various HLA-A2 ivt-RNA species are used 24 h after electroporation for co-incubation with the HLA-A2 allo-reactive CTL clone JB4 to assess their stimulatory capacity. IFNγ is quantified in culture supernatants by ELISA and presented as pg/ml. Mean values and mean deviations are derived from duplicate measurements.
Various forms of ivt-RNA can be used to transfect mDC for allogeneic MHC expression. We analyzed four sources of ivt-RNA that contained α-globin or β-globin domains in the 3′ region, introduced to increase mRNA stability. The 3′ untranslated region of both the human α-globin and β-globin-gene was linked to the HLA-A2 construct in different ways leading to α-globin stabilized and β-globin stabilized HLA-A2 ivt-RNA. These four globin-stabilized ivt-RNA were compared with a standard ivt-RNA that was generated according to published procedures using commercially-available kits and reagents.15,16 This ivt-RNA carried a poly-A tail but did not contain any extra globin domain. When mDC were loaded with the different sources of RNA and analyzed by flow cytometry, a similar kinetics of HLA-A2 surface expression was observed, but use of ivt-RNA carrying a globin domain to improve mRNA stability did not increase the levels of surface HLA-A2. In fact, substantially higher levels of HLA-A2 were seen in mDC expressing the standard ivt-RNA (Fig. 2B). The differences in levels of HLA-A2 surface expression imbued by the various RNA also impacted on the capacity of the mDC to stimulate cytokine secretion by a T cell clone (JB-4) that recognizes HLA-A2 as an alloantigen.17 JB-4 cells released a far greater amount of IFNγ after stimulation with standard ivt-RNA (Fig. 2C).
Co-expression of ivt-RNA encoding MHC and antigen creates new peptide-MHC ligands
New pMHC ligands of defined specificity can be created on mDC by introducing two different species of ivt-RNA, one encoding an allogeneic HLA and the second encoding a new antigen, for example a defined TAA. To demonstrate that both proteins, encoded by the two co-transfected ivt-RNA species, are expressed in the mDC after electroporation, we used ivt-RNA encoding HLA-A2 combined with ivt-RNA encoding tyrosinase, which is expressed in many melanomas, as a model TAA. As shown above, HLA-A2 expression was easily monitored by surface staining of mDC. Tyrosinase expression was monitored by intracellular staining using tyrosinase-specific antibody since it is not expressed as a membrane protein.
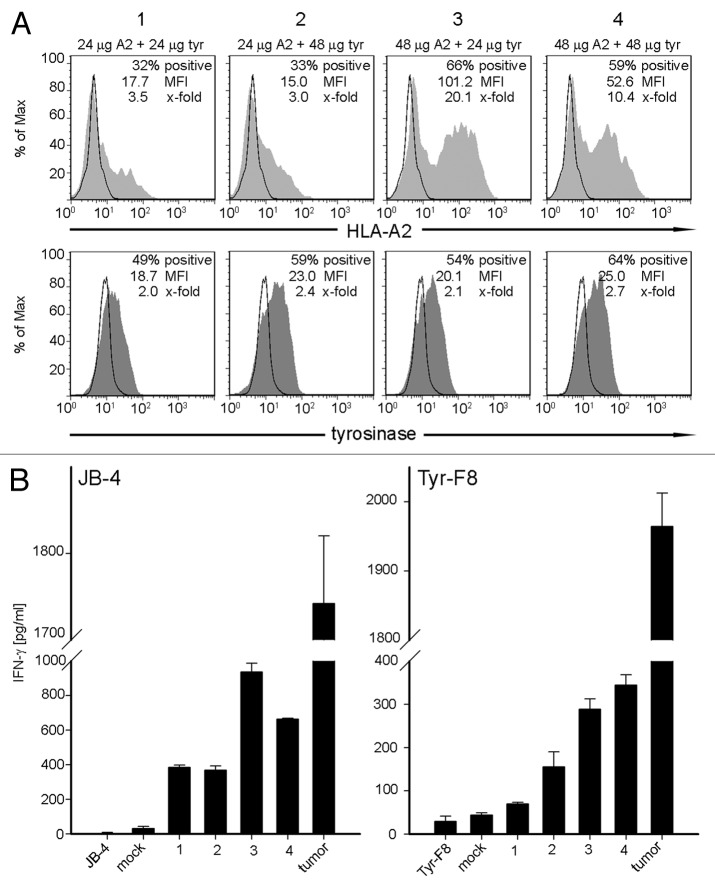
In this analysis, two concentrations (24 μg and 48 μg) of ivt-RNA encoding HLA-A2 and tyrosinase were introduced simultaneously by electroporation into mDC prepared from an HLA-A2- donor (Fig. 3A). The intracellular expression of tyrosinase was analyzed after 3 h and surface expression of HLA-A2 was assessed at 6 h, based on preliminary studies of each individual protein (data not shown). The percentages of HLA-A2+ mDC increased from around 30% using 24 μg ivt-RNA (Fig. 3A, panels 1 and 2) to around 60% using 48 μg ivt-RNA (Fig. 3A, panels 3 and 4). Only a small but non-significant increase in the percentages of mDC expressing intracellular tyrosinase protein was observed when the concentration of tyrosinase ivt-RNA was doubled (Fig. 3A, panels 1 and 3 vs. 2 and 4). A paradoxical decrease in HLA-A2 expression was seen when the concentration of tyrosinase ivt-RNA was increased, leading to a decrease in MFI from 101 when 24 μg of tyrosinase ivt-RNA (Fig. 3A, panel 3) was used to a MFI of only 53 when 48 μg of tyrosinase ivt-RNA was co-expressed in the mDC (Fig. 3A, panel 4). This suggested that the total amount of ivt-RNA expressed in the mDC might be inhibitory. This observation was similar to effects we observed when multiple sources of ivt-RNA that encode different TAA were introduced into mDC.
Figure 3. Co-expression of two different ivt-RNA species encoding HLA-A2 and tyrosinase. mDC are electroporated with four different ivt-RNA combinations: (1) 24 µg HLA-A2 and 24 µg tyrosinase ivt-RNA, (2) 24 µg HLA-A2 and 48 µg tyrosinase ivt-RNA, (3) 48 µg HLA-A2 and 24 µg tyrosinase ivt-RNA and (4) 48 µg HLA-A2 and 48 µg tyrosinase ivt-RNA are mixed and co-transfected into mDC of an HLA-A2- donor. (A) Surface staining of HLA-A2 is performed at 6 h and intracellular staining of tyrosinase at 3 h after transfection and analyzed by flow cytometry. HLA-A2 expression is represented by filled curves, corresponding tyrosinase expression of the same sample is shown beneath by filled curves. Mock-control mDC are included in the respective histograms by open curves. Percent positive cells, MFI and x-fold expression are shown in the upper right corner of each histogram. (B) Stimulatory capacity of the mDC loaded with different concentrations of HLA-A2 and tyrosinase ivt-RNA is analyzed by induction of IFNγ secretion of specific T cell clones. Columns represent the amount of IFNγ (pg/ml) secreted by a tyrosinase-independent HLA-A2 allo-reactive CTL (JB4) and an HLA-A2-restricted tyrosinase peptide-specific CTL (Tyr-F8) after co-incubation with ivt-RNA-pulsed DC, 24 h after electroporation. IFNγ is quantified in culture supernatants by ELISA. Mean values and mean deviations are derived from duplicate measurements. IFNγ secretion of T cells alone and in co-culture with non-transfected mock mDC serve as negative controls. Maximal IFNγ release of the CTL is observed in co-cultures with the tumor cell line Mel-93.04A12 as the positive control.
The response of a T cell clone (Tyr-F8) that is specific for a new pMHC ligand comprised of tyrosinase peptide369–377 presented by HLA-A2 was assessed to determine whether peptides were processed from the tyrosinase protein and presented on the mDC surface in association with transgenic HLA-A2 molecules.18 First, T cell recognition of HLA-A2 as an alloantigen expressed by the mDC, irrespective of specific peptide, was seen with clone JB-4. This clone responded with the highest level of IFNγ secretion after 24 h co-culture with mDC loaded with 48 μg of HLA-A2 and 24 μg of tyrosinase ivt-RNA (Fig. 3A, panel 3), which corresponded to the highest MFI of HLA-A2 on the mDC (Fig. 3B). As shown in Figure 2, JB-4 cells also recognized transgenic HLA-A2 molecules on mDC in the absence of tyrosinase protein, demonstrating their independence from tyrosinase peptide. Clone Tyr-F8 responded to all four mDC populations, with the greatest response seen when 48 μg of each ivt-RNA was used (Fig. 3A, panel 4 and Fig. 3B). Published results demonstrated that activation of this clone was dependent upon the co-expression of HLA-A2 and tyrosinase protein in the mDC.
When co-expression was analyzed using independent sources of mDC, the lower concentrations of ivt-RNA depicted in panels 1 and 2 (Fig. 3A) were generally found to stimulate weaker responses of Tyr-F8 cells than mDCs loaded with higher concentrations of ivt-RNA, as depicted in Figure 3A, panels 3 and 4. This revealed that both the percentages of HLA-A2+ mDC as well as the level of HLA-A2 expression were important. The small variations in tyrosinase protein expression seemed not to be relevant (Table 1).
Table 1. Stimulatory capacity of mDC loaded with different amounts of ivt-RNA.
| |
|
|
IFN-γ release relative to tumor [%] |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA-A2 tyrosinase | Exp. | 1 | 2 | 3 | mean | ||||||
| Tyr-F8 |
24 µg |
24 µg |
|
0.0 |
1.5 |
4.1 |
1.9 |
||||
| mDC* mock |
24 µg |
48 µg |
|
3.6 |
2.2 |
4.0 |
3.3 |
||||
| mDC 1 |
48 µg |
24 µg |
|
14.0 |
3.6 |
9.9 |
9.2 |
||||
| mDC 2 |
48 µg |
48 µg |
|
8.6 |
7.9 |
7.5 |
8.0 |
||||
| mDC 3 |
|
|
|
14.0 |
14.7 |
12.1 |
13.6 |
||||
| mDC 4 |
|
|
|
7.4 |
17.5 |
7.8 |
10.9 |
||||
| tumor cells | 100.0 | 100.0 | 100.0 | 100.0 | |||||||
mDC were electroporated in absence of ivt-RNA
Co-expression of other ivt-RNA combinations allows HLA and TAA expression on mDC
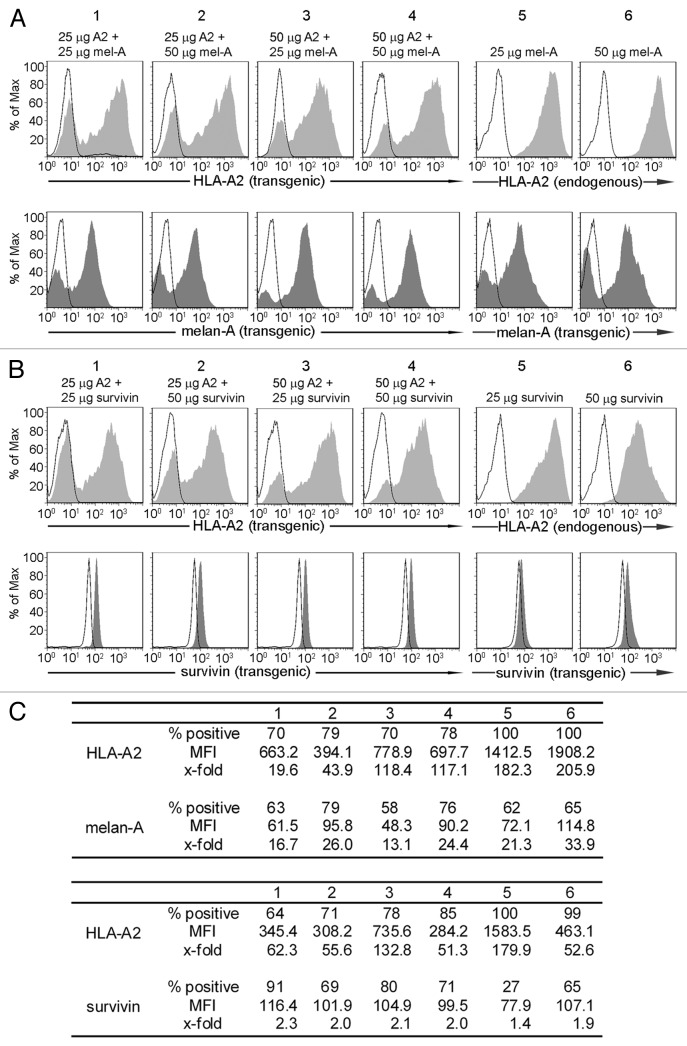
To establish that other combinations of ivt-RNA also led to strong co-expression of MHC and TAA in mDC, we analyzed melan-A and survivin as TAA, together with HLA-A2. Again, transgenic expression of HLA-A2 was monitored by surface staining and TAA protein expression was detected by intracellular staining. The amounts of ivt-RNA introduced into the mDC were also varied. With both combinations, it was also possible to detect high percentages of HLA-A2+ mDC and TAA+ mDC with substantial levels of protein expression (Fig. 4). These results demonstrated that this approach provides a robust method to efficiently generate mDC that co-express new MHC and TAA. Furthermore, the same method could be easily used to introduce ivt-RNA only encoding a TAA into mDC that naturally expressed the desired HLA restriction element due to presence of the endogenous class I allele, as shown in Figure 4, panels 5 and 6 for both melan-A and survivin (Fig. 4).
Figure 4. Co-expression of additional combinations of ivt-RNA encoding TAA and MHC. (A) mDC of an HLA-A2- donor are electroporated with four different RNA combinations: (1) 25 µg HLA-A2 and 25 µg melan-A ivt-RNA, (2) 25 µg HLA-A2 and 50 µg melan-A ivt-RNA, (3) 50 µg HLA-A2 and 25 µg melan-A ivt-RNA and (4) 50 µg HLA-A2 and 50 µg melan-A ivt-RNA. mDC of an HLA-A2+ donor are transfected with (5) 25 µg melan-A or (6) 50 µg melan-A ivt-RNA. Surface staining of HLA-A2 and intracellular staining of melan-A is performed 6 h after transfection and analyzed by flow cytometry. HLA-A2 expression is represented by filled curves in the upper panels; the corresponding melan-A expression of the same sample is shown beneath by filled curves. Corresponding controls are depicted in the respective histograms by open curves. (B) mDC of an HLA-A2- donor are electroporated with four different RNA combinations: (1) 25 µg HLA-A2 and 25 µg survivin ivt-RNA, (2) 25 µg HLA-A2 and 50 µg survivin ivt-RNA, (3) 50 µg HLA-A2 and 25 µg survivin ivt-RNA and (4) 50 µg HLA-A2 and 50 µg survivin ivt-RNA. DC of an HLA-A2+ donor are transfected with (5) 25 µg survivin or (6) 50 µg survivin ivt-RNA. Surface staining of HLA-A2 is performed 6 h and intracellular staining of survivin 3 h after transfection and analyzed by flow cytometry. HLA-A2 expression is represented by filled curves; corresponding survivin expression of the same sample is shown beneath by filled curves. Corresponding controls are depicted in the respective histograms by open curves. (C) The percent positive cells, MFI and x-fold expression of all stainings in (A and B) are shown.
Once again, some decrease in the MFI of HLA-A2 expression was detected in the presence of melan-A or survivin ivt-RNA. This was observed for melan-A (Fig. 4, panel 2 vs. 1) when the amount of melan-A ivt-RNA was doubled. Although not as dramatic, a small change in MFI of melan-A was seen when 25 μg of TAA ivt-RNA was used and the concentration of HLA-A2 ivt-RNA was increased from 25 μg to 50 μg (Fig. 4, panel 1 vs. 3) (Fig. 4A). In mDC co-expressing HLA-A2 and survivin, the decrease in HLA-A2 expression was greatest when 50 μg of survivin ivt-RNA was combined with 50 μg of HLA-A2 ivt-RNA (Fig. 4B, panel 4). A reciprocal impact on survivin expression was not observed. Thus, expression of HLA-A2 seemed to be more susceptible to interference, although impacts on TAA expression may be more difficult to precisely quantify because of the more complex method used to detect intracellular proteins by antibody staining. The reasons for inhibition are not clear since higher amounts of ivt-RNA were not necessarily detrimental; it appears that the ratio of the two species of RNA may also play some role. Nevertheless, all of the mDC populations still showed high percentages of positive cells with substantial MFI of both HLA and TAA protein expression, indicating the suitability of using ivt-RNA to enable good co-expression of these protein combinations in mDC.
Induction of allo-restricted peptide-specific T cell lines using ivt-RNA-pulsed mDC
The preceding studies established the parameters for creating mDC that can be used as stimulating cells for the induction of allo-restricted peptide-specific T cells using CD8-enriched T cells that are prepared from the same donor as the mDC. An experiment demonstrating the application of these ivt-RNA-pulsed DC for T cell stimulation is shown in Figure 5. Here two comparisons are included: mDC were prepared from HLA-A2+ donors who carried an endogenous HLA-A*0201 allele and thereby received only tyrosinase-encoding ivt-RNA while the other mDC were prepared from HLA-A2- donors that were pulsed with ivt-RNA encoding both tyrosinase and HLA-A2.
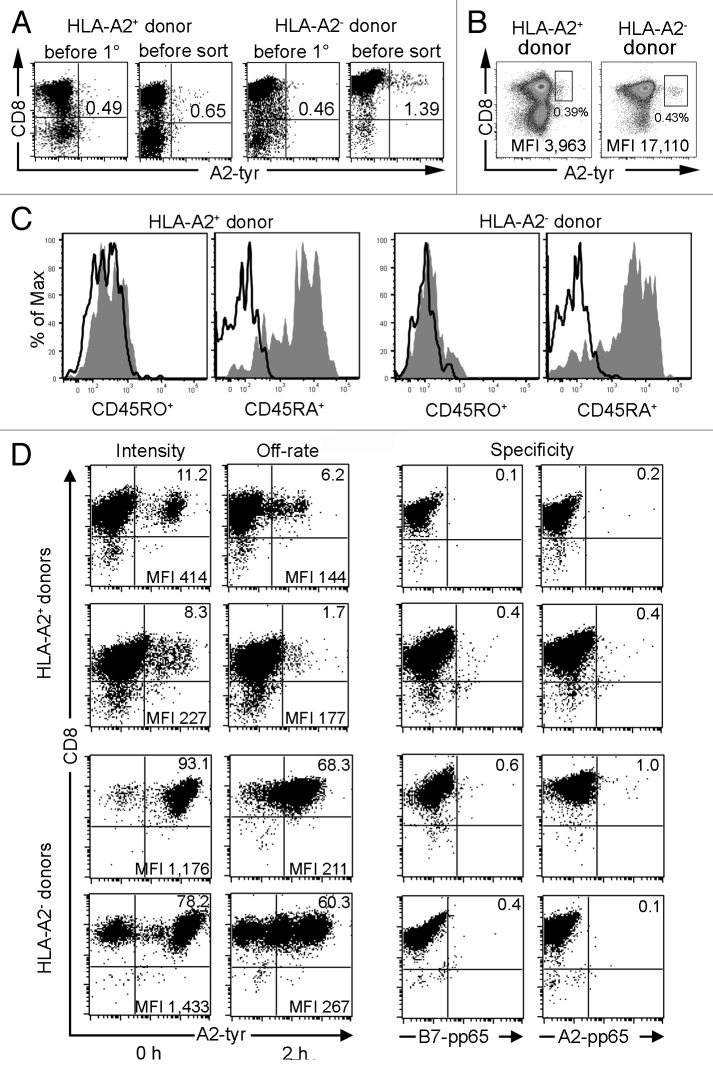
Figure 5. De novo priming of CD8+ T cells with RNA-pulsed DC. mDC derived from an HLA-A2+ donor transfected with 24 µg tyrosinase ivt-RNA and mDC derived from an HLA-A2- donor transfected with 48 µg HLA-A2 and 24 µg tyrosinase ivt-RNA are used for a de novo T cell priming. (A) A2-tyr multimer staining (x-axis) of CD8-enriched T cell populations are determined for cells obtained from an HLA-A2+ donor and an HLA-A2- donor before initiation of primary cultures with ivt-RNA-loaded autologous mDC and one day prior to cell sorting (i.e., 6 d after the 2° stimulation). Staining for CD8 is shown on the y-axis. The percentage of multimer+CD8+ T cells is displayed in the upper right quadrant, showing an increase after mDC stimulation. (B) Sorting scheme for two T cell cultures derived from an HLA-A2+ (left diagram) and an HLA-A2- donor (right diagram). A2-tyr multimer staining is shown on the x-axis and CD8 staining on the y-axis. The boxed gates represent the multimer+CD8+ T cell populations that are selected for fluorescence-activated cell sorting. Dead cells are excluded by propidium iodide staining (data not shown). MFI of multimer staining is given for the gated cells in the lower right corner and the percentage of gated cells of total T cells is indicated beneath the gates. (C) Before initiation of primary cultures, CD8-enriched T cells are stained simultaneously for A2-tyr multimer, CD8 and either CD45RA or CD45RO. Histogram analysis for CD45RA+ and CD45RO+ cells is performed on gated multimer+CD8+ cells of both donors. (D) Sorted T cells are expanded as bulk lines and further re-analyzed after 7–22 d of culture via A2-tyr multimer (x-axis) and CD8 (y-axis) staining. T cell lines of two HLA-A2+ and two HLA-A2- donors are depicted. Percentages of multimer+CD8+ T cells are displayed in upper right quadrant and MFI for this population is presented in lower right corner. T cells are labeled for 45 min with the multimer and either fixed immediately with 1% paraformaldehyde (0 h) to determine MFI of multimer staining (i.e., Intensity) or 2 h after washing of cells to remove unbound multimers with addition of HLA-A2-specific antibody to prevent rebinding of dissociated multimers to assess loss of multimer binding (i.e., Off-rate). Specificity is assessed with control HLA multimers, utilizing peptides from the pp65 protein of cytomegalovirus bound either to HLA-B7 or HLA-A2 molecules (i.e., Specificity).
These DC were used to directly prime enriched, autologous CD8+ T cells, using two rounds of in vitro stimulation with freshly prepared mDC. Prior to activation and after the second stimulation, the frequency of CD8+ T cells bearing TCR specific for HLA-A2-tyrosinase369–377-peptide ligand was measured using anti-CD8 antibody and a tyrosinase369–377/HLA-A*0201-multimer. Frequencies of multimer+CD8+ T cells were approximately equal (0.46% and 0.49%) in enriched CD8+ T cells before DC stimulation (Fig. 5A). Multimer+CD8+ cells that were detected prior to DC stimulation revealed a phenotype of naïve T cells since they were positive for CD45RA and negative for CD45RO surface markers19 (Fig. 5C). The multimer+ cell frequencies increased to 0.65% of HLA-A2+ and 1.39% of HLA-A2- primed T cells, based on total T cells at the end of the 2° co-culture period (Fig. 5A). One day later, cultures were harvested and labeled with anti-CD8-antibody and A2-tyr multimer to perform fluorescence-activated cell sorting. Multimer+ cells were gated as shown and isolated (Fig. 5B). Distinct populations of multimer+ T cells were more difficult to identify in cultures from HLA-A2+ donors but were easy to delineate in cultures from HLA-A2- donors. The double-positive cells from the HLA-A2+ donor shown here had a relatively low mean fluorescence intensity (MFI) of multimer binding (MFI = 3,963). In contrast, substantially higher multimer staining intensity (MFI = 17,110) was seen on gated cells of the HLA-A2- donor. Sorted cells were cloned in limiting dilution cultures and the remaining cells were expanded as bulk cultures using antigen-independent stimulation.19
Bulk cultures of multimer-sorted T cells were re-analyzed after 7–22 d of in vitro expansion. Results are presented from four different donors, primed and sorted according to the full experimental procedure depicted in Figure 1. As expected, the majority of cells were CD8+ in all four T cell lines (Fig. 5D). MFI of multimer binding was used as a first estimate of structural TCR-pMHC binding affinity since higher multimer staining intensity has been shown to indicate a stronger interaction of TCR with their ligands.20 Multimer+ cells were present in the cultures of HLA-A2+ donors at frequencies of 11.2% and 8.3%, respectively. These T cells had low-intermediate intensities of multimer binding (MFI = 414 and 227). Apparently, substantial numbers of non-specific T cells were gated and sorted. In contrast, 93.1% and 78.2% of sorted cells of the two HLA-A2- donors bound HLA-multimers with higher intensities (MFI = 1,176 and 1,433). We also measured loss of HLA-multimer binding over time (i.e. HLA-multimer off-rate) as a second parameter of structural TCR-pMHC binding affinity. A slower off-rate indicates that the TCR-ligand interactions are more stable and thereby of higher structural affinity.21 After initial incubation with HLA-multimer and washing, the T cells were incubated for 2 h at 4°C in the absence of multimer and the presence of antibody specific for HLA-A2 molecules to prevent cellular re-association of released multimers. The rapid loss of multimer binding in T cells of HLA-A2+ donors indicated that the self-MHC-restricted T cells were of lower TCR affinity. After 2 h only 6.2% and 1.7% of the T cells were multimer+, accounting for losses of 45% and 79%, respectively. Multimer binding of T cells from HLA-A2- donors was more stable over time, indicating a higher structural TCR binding affinity for the same pMHC ligands. After 2 h, 68.3% and 60.3% of the cells were multimer+, accounting for losses of only 27% and 23%, respectively. Specificity of multimer binding was confirmed using two control multimers, containing peptides derived from cytomegalovirus protein pp65. Both the HLA-B7-pp65 and HLA-A2-pp65 multimers showed negligible binding to the four T cell lines (0.1–1.0%). These results revealed that bulk T cell cultures containing highly enriched numbers of multimer+ T cells could be readily generated using the detailed experimental procedure. After expansion, the T cell lines can be further enriched by multimer sorting or they can serve as a source of T cell clones that can be isolated by single cell sorting or limiting dilution cultures. We have found that it is equally feasible to establish limiting dilution cultures directly from the multimer-sorted T cells on day 14 of the experimental procedure.22
Extensive analysis of tyrosinase-specific T cell clones derived from both types of priming cultures have been published elsewhere. These studies demonstrated that substantial numbers of allo-restricted tyrosinase-peptide specific T cell clones could be established by this procedure. They represented about 30% of the clones isolated from the HLA-A2- donors. They were able to secrete IFNγ and mediated strong killing of peptide-pulsed antigen-presenting cells, but most importantly, they also secreted high levels of cytokine and showed strong specific killing of HLA-A2+ melanoma tumor cells that also expressed tyrosinase. T cell clones were also isolated from the HLA-A2+ donors but they showed lower levels of cytokine secretion and tumor cell killing.23
Discussion
The experimental procedure described here provides a highly flexible method to generate allo-restricted peptide-specific T cell lines and clones. We have applied this experimental approach to generate HLA-A2-allo-restricted T cell clones specific for tyrosinase, survivin,22 melan-A (unpublished observations), hyaluronan-mediated motility receptor (HMMR), Wilm’s tumor -1 (WT-1), NY-ESO among others. Two specificities of T cells arise in the priming cultures using mDC expressing new allogeneic MHC molecules and TAA. Since the responding cells and DC are derived from HLA-A2- donors, one fraction of T cell clones recognizes the transgenic HLA-A2 as an alloantigen, irrespective of specific TAA-derived peptide. The second fraction recognizes a TAA-derived peptide in association with allogeneic HLA-A2 molecules. These two types of response must be separated at the level of individual clones. We observed that about 50% of the multimer-sorted cells arising in the allogeneic co-cultures recognized HLA-A2 as an alloantigen, independent of TAA-derived peptide. Fortunately, after multimer sorting and cloning at the end of the 2° co-culture period around 30% of the T cell clones were allo-restricted and peptide-specific. As expected, none of the T cell clones derived from HLA-A2+ donors showed HLA-A2 alloreactivity. Thus, this experimental procedure provides a robust method for efficiently obtaining a variety of different T cell clones for further study.
The preparation of mDC as stimulating cells is a particularly critical component of the experimental procedure. The use of different combinations of ivt-RNA encoding MHC and TAA showed that the method is reliable for obtaining good co-expression of both proteins. In essence it can be used for any MHC allele and TAA for which cDNA are available to generate ivt-RNA. However it is important to track the expression of both the MHC and the TAA in mDC to assure adequate co-expression of the proteins. In the first instance, this can be done using flow cytometry to detect expression of the new proteins. Ideally, detection of a new pMHC ligand through recognition of a specific T cell clone is useful to demonstrate that the functions of antigen processing and presentation are intact in the mDC. This may not always be feasible due to lack of specific T cell clones.
Because the entire TAA appears as an intracellular protein, it can be processed and presented by the natural antigen processing machinery of the mDC, thereby creating multiple new epitopes at the cell surface, often of unknown sequence. For known pMHC ligands, specific T cells can be sorted using multimers. For T cells of unknown peptide specificity, other isolation procedures can be employed, such as cytokine capture or CD137 expression (unpublished observations) after APC stimulation.24 Thereby, this method opens the possibility to tap T cells with high functional avidities for many new TAA specificities.
The major advantage of this experimental procedure is that responding CD8+ T cells and mDC can be used from any healthy donor who does not carry the HLA-allotype that is selected as the molecule for allo-restriction. This eliminates the need for extensive searches of HLA-typed donors to identify partial HLA-mismatched pairs that differ by only one HLA-allotype. Because an extensive bank of cDNA for different HLA alleles is available, or they can be quickly isolated from any donor of choice, studies of TAA presentation by HLA molecules other than HLA-A2 are now much easier to perform. Furthermore, it is possible to extend the studies to TAA presentation by MHC class II molecules using the same experimental procedure, with the modification that the TAA-encoding ivt-RNA should be modified to target the protein to the class II presentation pathway.25
Materials and Methods
Culture of cell lines and effector T cell clones
The human melanoma cell line Mel-93.04A12 (HLA-A2+ tyrosinase+ melan-A+; gift of P. Schrier, Department of Immunohematology, Leiden University Hospital, The Netherlands) is cultured in RPMI 1640 medium (Invitrogen, 318725) supplemented with 12% fetal bovine serum (FBS) (Invitrogen, 10091148), 2 mM L-glutamine (Invitrogen, 25030-024) and 1 mM sodium-pyruvate (Invitrogen, 11360039) and non-essential amino acids (Invitrogen, 11140035).
For pre-testing the stimulatory capacitiy of ivt-RNA-pulsed mDC, the HLA-A2 allo-specific CTL JB417 and the HLA-A*0201-restricted tyrosinase369–377 peptide-specific CTL Tyr-F818 (gift of P. Schrier, Department of Clinical Oncology, Leiden University Hospital, The Netherlands) are cultured as described.
Generation of DC
Blood samples from healthy donors are collected after informed consent, in accordance with the Declaration of Helsinki and approval of the Institutional Review Board of the University Hospital of the Ludwig-Maximilians-University, Munich, Germany. Peripheral blood mononuclear cells (PBMC) are isolated by Ficoll density gradient centrifugation. PBMC are resuspended in 15 ml VLE (very low endotoxin) RPMI 1640 medium (Biochrom, FG1415) supplemented with 1.5% human serum (DC medium) at 7.5′107 cells per 75 cm2 culture flask (NUNC, 178905) and incubated at 37°C and 5% CO2 for 1 h. Non-adherent cells are carefully removed by washing. Adherent monocytes are cultured in medium containing 100 ng/ml GM-CSF (Leukine® by Berlex, NDC50419-050-30) and 20 ng/ml interleukin-4 (R&D Systems, 104-IL-050-CF) and fed with the same medium on days 3 and 6. On day 6 of culture, the immature DC are differentiated into mDC by addition of medium containing 10 ng/ml IL-1β (R&D Systems, 201-LB-025-CF), 15 ng/ml IL-6 (R&D Systems, 206-IL-050-CF), 10 ng/ml TNFα (R&D Systems, 210-TA-050-CF) and 1 µg/ml PGE2 (Sigma-Aldrich, p5640-10MG) for 2 d.
Production of HLA-A2, tyrosinase, melan-A and survivin ivt-RNA
The following linearized plasmids were used as in vitro transcription templates to produce single-species ivt-RNA with the aid of the mMESSAGE mMACHINE T7 kit and Poly(A) tailing kit (both Ambion, AM1344 and AM1350), according to the manufacturer’s instructions: pCDM8-HLA-A2 plasmid with HLA-A*0201 cDNA (gift of E. Weiß, Department Biology II, Ludwig-Maximilian-University, Munich, Germany), pZeoSV2+/huTyr with tyrosinase cDNA (gift of I. Drexler, Institute of Molecular Virology, Helmholtz Zentrum München, Munich, Germany), pcDNAI/Amp/Aa1 with melan-A cDNA (gift of T. Wölfel, Third Department of Medicine, Hematology and Oncology, Johannes-Gutenberg University of Mainz, Mainz, Germany) and pGEM4Z/survivin/A64 plasmid (cloned by S. Milosevic, Institute of Molecular Immunology, Helmholtz Zentrum München).
Also different modified mRNA molecules (RNActive®, CureVac) were used that encode HLA-A2 molecules. Four different HLA-A2 mRNA are compared, here whereby the 3′ untranslated region of either the human α-globin or β-globin gene is linked to the HLA-A2 construct in different ways, leading to α-globin or β-globin stabilized HLA-A2 mRNA.
Electroporation of DC
On day 8, mDC are harvested 48 h after addition of maturation cytokines. Electroporation is performed as previously described.16 In brief, 2 to 3x106 cells are resuspended in approximately 200–300 µl OptiMEM I medium (Invitrogen, 31985047), placed in a 0.4-cm electroporation cuvette (Bio-Rad, 5000835) and incubated for 3 min on ice. Electroporation is performed with the Gene Pulser Xcell (Bio-Rad) using the exponential protocol at 250 V and 150 µF. RNA species are transfected using different amounts of individual RNA, between 24–50 µg per electroporation.
FACS analysis
HLA-A2 molecules are stained on the surface of DC with BB7.2 monoclonal antibody (HB82, ATCC) for 45 min on ice followed by a secondary antibody conjugated with phycoerythrin (PE) (goat anti-mouse IgG; Jackson ImmunoResearch, 115-116-146) for 45 min. The intracellular protein expression in ivt-RNA-transfected mDC is detected using the following primary antibodies: tyrosinase-specific primary monoclonal antibody (clone T311; diluted 1:10; Novocastra Laboratories Ltd., 600741), melan-A-specific primary monoclonal antibody (clone A103; diluted 1:20; DakoCytomation, M7196) and survivin-specific primary monoclonal antibody (clone 91630; diluted 1:2.5; R&D Systems, MAB886) followed by staining with Cy5-conjugated secondary antibody (rat anti-mouse IgG; Jackson ImmunoResearch, 415-176-166). The intracellular staining is performed as follows. The mDC samples are first fixed in FACS buffer containing 1% paraformaldehyde (Sigma, P6148-1KG) for 30 min on ice and washed twice in buffer without additives and once in buffer containing 0.1% saponin (Sigma, S4521-25G). Samples are then incubated with primary antibody in buffer containing 0.25% saponin for 1 h at room temperature, washed once in buffer containing 0.1% saponin and stained for 30 min at room temperature in the dark with the Cy5-conjugated secondary antibody, diluted in buffer containing 0.25% saponin. Cells are washed once and resuspended in buffer with 0.1% saponin. Untransfected mDC are treated in the same manner and used as negative controls. Data acquisition and analysis are done on a FACSCalibur (BD Biosciences) using CellQuest Pro software or FloJo software (Tree Star). X-fold expression is calculated by dividing the mean fluorescence of ivt-RNA-transfected mDC by the mean fluorescence of untransfected mDC.
To determine the phenotype of unprimed multimer+CD8+ T cells, cells are double-stained with PE-labeled HLA-A2 tyrosinase369–377 (YMDGTMSQV) (A2-tyr) multimer,26 CD8-specific antibody (clone RPA-T8, BD PharMingen, 555369) and simultaneously with either CD45RO- (clone UCHL1, Immunotech, IM1247) or CD45RA-specific (clone HI100, BD PharMingen, 550855) antibody. Histogram analysis for CD45RO and CD45RA staining is performed on gated multimer+CD8+ T cells. Data acquisition and analysis are made with an LSRII instrument (BD Biosciences) using FloJo software (Tree Star).
IFNγ release assay
Stimulatory capacity of RNA-pulsed mDC is analyzed 24 h after electroporation. JB4 or Tyr-F8 CTL suspensions (2 × 104 cells in 100 µl) are added to the RNA-loaded mDC (4 × 104 cells in 100 µl) in round-bottom 96-well plates. T cells without stimulator cells and with mock-transfected mDC serve as negative controls and show almost no IFNγ secretion. T cells are co-cultured with the HLA-A2+, tyrosinase+, melan-A+ tumor cell line Mel-93.04A12, as positive control. Culture supernatants are harvested after 24 h co-culture and assessed by a standard ELISA using the OptEIA™ Human IFNγ Set (BD Biosciences, 555142). Data represent mean values with corresponding mean deviations calculated from duplicate determinations.
De novo priming of T cells with ivt-RNA-pulsed DC
mDC are transfected with pre-selected optimal ivt-RNA concentrations via electroporation as described above. mDC derived from HLA-A2+ donors are loaded with 24 µg tyrosinase ivt-RNA and mDC prepared from HLA-A2- donors are co-transfected with 24 µg tyrosinase and 48 µg HLA-A2 ivt-RNA. Untouched autologous CD8+ T lymphocytes are enriched from PBMC via negative selection using a commercial kit, according to the manufacturer’s instructions (CD8+ T Cell Isolation Kit, Miltenyi Biotec, 130-094-156) on the same day. Co-cultures are initiated 10 h after electroporation in 24-well plates (TPP, Z707805-64EA). 1 × 105 ivt-RNA-pulsed mDC are added to 1 × 106 CD8+ T cells, yielding a mDC:T cell ratio of 1:10 in T cell medium consisting of RPMI 1640 (Invitrogen, 318725), supplemented with 10% heat inactivated human serum, 4 mM L-glutamine (Invitrogen, 25030-024), 12.5 mM HEPES (Invitrogen, -15630049), 50 µM β-mercaptoethanol (Sigma, -63700-50ML-F) and 100 U/ml penicillin/streptomycin (Invitrogen, -15070063). On day 0, 5 ng/ml IL-7 (Promokine, -C-61712) is added. Addition of IL-2 (Proleukin®S, Novartis) is delayed for 2 d to decrease proliferation of non-specific CD8+ T cells,19 then it is added every 3rd day. The 2nd stimulation of the primed T cells is made seven days later using freshly generated ivt-RNA-pulsed mDC, prepared as above.
HLA-multimer staining and sorting
Prior to priming and six days after the 2nd stimulation of CD8-enriched T cells with ivt-RNA-pulsed mDC, HLA-A2-restricted tyrosinase-specific T cells are detected by staining with PE-labeled HLA-A2 tyrosinase369–377 (YMDGTMSQV) (A2-tyr) multimer,26 CD8-specific antibody (clone RPA-T8, BD PharMingen) and propidium iodide (PI: 2 µg/ml). Up to 1 × 106 cells are incubated in 50 µl volume for 25 min with 4 µg PE-labeled multimer on ice in the dark. For sorting, multiple samples of up to 5 × 106 cells are incubated with 12 µg multimer in 100 µl PBS + 0.5% human serum. APC-labeled, CD8-specific antibody is then added for an additional 25 min. After staining, an aliquot of cells is washed and fixed for analysis by flow cytometry using a FACSCalibur (BD Biosciences) and the remaining cells are pooled and sorted on a FACS Aria instrument (BD Biosciences) as described.27
To observe HLA-multimer dissociation (off-rate), cells are washed after multimer binding and resuspended in FACS buffer containing saturating amounts of monoclonal HLA-A2-specific BB7.2 antibody to capture detached multimers and prevent rebinding to T cells. After 2 h, samples are fixed and analyzed by flow cytometry.21 PE-labeled HLA-B7/pp65417–427 (TPRVTGGGAM) peptide/human β2m multimers serve as the HLA control and HLA-A*0201/pp65495–503 (NLVPMVATV) peptide/human β2m multimers serve as a peptide-specificity control. Peptides for these multimers are derived from the pp65 protein of human cytomegalovirus.
Culture of peptide-specific T cell lines
Multimer-sorted T cells are expanded as bulk T cell lines in 96-well round-bottom plates (TPP, 5000773) in 200 µl/well T cell medium (600 cells per well). 50 IU/ml IL-2 is supplemented every three days with 5 ng/ml IL-7 (Promokine, C-61712) and 10 ng/ml IL-15 (PeproTech Inc., AF-200-15) every seven days. Every 14 d T cell lines are restimulated non-specifically with anti-CD3 antibody (0.1 µg/ml) (OKT-3; gift of E. Kremmer, Institute of Molecular Immunology, Helmholtz Zentrum München) and provided with 1 × 105 feeder cells per 96-well (TPP, 92097), consisting of irradiated (50 Gy) PBMC derived from a pool of five unrelated donors. Proliferating T cells are transferred into 24-well plates (TPP, Z707805-64EA) and cultured in 1.5 ml T cell medium plus cytokines. 1 × 106 allogeneic irradiated PBMC are added per well as feeder cells in 24-well plates.
Acknowledgments
The authors would like to thank A. Zobywalski for advice on DC; D.H. Busch and M. Schiemann for providing the multimer technology; J. Ellwart and J. Mysliwietz for help with flow cytometry; B. Frankenberger, E. Noessner and M. Javorovic for technical advice. This work was supported by grants of the German Research Council (SFB-TR36 and SFB-455) and the Helmholtz Society (Alliance for Immunotherapy of Cancer, HA-202). A patent has been granted to the Helmholtz Zentrum München, German Research Center for Environmental Health, Munich, Germanshangrilay.
Glossary
Abbreviations:
- APC
antigen-presenting cells
- DC
dendritic cell
- DLI
donor leukocyte infusion
- HSCT
hematopoietic stem cell transplantation
- mDC
mature DC
- MFI
mean fluorescence intensity
- MHC
major histocompatibility complex
- TAA
tumor-associated antigen
- TCR
T cell receptor
- tg-TCR transgenic
TCR
- TIL
tumor-infiltrating lymphocytes
- PBMC
blood mononuclear cells
- pMHC
peptide-MHC
Declaration of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/18216
References
- 1.Riddell SR, Berger C, Murata M, Randolph S, Warren EH. The graft versus leukemia response after allogeneic hematopoietic stem cell transplantation. Blood Rev. 2003;17:153–62. doi: 10.1016/S0268-960X(03)00007-9. [DOI] [PubMed] [Google Scholar]
- 2.Wölfel C, Lennerz V, Lindemann E, Hess G, Derigs HG, Huber C, et al. Dissection and molecular analysis of alloreactive CD8+ T cell responses in allogeneic haematopoietic stem cell transplantation. Cancer Immunol Immunother. 2008;57:849–57. doi: 10.1007/s00262-007-0421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishida T, Hudecek M, Kostic A, Bleakley M, Warren EH, Maloney D, et al. Development of tumor-reactive T cells after nonmyeloablative allogeneic hematopoietic stem cell transplant for chronic lymphocytic leukemia. Clin Cancer Res. 2009;15:4759–68. doi: 10.1158/1078-0432.CCR-09-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolb HJ, Schmid C, Barrett AJ, Schendel DJ. Graft-versus-leukemia reactions in allogeneic chimeras. Blood. 2004;103:767–76. doi: 10.1182/blood-2003-02-0342. [DOI] [PubMed] [Google Scholar]
- 5.Kolb HJ. Graft-versus-leukemia effects of transplantation and donor lymphocytes. Blood. 2008;112:4371–83. doi: 10.1182/blood-2008-03-077974. [DOI] [PubMed] [Google Scholar]
- 6.Jäger D, Jager E, Knuth A. Immune responses to tumour antigens: implications for antigen specific immunotherapy of cancer. J Clin Pathol. 2001;54:669–74. doi: 10.1136/jcp.54.9.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Visser KE, Schumacher TN, Kruisbeek AM. CD8+ T cell tolerance and cancer immunotherapy. J Immunother. 2003;26:1–11. doi: 10.1097/00002371-200301000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. Gene therapy with human and mouse T cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–46. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Offringa R. Antigen choice in adoptive T-cell therapy of cancer. Curr Opin Immunol. 2009;21:190–9. doi: 10.1016/j.coi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–37. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker LS, Abbas AK. The enemy within: keeping self-reactive T cells at bay in the periphery. Nat Rev Immunol. 2002;2:11–9. doi: 10.1038/nri701. [DOI] [PubMed] [Google Scholar]
- 12.Morris E, Hart D, Gao L, Tsallios A, Xue SA, Stauss H. Generation of tumor-specific T-cell therapies. Blood Rev. 2006;20:61–9. doi: 10.1016/j.blre.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Savage P, Gao L, Vento K, Cowburn P, Man S, Steven N, et al. Use of B cell-bound HLA-A2 class I monomers to generate high-avidity, allo-restricted CTLs against the leukemia-associated protein Wilms tumor antigen. Blood. 2004;103:4613–5. doi: 10.1182/blood-2003-11-3903. [DOI] [PubMed] [Google Scholar]
- 14.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 15.Javorovic M, Pohla H, Frankenberger B, Wolfel T, Schendel DJ. RNA transfer by electroporation into mature dendritic cells leading to reactivation of effector-memory cytotoxic T lymphocytes: a quantitative analysis. Mol Ther. 2005;12:734–43. doi: 10.1016/j.ymthe.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 16.Javorovic M, Wilde S, Zobywalski A, Noessner E, Lennerz V, Wolfel T, et al. Inhibitory effect of RNA pool complexity on stimulatory capacity of RNA-pulsed dendritic cells. J Immunother. 2008;31:52–62. doi: 10.1097/CJI.0b013e31815a1202. [DOI] [PubMed] [Google Scholar]
- 17.Milani V, Frankenberger B, Heinz O, Brandl A, Ruhland S, Issels RD, et al. Melanoma-associated antigen tyrosinase but not Melan-A/MART-1 expression and presentation dissociate during the heat shock response. Int Immunol. 2005;17:257–68. doi: 10.1093/intimm/dxh203. [DOI] [PubMed] [Google Scholar]
- 18.Visseren MJ, van Elsas A, van der Voort EI, Ressing ME, Kast WM, Schrier PI, et al. CTL specific for the tyrosinase autoantigen can be induced from healthy donor blood to lyse melanoma cells. J Immunol. 1995;154:3991–8. [PubMed] [Google Scholar]
- 19.Ho WY, Nguyen HN, Wolfl M, Kuball J, Greenberg PD. In vitro methods for generating CD8+ T-cell clones for immunotherapy from the naive repertoire. J Immunol Methods. 2006;310:40–52. doi: 10.1016/j.jim.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 20.Yee C, Savage PA, Lee PP, Davis MM, Greenberg PD. Isolation of high avidity melanoma-reactive CTL from heterogeneous populations using peptide-MHC tetramers. J Immunol. 1999;162:2227–34. [PubMed] [Google Scholar]
- 21.Palermo B, Garbelli S, Mantovani S, Scoccia E, Da Prada GA, Bernabei P, et al. Qualitative difference between the cytotoxic T lymphocyte responses to melanocyte antigens in melanoma and vitiligo. Eur J Immunol. 2005;35:3153–62. doi: 10.1002/eji.200535110. [DOI] [PubMed] [Google Scholar]
- 22.Leisegang M, Turqueti-Neves A, Engels B, Blankenstein T, Schendel DJ, Uckert W, et al. T-cell receptor gene-modified T cells with shared renal cell carcinoma specificity for adoptive T-cell therapy. Clin Cancer Res. 2010;16:2333–43. doi: 10.1158/1078-0432.CCR-09-2897. [DOI] [PubMed] [Google Scholar]
- 23.Wilde S, Sommermeyer D, Frankenberger B, Schiemann M, Milosevic S, Spranger S, et al. Dendritic cells pulsed with RNA encoding allogeneic MHC and antigen induce T cells with superior antitumor activity and higher TCR functional avidity. Blood. 2009;114:2131–9. doi: 10.1182/blood-2009-03-209387. [DOI] [PubMed] [Google Scholar]
- 24.Becker C, Pohla H, Frankenberger B, Schuler T, Assenmacher M, Schendel DJ, et al. Adoptive tumor therapy with T lymphocytes enriched through an IFN-gamma capture assay. Nat Med. 2001;7:1159–62. doi: 10.1038/nm1001-1159. [DOI] [PubMed] [Google Scholar]
- 25.Nair SK, Hull S, Coleman D, Gilboa E, Lyerly HK, Morse MA. Induction of carcinoembryonic antigen (CEA)-specific cytotoxic T-lymphocyte responses in vitro using autologous dendritic cells loaded with CEA peptide or CEA RNA in patients with metastatic malignancies expressing CEA. Int J Cancer. 1999;82:121–4. doi: 10.1002/(SICI)1097-0215(19990702)82:1<121::AID-IJC20>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 26.Wölfl M, Schalk S, Hellmich M, Huster KM, Busch DH, Berthold F. Quantitation of MHC tetramer-positive cells from whole blood: evaluation of a single-platform, six-parameter flow cytometric method. Cytometry A. 2004;57:120–30. doi: 10.1002/cyto.a.10116. [DOI] [PubMed] [Google Scholar]
- 27.Schuster IG, Busch DH, Eppinger E, Kremmer E, Milosevic S, Hennard C, et al. Allorestricted T cells with specificity for the FMNL1-derived peptide PP2 have potent antitumor activity against hematologic and other malignancies. Blood. 2007;110:2931–9. doi: 10.1182/blood-2006-11-058750. [DOI] [PubMed] [Google Scholar]