Abstract
Background
Tissue imprinting can generate molecular marker maps of tumor cells at deep surgical margins. This study evaluates the feasibility of this method for detection of residual head and neck squamous cell carcinoma (HNSCC).
Methods
Paired fresh tissue and nitrocellulose membrane imprints of tumor and deep margins were collected from 17 HNSCC resections. DNA was amplified using quantitative methylation-specific PCR (qMSP) for p16, DCC, KIF1A, and EDNRB. Levels of methylation in tumors and deep margins were compared.
Results
DNA from imprints was adequate for qMSP. Hypermethylation of target genes was present in 12/17 tumors and in 8 deep margins. Methylation level was better from margin imprints than tissue. During follow-up (median 13 months), local or regional recurrences occurred in six cases of which five had molecularly positive margins.
Conclusion
Tissue imprinting is feasible for molecular detection of residual tumor at deep surgical margins and may correlate with locoregional recurrence.
Keywords: Tissue imprint, head and neck squamous cell carcinoma, deep surgical margin, molecular diagnosis, quantitative methylation-specific PCR, Imprint, Deep margin, Surgery, Recurrence
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide, comprising approximately 5% of all newly diagnosed cancer cases in USA.1,2 The overall 5-year survival rates of HNSCC are approximately 60% but have not substantially changed despite progress in understanding the molecular underpinnings of disease and therapeutic innovation over the last two decades.1–3 HNSCC patients experience a high rate of local recurrence (10–40%) even when the surgical margins are tumor-free by histopathological examination.4 Local recurrence is clinically defined as occurring within 2 cm of the index tumor within three years of the end of treatment and is attributed to regrowth of residual fully transformed cancer cells that survived primary and adjuvant treatment.5 Second primary or field tumors are relapses not fulfilling these criteria and may be derived from a surrounding mucosal field containing partially genetically altered cells.6,7 The field may give rise to new invasive carcinomas when these cells undergo additional genetic alteration.8,9,10
p53 mutations, present in 50–60% of HNSCC, have been used as a molecular marker for assessment of surgical margins demonstrating value in predicting local and regional control.11–13 Other molecular markers including microsatellite alterations, eIF4E overexpression, and quantitative methylation have also been used to detect minimal residual disease in surgical margins and predict local recurrence of HNSCC.14–16 Most reports have taken samples from the mucosal surgical margin but not deep tissues of surgical defects. However, some investigators have provided evidence suggesting that deep margins are more predictive of local recurrence.17 Because surgical deep margins are commonly extensive and only tested intraoperatively by harvesting small representative tissue pieces for frozen section, residual tumor cells at the cut surface may be left undetected in surgical fields and only discovered on processing of the bulk resection specimen. This is done by sampling representative sections from key areas of the tumor specimen after formalin fixation and is reported days later after wound reconstruction and initial healing has occurred. Proper orientation of the specimen is dependent on communication between the surgeon and pathologist, and precise relocation of any close or positive final margins is hampered by the lack of landmarks within deep tissues such as the intrinsic tongue musculature to guide efforts at additional resection or targeted radiation.
Tissue imprinting techniques have been described for assessment of residual tumor at the prostate capsule.18 The tissue imprints collected from the cut surface of specimens may permit tissue-specific molecular profiling that allows the detection of residual tumor cells from the entire deep margin, at a concentration below that detected by light microscopy. This technique has not been tested for HNSCC. This study examined the feasibility of tissue imprinting and molecular mapping for detection of residual tumor cells at deep surgical margins in patients with HNSCC.
MATERIALS AND METHODS
Tissue and imprint collection
Tissue imprints and tissues were collected from tumor and grossly tumor-free deep margins in 17 patients undergoing surgery for HNSCC. The study protocol was approved by the Institutional Review Board of Johns Hopkins Medical Institution and informed consent was obtained from each patient. ligibility criteria included planned surgical resection of HNSCC with disease deemed resectable with curative intent, location of tumor associated with substantial deep soft tissue – tumor interface and lack of distant metastasis. Because we wanted to ensure that some cases would have positive deep margins and a high likelihood of recurrence, patients with advanced or recurrent cancer (high level of concern for treatment failure) were chosen. Patient and tumor characteristics are found in Table 1. One case was eliminated from further study because of inability to follow clinical course. The tumor from four other cases did not display methylation of any of the markers. The cases without methylation of markers in tumor serve as negative controls for analysis of methylation in imprint DNA. Five subjects had had prior treatment with radiation or chemotherapy and 5 had had prior surgery. Samples were obtained from primary tumors of oral cavity (n = 8), oropharynx (n = 1), hypopharynx (n = 1), and neck skin (n = 1), and from bulky neck nodes (n = 1) after radiotherapy for nasopharyngeal carcinoma.
Table 1.
Clinical characteristics, treatment profiles, pathologic and molecular margin status, and follow-up outcomes of cases
| Case | Sex | Age | Site | pTNM | Prior Rx | Pathology
|
No. of hypermethylated genes in tumor* | Molecular deep margin
|
Postop Rx | Site of recurrence | Follow-up months (time to recurrence) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intraop | Imprint (any) | Tissues | ||||||||||||
| Muc | Deep | Final | ||||||||||||
| 1 | M | 59 | OC/Tongue | rT4aN2b | S | (−) | (−) | D close | 3 | + | NE | CR | R | 13(4) |
| 2 | M | 68 | OC/Tongue | T1N1 | Dys | (−) | D close | 4 | + | NE | RT | LR (m) | 13 (5) | |
| 3 | F | 55 | OP | rT2N0 | RT | (−) | (−) | D close | 2 | + | NE | L(u)†+DM | 22 (5) | |
| 4 | M | 34 | OC/Tongue | T3N2b | (−) | (−) | D close | 2 | + | − | CR | L(d) (D) | 18 (8) | |
| 5 | F | 64 | N | rTxN2b | RT | na | (−) | (−) | 2 | − | − | DM | 17 (12) | |
| 6 | M | 68 | HP | T4aN2b | Dys | (−) | D+ | 1 | − | − | RT | R | 14 (5) | |
| 7 | M | 60 | OC/RMT | rT1N3 | S | (−) | (−) | (−) | 2 | + | + | CR | LTF | |
| 8 | M | 75 | OC/Tongue | rT4aN1 | S | (−) | (−) | D close | 4 | + | + | L (d + m) | 8 (3) | |
| 9 | M | 98 | Neck skin | T3N0 | S | (−) | (−) | (−) | 1 | − | − | LTF | ||
| 10 | M | 54 | OC/buccal | rT4aN2b | S | (−) | (−) | M+ | 2 | + | + | DM | LTF | |
| 11 | M | 84 | OC/FOM | T3N1 | (−) | (−) | (−) | 2 | + | + | 12 | |||
| 12 | M | 66 | OC/Tongue | T2N0 | (−) | (−) | Muc dys | 2 | − | − | 10 | |||
Abbreviations CR, chemoradiotherapy; D, deep margin; D close, tumor-close deep margin;; DM, distant metastasis; Dys, dysplasia; F, female; FOM, floor of mouth; L, local; LR, locoregional; LTF, Lost to follow-up; M, male; Muc, mucosal margin; M+, tumor-positive mucosal margin; (−), tumor-negative; na, not applicable; N, neck nodes; NE, not existent; OC, oral cavity; OP, oropharynx; R, regional; Rx, treatment; RT, radiotherapy; RMT, retromolar trigone; S, surgery; T, tumor. (d) recurrence in deep soft tissue; (m) recurrence at mucosal surface;
Of four genes tested (p16, DCC, KIF1A, and EDNRB)
(u) uncertain site of origin- Impossible to distinguish between mucosal and deep local recurrence
After tumor resection, the specimen was immediately placed in saline on a back table. Separate sampling from mucosal margins and representative deep tissue was sent for analysis to pathology for frozen section following standard practice to ensure completeness of extirpation. The specimen was blotted with clean surgical towels to remove excess blood and debris. It was placed tumor side down on a fresh towel, and nitrocellulose imprints were taken first from the deep margin. The specimen was then turned to expose the visible tumor on the mucosal surface, and an additional imprint taken from the tumor as a positive control for cancer cell molecular detection. Each tissue imprint was taken by applying separate nitrocellulose membranes (Hybond-C Extra, Amersham Biosciences/GE Healthcare) to the selected surfaces of surgical specimen with approximately 2.5 kg of manual force for 30 seconds. The imprint from the deep margin was cut into 2–4 pieces or quadrants that were mapped according to resection regions. Paired tissue samples were then taken from each quadrant of the surgical deep margin and tumor to serve for comparison with imprinted DNA quality and quantity. The imprints were immediately soaked in the DNA elution buffer and the tissue was stored at −80°C until microdissection.
DNA extraction and bisulfite treatment
Frozen tumor specimens were microdissected to ensure tumor purity while focal deep margin samples were processed for light microscopy to ensure absence of tumor cells. Imprint and tissue samples were digested with DNA elution buffer containing 50 μg/mL proteinase K (Boehringer, Mannheim, Germany) and 1% SDS at 48°C for 2–3 days. Genomic DNA was extracted with phenol/chloroform and ethanol precipitation, dissolved in low-salt 10 mM Trix-HCl and 2.5 mM EDTA (LoTE) buffer and stored at −20°C.
DNA (2 μg) extracted from imprints and tissues was subjected to bisulfite treatment, which modifies CpG islands, using the EpiTect® Bisulfite Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The bisulfite-treated DNA was resuspended in 50 uL of H2O and stored at −80°C.
Real-time quantitative methylation-specific PCR
Bisulfite-modified DNA was used as a template for fluorescence-based real-time polymerase chain reaction (PCR) and was amplified using quantitative methylation-specific PCR (qMSP) primer and probe sets of the four gene promoters (Table 2). For all qMSP reactions, 3 μL of bisulfite-treated DNA were added to a final volume of 20 μL consisting of each primer (600 nM) and probe (200 μM), platinum Taq polymerase (0.75 U, Invitrogen, Carlsbad, CA), ROX reference dye (200 nM, Invitrogen), dNTP (200 μM each), ammonium sulfate (16.6 mM), Trizma (67 mM, Sigma, St Louis, MO), magnesium chloride (6.7 mM), mercaptoethanol (10 mM) and 0.1% dimethylsulfoxide. All reactions were carried out in triplicate to ensure consistent results in 384-well plates using a 7900 Sequence Detector System (Perkin-Elmer Applied Biosystems, Norwalk, CT). Thermal cycling included a first denaturation step at 95°C for 2 min and followed by 50 cycles of 95°C for 15 sec and 60°C for 1 min.
Table 2.
Primer and probe sequences used for quantitative methylation-specific PCR
| Gene | Forward primer | Reverse primer | Probe (5′Fam-3′Tamra) |
|---|---|---|---|
| p16 | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA | AGTAGTATGGAGTCGGCGGCGGG |
| DCC | TTGTTCGCGATTTTTGGTTTC | ACCGATTACTTAAAAATACGCG | GCGCTAAACAAAAAAACTCCGAAAA |
| KIF1A | GCGCGATAAATTAGTTGGCGATT | CTCGACGACTACTCTACGCTAT | CCTCCCGAAACGCTAATTAACTACGCG |
| EDNRB | GGGAGTTGTAGTTTAGTTAGTTAGGGAGTAG | CCCGCGATTAAACTCGAAAA | TTTTTATTCGTCGGGAGGAG |
| β-actin | TGGTGATGGAGGAGGTTTAGTAAGT | AACCAATAAAACCTACTCCTCCCTTAA | ACCACCACCCAACACACAATAACAAACACA |
As a standard reference for qMSP, leukocyte DNA from a healthy individual was completely methylated using excess SssI methyltransferase (New England Biolab Inc, Beverly, MA) and methylated DNA were serially diluted. The methylation level of each gene in each DNA sample was normalized to β-actin and calculated as an equation of (gene of interest/β-actin) × 100. DNA- and RNA-free water was used for negative control for each qMSP run. The cases in which microdissected tumor did not display any of the four methylation markers served as additional negative controls, while the microdissected tumor with methylation markers served as positive controls for analysis of deep margin tissue and all imprints.
Statistical Analysis
The methylation level of each gene of interest above established normal thresholds for imprint and tissue DNAs was plotted and the correlation was analyzed by Spearman’s rho correlation coefficients (r) using SPSS 12.0 for Windows (SPSS Inc, Chicago, USA). The statistical significance was accepted at P < 0.05.
RESULTS
Tissue imprinting from surgery for HNSCC
We performed the tissue imprinting procedure for total 17 subjects from May 2009 to June 2010. One case was excluded from further analysis because of being unavailable for follow-up having died in the immediate postoperative period. Four had no significant methylation signal of any of the four genes in the tumor (two of four cases had no viable tumor cells due to extensive necrosis). The margin and imprint samples from these four cases were tested for methylation as negative controls, but the cases are not included in assessment of the clinical correlation of deep margin imprints and disease recurrence;. The 12 remaining subjects who were followed for 10–22 months after surgery were included in margin-outcome analysis. We collected 12 tumor imprints and a total of 34 tissue imprints from quadrants of deep margins from these cases. Standard histologic examination confirmed that all tumors were squamous cell carcinoma and that frozen tissue samples taken from deep margins had no visible residual tumor. On final routine clinical surgical pathology analysis of the main resection specimen, however, one case had dysplasia at a mucosal margin, five had close deep margins (< 2 mm), and two had tumors at a mucosal or deep margin (Table 1).
DNA was successfully extracted from tumor and deep margin imprints of all cases. The mean total concentration of DNA extracted was measured as 10.4±5.6 μg/μL (range 4.6–25.2) from tumor imprint and 5.9±3.7 μg/μL (range 2.1–18.0) from deep margin imprint. The quantity and quality of imprint DNA was fit for bisulfite treatment and qMSP as was DNA extracted by tissue microdissection.
Detection of residual tumor cells at deep margisn with tissue imprinting and qMSP techniques
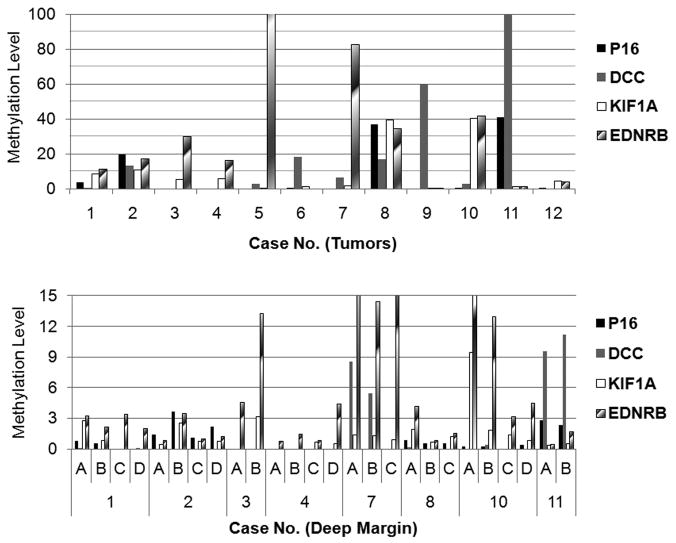
Hypermethylation of one or more genes was found in 12 tumors and in 21/34 tissue imprints taken from deep margins of 8/12 cases (Table 1 and Fig. 1). The four negative control cases (no markers in microdissected tumor) had no hypermethylation of any of the four genes in deep margin tissues or imprints. The specific methylation pattern of the four genes in tumor imprints differed among cases. The methylation patterns in tumors generally matched those at deep margins in the eight cases with molecularly positive margins. For example, case 3 had hypermethylation of KIF1A and EDNRB in primary tumor and the same methylation pattern was found in one quadrant of deep margin.
Figure 1.
Methylation analyses in tumors and deep margin. Imprints and tissues were obtained from tumors and each quadrant of deep margin of surgical specimens and qMSP was performed in bisulfite-treated DNAs. The methylation levels of four genes were calculated as indicated in Methods. The cases in the lower panel were chosen from patients with hypermethylated genes at deep margin imprints.
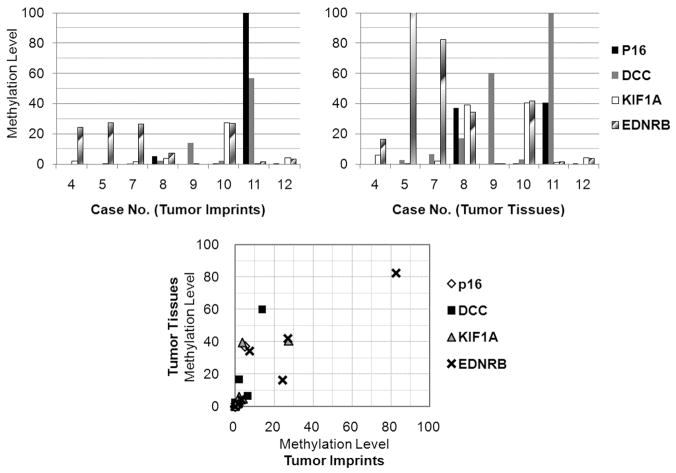
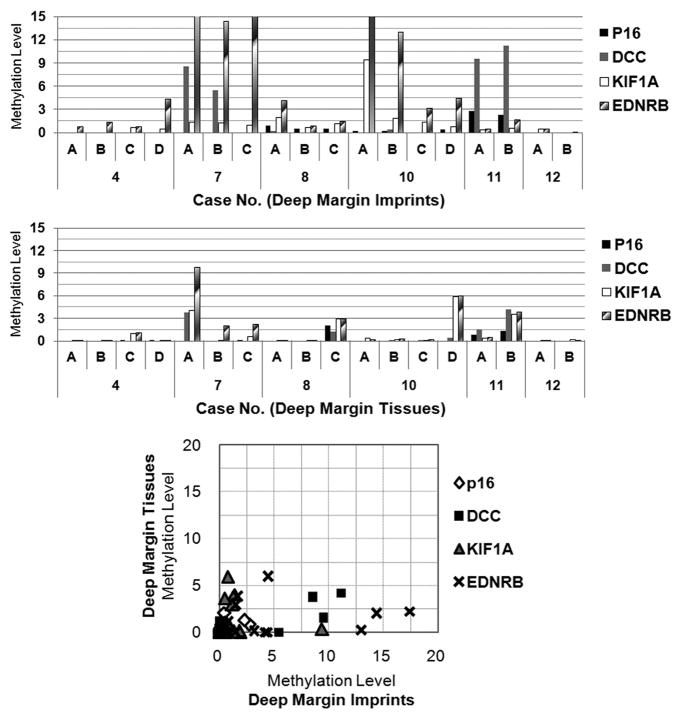
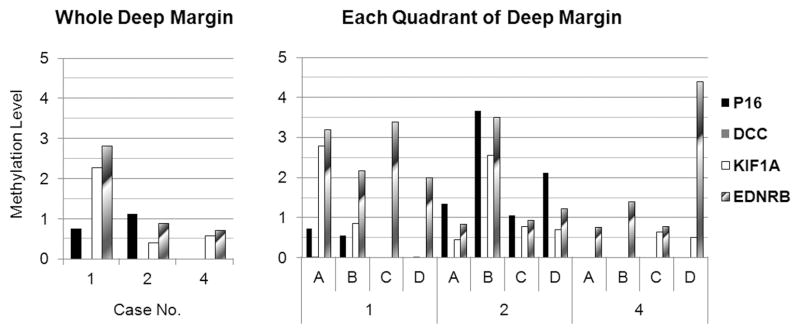
Tissue imprinting improves detection of residual tumor cells at deep margin
The qMSP results of imprint and tissue DNAs were compared. The methylation levels of genes were commonly higher in tumor tissues but were well correlated between tumor imprints and tissues (r > 0.85, P < 0.01) (Fig. 2). In contrast, the methylation levels of genes were higher in imprints from deep margin than from tissue samples and were not well correlated between margin tissue samples and imprints (P > 0.05) with the exception of DCC (r = 0.59, P = 0.01) (Fig. 3). Residual tumor DNA was more frequently detected by margin imprinting than biopsy of small tissue pieces. For example, hypermethylation of EDNRB was found in imprints of tumor and quadrant D of the deep margin but not found in any tissue samples from the deep margin in case 4. When the qMSP results of imprint DNAs from whole area and each quadrant of deep margin were compared, quadrant margin imprinting appeared to provide additional spatial information of residual tumor cells (Fig. 4).
Figure 2.
Comparison of methylation levels between imprints and tissues of tumors. Paired imprint and tissue samples were obtained from eight cases. The methylation levels of four genes were compared between imprint and tissue DNA eluted from tumors. The lower panel shows correlation of the gene methylation levels of tumor tissue and imprint DNA.
Figure 3.
Methylation levels of imprint and tissue DNA taken from deep margin. Paired imprint and small tissue samples were obtained from each quadrant of deep margin. The methylation levels of four genes were plotted and compared between imprint and tissue DNA.
Figure 4.
Methylation levels of imprint DNAs from whole area and each quadrant of deep margin. Left and right panels show the methylation levels of four genes in imprint DNA extracted from whole deep margin and each quadrant from three cases.
Molecular diagnosis of resection bed and clinical follow-up
During follow-up of median 13 months (range, 10–22), six had local and/or regional recurrences (Table 1) and 4 developed distant (pulmonary) metastases. Of eight cases with a molecularly positive deep margin, four had local recurrences during the follow-up period (median 13 months). Of these four cases of local recurrence, one involved deep soft tissue only, one was located at the mucosal surface and displayed dysplasia at the edges when re-resected, one displayed both deep and mucosal recurrence, and in the final case, the precise location of recurrence could not be discerned (table 1). Of four cases with molecularly negative deep margin, one patent (case 6) with a tumor-negative deep margin on both pathology and methylation examinations had regional nodal recurrence (retropharyngeal and paratracheal) 5 months after surgery. Another (case 5) had distant metastasis 12 months after surgery.
DISCUSSION
We have shown that tissue imprinting of specimens from resection of HNSCC is feasible for the detection of residual tumor cells at the deep surgical margin. All of our cases had tumor-free resection mucosal and deep margins on intraoperative frozen section (intraoperative) histopathologic examination. On final fixed tissue deep margin analysis some cases were judged as “close” or even “positive” (case #6). This is illustrative of the surgeon’s dilemma with the difficult task of assessing the adequacy of resection of deep margins during the case, and with determining with the pathologist the precise location of close margins on the final fixed resection specimen. The presence of hypermethylated genes in DNA taken from tumors and deep margin imprints was detected in about half of the cases and appears to correlate with locoregional recurrence. One case (#6) had no hypermethylated DNA detected on imprint sampling of the deep margin despite the final pathology report of a positive deep margin on the main specimen. Both imprint and histologic evaluation of tumor margins by the surgical pathologist are sampling and do not correspond 100% with clinical outcome. Subject #6 has not had a local recurrence during the follow-up period, but did re-present with tumor in a retropharyngeal node. It is possible that the imprint technique missed tumor seen by the pathologist particularly with the availability of only one methylation marker. It is also possible that the pathologist mis-judged the deep margin on the fixed specimen because of an artifact of handling. The sample size and follow-up period are not sufficient for rigorous assessment of the clinical value of molecular probing of deep surgical margins. The imprint method needs further refinement with identification of more robust methylation markers and testing on deep tissues taken from cancer-free individuals. However, even this preliminary evaluation provides provocative results suggesting that tissue imprinting at deep resection margin and qMSP detection of candidate genes may be useful to predict recurrence in head and neck cancer surgery.11–17,19
Transcriptional silencing of tumor suppressor genes by promoter CpG island hypermethylation has been identified as an important mechanism contributing to tumorigenesis in a variety of malignancies.20 Because cancer-specific DNA methylation can be detected in epithelial tumor cells, DNA methylation biomarkers have been used to probe for tumor-derived free DNA in body fluids for early detection of cancer.20,21 In addition, a few reports have described methylation analyses of resection margins in head and neck surgery.16,19,22,23 Two early reports failed to demonstrate an association between molecularly positive margins and local recurrence perhaps because of small sample size, limited methylation panels, and short follow-up period.22,23 Shaw, et al. did look at deep margins, but used a technique that may have introduced contamination from the adjacent tumor.23 Two reports identified similar methylation profiles between tumors and margins and an association between margin methylation results and relapse or cancer-specific deaths.16,19 One of these demonstrated that hypermethylation of promoters (CDKN2A, CCNA1, and DCC) in primary tumors was found in 27 of 42 eligible patients and promoter hypermethylation at surgical margins predicted all 5 disease-specific events (local recurrence or metastases) in 11 patents with molecularly positive margins.16 Tumor hypermethylation of one or more genes tested in our study was detected in 13 (76%) of all consecutive 17 cases. This is higher than the detection rates (50%) of HNSCC in previous reports using molecular markers such as p53 mutations and microsatellite instability11–24 but less than that of eIF4E.15 The percentage of hypermethylated tumors may be increased by including additional suitable candidate genes. The panel of markers used in our study has been shown to be altered in most HNSCC and are specific to the cancer phenotype. Tumor hypermethylation of the four genes has been validated with comparison to methylation levels of the genes in normal mucosal cells or saliva.22,24,25 These previous studies established threshold cutoffs above levels seen in normal blood and mucosa for each marker. The identical thresholds were used to score the margin imprints for this report. Assessment of the incremental contribution of each marker in the panel, that is the relative value of the panel over fewer selected markers is beyond the scope of this report.
The impetus of this study is to develop a method to test the proposed clinical importance of assessing surgical deep margin. This hypothesis is supported by a recent paper analyzing molecular margin from mucosal edges and deep sites in surgical defects which showed that p53 mutations in deep tissues was associated with treatment failure.17 The study revealed local recurrences in 10 of 16 patients with p53 mutation positive margin (5 at mucosal margin, 7 at deep margin, and 4 at both margins) and in 2 of 9 patients without p53-mutation positive margin. Of 10 true-positive cases, six were p53-mutation positive at a deep margin, one at a mucosal margin and three at both margins. The authors observed that p53 mutations in deep tissues are more strongly associated with recurrence than those at mucosal margin. They called for improved sampling techniques in order to enhance their results.
The present study focused on deep margin sample collection in order to assess the feasibility of the imprint technique. Mucosal margins were not tested, so we cannot comment on the relative value of deep vs. mucosal margins. We have demonstrated that residual tumor cells unnoticed by light microscopic evaluation of deep margin tissues are detected by the molecular assessment of DNA collected by simple nitrocellulose imprints that assess the entire resection margin. This approach samples the entire cut (true) deep margin, rather than a volume of tissue as is possible when a block of tissue is submitted for molecular analysis. There are hypothetical advantages to this and alternate approaches. Only rigorous testing of this approach in a larger, prospective sample will provide data to judge its full clinical relevance and allow comparison with other methods.
Imprinting of biological macromolecules blotted onto nitrocellulose membranes has been used for molecular profiling associated with biological changes that characterize specific cancers.18,26 Our technique differs from those published previously in that the DNA of adherent cells was eluted and processed for QMSP rather than analyzed directly on the nitrocellulose. While this approach sacrifices some of the precise topographical localization of tumor signal, it allows for rapid processing with a highly sensitive PCR based method that requires bisulfate treatment. Our approach allows for positive signal mapping as demonstrated by dividing the nitrocellulose imprint into quadrants. We found that some quadrants displayed tumor signal, while others from a neighboring region of the same specimen did not, indicating proof of principle for the ability of the method to pinpoint to regions of greatest concern for minimal residual tumor.
Tissue imprinting of head and neck specimens may have other potential benefits. Imprint and DNA elution procedures were very simple when compared with conventional tissue processing because imprinting can bypass the steps of tissue preparation and microdissection. The blotted macromolecules from tissues can be also tested or visualized by other molecular techniques, such as immunohistochemistry, immunoblotting, DNA-RNA hybridization, or other affinity-based techniques, for detection of tumor invasion or residual tumor cells at resection margins and deep tissues.18
This study has shown the potential technical advantages of tissue imprinting and molecular mapping of deep surgical margins. However, adequate assessment of predictive value of disease-specific survival data will require longer follow-up of a larger case series. Novel methylation markers may be also added in future studies. The data from tissue imprinting and methylation analyses at surgical margins may be combined with imaging studies or other molecular surveillance methods using body fluids for early detection of tumor recurrences. Tissue imprinting may also be adapted for rapid detection of residual tumor cells during head and neck cancer surgery.
In conclusion, our data suggest that the imprinting technique is a feasible method for molecular diagnosis of residual tumor cells at deep margins in head and neck surgery. The combination of tissue imprinting and methylation analyses at surgical deep margin may accurately predict locoregional recurrence in HNSCC patients undergoing surgery.
Translational Relevance.
Locoregional recurrence occurs in as many as 40% of patients undergoing surgery for head and neck squamous cell carcinoma (HNSCC) and contributes to poor survival. Positive surgical margins are a well established predictor of local recurrence. In head and neck cancer resection, the deep margins are more difficult to evaluate than are mucosal margins. This is true in part because of the larger surface area involved in the interface between infiltrating tumor and the muscles and other connective tissues below the surface compared to the linear cut edges of the mucosa. In addition, the deep margin tumor-host interface has few landmarks that can be used to orient specimens for re-localization of positive margins intra-operatively. Yet, the deep margin may be of greater clinical importance than the mucosal margins since residual cancer deep in the wound has ready access to vascular and neural pathways for metastasis, little to inhibit direct extension, and may be hidden from view beneath reconstruction or repair of the surgical defect. Studies that focus on the status of HNSCC surgical margins rarely consider deep margins with appropriate rigor.
Molecular diagnostic methods have been described to enhance detection of residual tumor cells at surgical margins that appear to be clear on routine histopathology. Tissue imprinting from surgical specimens with tissue-specific molecular profiling has the potential to improve detection of residual tumor cells at deep margins. In this study, we have shown that tissue imprinting and quantitative methylation-specific PCR can detect residual tumor cells and generate molecular maps of deep margins. This technique appears promising for molecular assessment of deep surgical margin to predict outcome (survival and locoregional recurrence).
Acknowledgments
Grant support: NIH/NIDCR R01 DE013152-11
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest are disclosed.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143–1154. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 4.Leemans CR, Tiwari R, Nauta JJ, van der Waal I, Snow GB. Recurrence at the primary site in head and neck cancer and the significance of neck lymph node metastases as a prognostic factor. Cancer. 1994;73:187–190. doi: 10.1002/1097-0142(19940101)73:1<187::aid-cncr2820730132>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 5.Warren S, Gates O. Multiple primary malignant tumors. A survey of the literature and statistical study. Am J Cancer. 1932;16:1358–1414. [Google Scholar]
- 6.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 7.Braakhuis BJ, Tabor MP, Leemans CR, van der Waal I, Snow GB, Brakenhoff RH. Second primary tumors and field cancerization in oral and oropharyngeal cancer: molecular techniques provide new insights and definitions. Head Neck. 2002;24:198–206. doi: 10.1002/hed.10042. [DOI] [PubMed] [Google Scholar]
- 8.Tabor MP, Brakenhoff RH, van Houten VM, et al. Persistence of genetically altered fields in head and neck cancer patients: biological and clinical implications. Clin Cancer Res. 2001;7:1523–1532. [PubMed] [Google Scholar]
- 9.Hermanek P, Hutter RV, Sobin LH, Wittekind C. Classification of isolated tumor cells and micrometastasis. Cancer. 1999;86:2668–2673. [PubMed] [Google Scholar]
- 10.van Houten VM, Tabor MP, van den Brekel MW, et al. Mutated p53 as a molecular marker for the diagnosis of head and neck cancer. J Pathol. 2002;198:476–486. doi: 10.1002/path.1242. [DOI] [PubMed] [Google Scholar]
- 11.Brennan JA, Mao L, Hruban RH, et al. Molecular assessment of histopathological staging in squamous-cell carcinoma of the head and neck. N Engl J Med. 1995;332:429–435. doi: 10.1056/NEJM199502163320704. [DOI] [PubMed] [Google Scholar]
- 12.Partridge M, Li SR, Pateromichelakis S, et al. Detection of minimal residual cancer to investigate why oral tumors recur despite seemingly adequate treatment. Clin Cancer Res. 2000;6:2718–2725. [PubMed] [Google Scholar]
- 13.van Houten VM, Leemans CR, Kummer JA, et al. Molecular diagnosis of surgical margins and local recurrence in head and neck cancer patients: a prospective study. Clin Cancer Res. 2004;10:3614–3620. doi: 10.1158/1078-0432.CCR-03-0631. [DOI] [PubMed] [Google Scholar]
- 14.Sardi I, Franchi A, Ferriero G, et al. Prediction of recurrence by microsatellite analysis in head and neck cancer. Genes Chromosomes Cancer. 2000;29:201–206. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1031>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 15.Nathan CO, Franklin S, Abreo FW, Nassar R, De Benedetti A, Glass J. Analysis of surgical margins with the molecular marker eIF4E: a prognostic factor in patients with head and neck cancer. J Clin Oncol. 1999;17:2909–2914. doi: 10.1200/JCO.1999.17.9.2909. [DOI] [PubMed] [Google Scholar]
- 16.Tan HK, Saulnier P, Auperin A, et al. Quantitative methylation analyses of resection margins predict local recurrences and disease-specific deaths in patients with head and neck squamous cell carcinomas. Br J Cancer. 2008;99:357–363. doi: 10.1038/sj.bjc.6604478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Pateromichelakis S, Hills A, et al. p53 mutations in deep tissues are more strongly associated with recurrence than mutation-positive mucosal margins. Clin Cancer Res. 2007;13:6099–6106. doi: 10.1158/1078-0432.CCR-07-1369. [DOI] [PubMed] [Google Scholar]
- 18.Gaston SM, Soares MA, Siddiqui MM, et al. Tissue-print and print-phoresis as platform technologies for the molecular analysis of human surgical specimens: mapping tumor invasion of the prostate capsule. Nat Med. 2005;11:95–101. doi: 10.1038/nm1169. [DOI] [PubMed] [Google Scholar]
- 19.Martone T, Gillio-Tos A, De Marco L, et al. Association between hypermethylated tumor and paired surgical margins in head and neck squamous cell carcinomas. Clin Cancer Res. 2007;13:5089–5094. doi: 10.1158/1078-0432.CCR-07-0119. [DOI] [PubMed] [Google Scholar]
- 20.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 21.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 22.Goldenberg D, Harden S, Masayesva BG, et al. Intraoperative molecular margin analysis in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2004;130:39–44. doi: 10.1001/archotol.130.1.39. [DOI] [PubMed] [Google Scholar]
- 23.Shaw RJ, Hall GL, Woolgar JA, et al. Quantitative methylation analysis of resection margins and lymph nodes in oral squamous cell carcinoma. Br J Oral Maxillofac Surg. 2007;45:617–622. doi: 10.1016/j.bjoms.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 24.Carvalho AL, Chuang A, Jiang WW, et al. Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res. 2006;66:9401–9407. doi: 10.1158/0008-5472.CAN-06-1073. [DOI] [PubMed] [Google Scholar]
- 25.Demokan S, Chang X, Chuang A, et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int J Cancer. 2010;127:2351–2359. doi: 10.1002/ijc.25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emmert-Buck MR, Strausberg RL, Krizman DB, et al. Molecular profiling of clinical tissues specimens: feasibility and applications. J Mol Diagn. 2000;2:60–66. doi: 10.1016/s1525-1578(10)60617-4. [DOI] [PMC free article] [PubMed] [Google Scholar]