Abstract
Autologous haematopoietic stem cell transplantation is highly efficient for the treatment of systemic autoimmune diseases, but its consequences for the immune system remain poorly understood. Here, we describe an optimized RNA-based technology for unbiased amplification of T cell receptor beta-chain libraries and use it to perform the first detailed, quantitative tracking of T cell clones during 10 months after transplantation. We show that multiple clones survive the procedure, contribute to the immune response to activated infections, and form a new skewed and stable T cell receptor repertoire.
Keywords: autoimmunity, ankylosing spondylitis, haematology, therapeutics, transplantation
INTRODUCTION
High-dose chemotherapy followed by autologous haematopoietic stem cell transplantation (HSCT), aiming to reset the dysregulated immune system, is becoming an established means of treating refractory autoimmune diseases, such as multiple sclerosis, rheumatoid arthritis, and systemic lupus erythematosus (Saccardi et al, 2008). However, few data are available regarding the specific changes in the immune system caused by HSCT, especially concerning the survival and re-expansion of T cell clones, which survive chemotherapy.
Several studies to characterize the changes in T cell receptor (TCR) repertoire after HSCT have been performed. The results of most reports indicate complete or nearly complete T cell repertoire ‘renovation’ and depletion of autoreactive clones (Alexander et al, 2009; Muraro et al, 2005), although some studies have indicated that pre-existing T cells can survive the conditioning regimen (Sun et al, 2004). However, in-depth comparative analysis of TCR repertoires before and after HSCT could not be performed, mainly because of the methodological limitations of the spectratyping and sequencing approaches employed for these studies (Rufer, 2005).
RESULTS
To overcome these limitations, we have developed an optimized TCR beta amplification technique and employed massive sequencing using the 454 Genome Sequencer FLX (Roche). The Genome Sequencer is preferable for this task since it generates a large number of reads of approximately 300–500 bp in length that is sufficient to acquire a barcode, TCR J beta segment, CDR3 region, and a significant portion of the TCR V beta segment sequences, allowing us to distinguish even highly homologous V beta genes. The latter is crucial for the rational analysis of TCR repertoires, since the V beta gene segment determines the complementarity determining region 1 (CDR1) and CDR2, which contribute to the TCR specificity.
To provide unbiased and representative TCR beta amplification without distorting the natural abundance of TCR clonal sequences that closely reflects the abundance of the corresponding T cell clones, we have developed an optimized technique (Supporting Information Fig 1), which:
Starts from RNA to exclude rearranged but non-functional TCR beta genes that are strongly downregulated by the nonsense-mediated mRNA decay mechanism (Bhalla et al, 2009; Wang et al, 2002).
Uses optimized TCR C beta-specific priming for cDNA synthesis, dramatically improving further amplification compared to oligo-dT or random hexamer priming.
Exploits the reverse transcriptase template switching effect (Douek et al, 2002; Matz et al, 1999; Zhu et al, 2001) to generate a universal primer at the 5′ end of the TCR beta chain and thus allows unbiased amplification using a single pair of universal primers.
Uses nested PCR to increase amplification specificity and step-out PCR (Matz et al, 1999) accompanied by the PCR-suppression effect (Siebert et al, 1995) to suppress products of non-specific primer annealing.
Introduces oligonucleotides for subsequent sequencing and DNA barcodes during amplification to avoid inefficient ligation of DNA adapters (Supporting Information Fig 1).
The combination of these technical solutions allowed us to specifically and uniformly amplify a ready-for-sequencing, rearranged TCR beta gene fragment library within 23 PCR cycles, when starting with approximately 2 µg of total RNA obtained from peripheral blood mononuclear cells (PBMC) purified from a 2 ml blood sample, containing approximately 2 million T cells (see Supporting Information Figs 1–3, Supporting Information Tables 1 and 2, and Supporting Information ‘Notes for details concerning technology development and verification’).
To analyse massive sequence data, we have developed a special software, named TCRbase, that has several capabilities including: extraction of the CDR3 regions; correction of typical Genome Sequencer errors; identification of TCR V beta and J beta gene families; clustering and ranging identical sequences; in silico spectratyping and clonotyping within specific TCR V beta and/or J beta gene families; and, importantly, cross analysis of multiple datasets to search for identical or homologous nucleotide or amino acid CDR3 sequences (see Supporting Information Figs 4 and 5 and Supporting Information ‘Notes for software details’).
This arsenal of tools was employed to perform the first deep, quantitative tracking of the fate of T cell clones in a 46-year-old patient with ankylosing spondylitis (a form of chronic, inflammatory arthritis) before and after autologous HSCT (see Supporting Information ‘Notes for the clinical details’). Samples of peripheral blood were collected 1 week before as well as 4 months and 10 months after the procedure and TCR beta amplification performed as described above. From these samples, TCRbase extracted approximately 11,500, 14,500, and 19,500 valid TCR beta sequences from the two small sequence runs performed (1/16 of a PicoTiterPlate) (see Supporting Information Data 1 for the list of clustered clonal sequences, raw data are available at NCBI SRA database, study accession number SRP005664). The three sequence sets were further analysed using TCRbase, which revealed the following:
At least 250 T cell clones survived HSCT, and we believe that deeper analysis would reveal many more clones that survived the procedure. This striking result demonstrates that effectiveness of HSCT therapy is based on reprogramming of re-expanding T cells, but not on the physical elimination of the ‘last autoimmune clone’ (de Kleer et al, 2006) (Supporting Information Table 3).
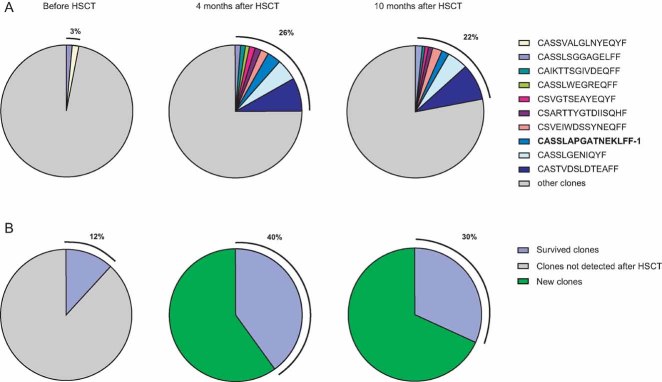
HSCT decreased overall diversity of T cell clones (Supporting Information Fig 6), while the number of ‘hyper-expanded’ clones (comprising >1% of all TCR beta sequences) increased, leading to the propagation of specific TCR V beta gene families (Supporting Information Fig 7). The cumulative contribution of ‘hyper-expanded’ clones increased from 3% before to 26% after HSCT and remained at this high level for at least 10 months after HSCT (Fig 1A).
A significant number of the clones, which survived the conditioning regimen, expanded after transplantation. Hence, most of the prominent expansions (Supporting Information Table 3) and as much as 40% of all T cells found after HSCT (Fig 1B) originated from the clones found before HSCT.
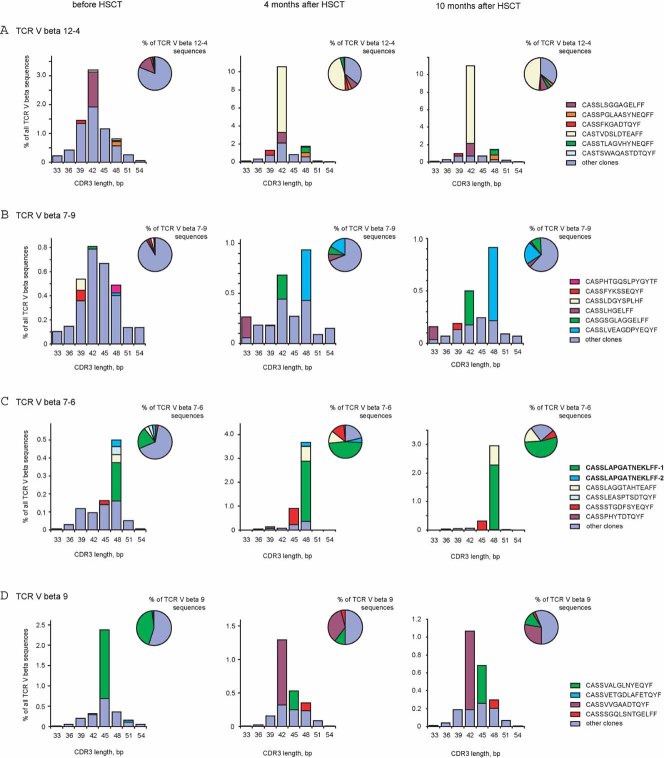
Clones, which survived, established a new balance that was unexpectedly stable, as revealed by in silico analysis, which demonstrates that the spectratypes and clonotypes skewed by HSCT remained intact over the next 6 months (Fig 2). Restoration of a full TCR beta repertoire which would re-produce the previous diversity and the clonal expansions normally present in healthy subjects, would probably take years, especially given the low abundance of naive T cells observed in the patient blood after HSCT (Supporting Information Fig 8).
At least some clones, which survived, expanded to provide specific immune defence early after transplantation. Using FACS sorting of T cells stained with an HLA-A2 MHC tetramer loaded with the immunodominant cytomegalovirus (CMV) peptide NLVPMVATV, we have identified two CMV-specific T cell clones that dramatically expanded after HSCT as shown by both mass sequencing (Fig 2 and Supporting Information Table 3) and flow cytometry (Supporting Information Fig 9) analysis. Both clones displayed the same TCR beta amino acid sequence identical to the CMV-specific sequences previously reported in other patients (V beta 7-6, J beta 1-4, CDR3: CASSLAPGATNEKLFF) (Price et al, 2005; Venturi et al, 2008). Expansion of these CMV-specific clones is consistent with clinical studies reporting the activation of persistent viral infections after HSCT (Afessa & Peters, 2006).
Stably expanded T cell clones have a high chance of surviving transplantation. Earlier, we described 11 T cell clones that were stably expanded in the peripheral blood of this patient for at least 3 years (Chkalina et al, 2010; Mamedov et al, 2009). Remarkably, all of these clones survived HSCT, although expansion of most of them was significantly suppressed (Supporting Information Tables 3 and 4).
Strikingly, more than 100 of the TCR beta CDR3 amino acid sequences from this one patient are 100% identical to CDR3 variants already referenced in the NCBI protein database (see Supporting Information Data 2 for the list of identified matches). Thus, despite the huge potential diversity of TCRs in humans (>1015, Davis & Bjorkman, 1988), the TCR sequence pool recognizing key antigenic peptides can appear to be rather restricted, at least in subjects sharing the same Class I MHC alleles.
Figure 1. Fate of the T cell clones that survived HSCT.
Each pie graph represents the total amount of TCR beta sequences obtained from a corresponding blood sample. Each slice represents the share of a clonal sequence or a group of clonal sequences, thus corresponding to abundance of specific T cell clones.
- Hyper-expanded clones (constituting >1% of all TCR beta sequences) before, 4 months after, and 10 months after transplantation are shown. Note that hyper-expanded clones constituted <3% of all T cells before and >20% after HSCT. Most of these clones were identified before HSCT at a low or medium level, but expanded dramatically after HSCT. CASSLSGGAGELFF was the only clone hyper-expanded in all three samples, while clone CASSVALGLNYEQYF was hyper-expanded before HSCT, but present at a relatively low level afterwards. The major CMV-specific clone CASSLAPGATNEKLFF-1 identified by MHC tetramer assay is shown in bold.
- Abundance of survived T cell clones within the overall T cell populations is shown. Those T cell clones, that survived HSCT initially, occupied only 12% of peripheral blood T cells before transplantation, but expanded to 40% of the population afterwards.
Figure 2. In silico CDR3 region spectratyping and clonotyping of selected TCR V beta gene segments before, 4 months after, and 10 months after HSCT.
CDR3 length is plotted along the x-axis. The 3 clones that were most abundant in at least one of the 3 blood samples are shown by individual colours. The abundance of the clonal TCR beta sequence, expressed as a percentage of total TCR beta sequences is plotted on the y-axis. Percentage of the clonal sequence among all sequences carrying the same TCR V beta gene is shown in pie graphs. CMV-specific clones CASSLAPGATNEKLFF-1 and -2 identified by MHC tetramer assay are shown in bold.
- TCR V beta 12-4
- TCR V beta 7-9
- TCR V beta 7-6
- TCR V beta 9
DISCUSSION
This work is the first deep analysis of the fate of clonal T cell populations after HSCT. It provides objective data to clarify the consequences of HSCT, which could previously only be assumed, and suggests the directions for further longitudinal studies of T cell repertoire and optimization of HSCT protocols. Importantly, intensive myeloablative regimens preceding HSCT were initially developed to eradicate tumour cells, which is critical for the successful treatment of malignant diseases (Dingli & Michor, 2006). However, satisfactory therapeutic effects using milder pre-transplant regimens were demonstrated for the treatment of autoimmune diseases (Hugle & van Laar, 2008). In our case, complete remission was observed for at least one year in spite of the fact that multiple T cell clones survived the procedure. On the other hand, although specific clones required to resist persistent infections and activated following immunosuppressive chemotherapy may successfully expand, as shown by the example of CMV-specific clones, the long term efficiency of adaptive immune response decreases (Kunisaki & Janoff, 2009). Thus, our data support the concept that chemotherapy regimens for the treatment of autoimmune diseases should be significantly moderated, both to reduce the toxicity of treatment and to preserve the clonal diversity of immune cells, which is otherwise dramatically altered.
The evolution of HSCT therapy protocols will require further studies of TCR repertoires before and after HSCT in order to understand the fate and the role of T cell clones in more detail. In particular, the fate of various subpopulations of T cells, including regulatory T and Th17 cells, is of significant interest. In addition, it is important to study whether changes in the T cell repertoire are dependent on different graft manipulation protocols, such as CD34+-positive selection or negative purging of lymphocyte subsets by monoclonal antibodies (Farge et al, 2010; Moore et al, 2002). While it was shown that CD34+-selected grafts do not contain large amounts of T cell clones after HSCT (Dubinsky et al, 2010), CD34+-positive selection was not performed in our case, and thus re-infusion of T cell clones with the autograft cannot be excluded.
Mass sequencing comparing TCR repertoires from a large number of healthy and diseased individuals, along with the progress in the analysis of TCR specificity (Bakker & Schumacher, 2005), should lead to the identification of common CDR3s that are generated in response to particular pathogens or to peptides from autoantigens. Along with the rapidly increasing cost efficiency of mass sequencing, these approaches might allow single-step diagnosis of particular infectious and autoimmune diseases, as well as deciphering an individual's history of diseases and vaccinations.
The paper explained
PROBLEM:
Autologous haematopoietic stem cell transplantation (HSCT) is successfully used to treat severe autoimmune diseases, but the mechanisms, by which it resets the dysregulated immune system, remain poorly understood.
RESULTS:
In this work, we report an optimized TCR profiling technology based on 454 massive sequencing and use it to track the fate of T cell clones at three time points: before, 4 months after and 10 months after high-dose chemotherapy and HSCT. We directly demonstrate that multiple T cell clones survive transplantation and that some of them expand and fight infection early after HSCT. We observe that, by 4 months after HSCT, a new balance is formed between the clonal T cell populations, which then remains stable at least for the next 6 months. Interestingly, we show that multiple TCR beta CDR3 amino acid sequences from one patient can be identical to CDR3 variants found in other patients, indicating that the actual TCR sequence pool recognizing key antigenic peptides can be rather restricted, in spite of their huge potential diversity.
IMPACT:
We anticipate that our work will intensify studies of human TCR repertoires and optimization of HSCT regimens aimed to reduce their toxicity and to improve long-term outcomes.
MATERIALS AND METHODS
HSCT
The study was approved by local ethical committee. Patient gave written informed consent prior to enrolling in the study. Stem cells were mobilized with G-CSF (10 µg/kg b wt.). A ‘Haemonetics MCS’ instrument was used for autologous stem cells aphaeresis. CD34+-positive selection was not performed. The patient received 200 mg/kg of cyclophosphamide over 4 days with autologous blood stem cells for rescue and infusion of antithymocyte globulin (ATG) for in vivo T cell depletion. On day 0 (June 6, 2009), the patient received 2.4 × 106 kg b wt. CD34+ stem cells, the number of which was determined by counting total number of cells and FACS analysis of an aliquot of cells using a CD34-specific antibody. No life-threatening events occurred during transplantation. Post-transplant toxicity included neutropenia with neutrophil count <0.5 × 109/L (from day +2 to day +9); thrombocytopenia, with thrombocyte count <50 × 109/L (from day +2 to day +12); fever (from day +4 to day +5); and stomatitis.
Flow cytometry
PBMCs were isolated from the peripheral blood samples by Ficoll-Paque (Paneco, Russia) density gradient centrifugation. Cells were washed twice with PBS and resuspended to a density of approximately 106 cells/ml. Cells were incubated with antibodies or MHC tetramer for 20 min at room temperature or at +8°C and washed twice with PBS. Phenotypic cell analysis was performed using a Cytomics FC 500 (Beckman Coulter) flow cytometer. Data were analysed using the program Cytomics RXP Analysis (Beckman Coulter).
Antibodies
Cells were stained with the following mouse anti-human antibodies: CD3-PC5 (clone UCHT1, Invitrogen), CD4-PC5 (clone S3.5, Invitrogen), CD8-FITC (clone YTC 182.20, Abcam, UK). The remaining antibodies were purchased from Beckman Coulter (USA): CD45RA-FITC (clone 2H4LDH11LDB9), CD27-PC5 (clone 1A4CD27), TCR-VB1(VB9)-PE (clone BL 37.2), TCR-VB7(VB4-1)-FITC (clone ZOE), TCR-VB16(VB14)-FITC (clone TAMAYA 1.2), CD8-PC7 (clone SFCI21Thy2D3).
Identification of CMV-specific T cell clones
PBMCs were incubated with CD8-specific antibody and MHC tetramer for 20 min at room temperature in PBS Ca2+, Mg2+ free/1 mM EDTA/1% BSA with added DNaseI (Promega, USA) and sorted on a MoFlo cell sorter (DakoCytomation, USA). RNA was extracted using TRIZOL reagent (Invitrogen). Fifty cloned TCR beta chains were sequenced.
Preparation of sample for mass sequencing
PBMCs were isolated by Ficoll-Paque (Paneco, Russia) density gradient centrifugation from fresh peripheral blood samples. RNA was isolated using Trizol reagent (Invitrogen, USA) according to manufacturer protocol. First strand cDNA was synthesized over a period of 2 h using Mint cDNA synthesis kit (Evrogen, Russia) and the specific primer BC1R, which binds to both constant regions of the human TCR beta transcript. Plug oligo (Evrogen) was added after the first 30 min of synthesis. The PCR amplification protocol was as follows: 94°C for 20 s; 65°C for 20 s; and 72°C for 50 s, 16 cycles. The PCR mixture (25 µl) contained 1× PCR buffer for Encyclo polymerase (Evrogen), 0.125 mM of each deoxyribonucleotide triphosphate (dNTP), 10 pmol of primers M1 and BC2R, 0.5 µl of Encyclo polymerase mix, and 1 µl of undiluted first-strand cDNA. After the first PCR step, the product was diluted 20× and 1 µl was used in the second 25-µl PCR reaction. This PCR reaction contained 1× PCR buffer, 0.125 mM of each dNTP, 10 pmol of each of the primers B-M1 and A-MID1-BC3R for the pre-HSCT sample, primer A-MID2-BC3R for post-HSCT sample, and B-MID3-BC3R, B-MID5-BC3R, B-MID6-BC3R for the 10 months post-HSCT sample along with 0.5 µl of Encyclo polymerase mix (Evrogen). The amplification protocol was as follows: 94°C for 20 s; 68°C for 20 s; and 72°C for 50 s, 12 cycles. After the second PCR, the amplicons were purified by agarose gel electrophoresis using a DNA gel extraction kit (Cytokin, Russia). The purified amplicons were collected, re-amplified for additional two cycles to polish the ends, and finally purified with QIAquick PCR purification kit (Qiagen, USA). Sequencing was performed using a Genome Sequencer FLX using a GS Em PCR Kit II (Roche Applied Science). For the oligonucleotides used, see Supporting Information Table 2.
Acknowledgments
We are grateful to Prof. J.S.H. Gaston for the valuable suggestions and comments on the manuscript. This work was supported by Molecular and Cell Biology program RAS, Rosnauka 02.512.12.2053, RFBR 10-04-01771-a, and Basic Research for Medicine RAS.
Supporting information is available at EMBO Molecular Medicine online.
The authors declare that they have no conflict of interest.
Author contributions
IZM, DAB, AVC, AAK, MAT, and IVZ performed DNA-technique optimization and prepared the samples for pyrosequencing. DAF and AAN performed HSCT. OVB, DBS, AAK, and GVS worked with blood samples and performed flow cytomerty analysis and sorting. DAB and DMC developed software. SL, YBL, and DMC contributed to the overall idea, organization and manuscript preparation.
For more information
Dmitriy Chudakov lab website:
http://www.ibch.ru/en/structure/groups/fluortools
Pirogov National Medical Surgical Center, Department of Haematology and Cellular Therapy:
http://www.gemclinic.ru/english.php
IMGT Repertoire, TCR and Ig database:
http://www.imgt.org/textes/IMGTrepertoire/
454 Sequencing:
Supplementary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Afessa B, Peters SG. Major complications following hematopoietic stem cell transplantation. Semin Respir Crit Care Med. 2006;27:297–309. doi: 10.1055/s-2006-945530. [DOI] [PubMed] [Google Scholar]
- Alexander T, Thiel A, Rosen O, Massenkeil G, Sattler A, Kohler S, Mei H, Radtke H, Gromnica-Ihle E, Burmester GR, et al. Depletion of autoreactive immunologic memory followed by autologous hematopoietic stem cell transplantation in patients with refractory SLE induces long-term remission through de novo generation of a juvenile and tolerant immune system. Blood. 2009;113:214–223. doi: 10.1182/blood-2008-07-168286. [DOI] [PubMed] [Google Scholar]
- Bakker AH, Schumacher TN. MHC multimer technology: current status and future prospects. Curr Opin Immunol. 2005;17:428–433. doi: 10.1016/j.coi.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Bhalla AD, Gudikote JP, Wang J, Chan WK, Chang YF, Olivas OR, Wilkinson MF. Nonsense codons trigger an RNA partitioning shift. J Biol Chem. 2009;284:4062–4072. doi: 10.1074/jbc.M805193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chkalina AV, Zvyagin IV, Mamedov IZ, Britanova OV, Staroverov DB, Lebedev YB. The oligoclonal expansion of T cells: the investigation of its stability over time. Russian J Bioorg Chem. 2010;36:191–198. doi: 10.1134/s1068162010020081. [DOI] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- de Kleer I, Vastert B, Klein M, Teklenburg G, Arkesteijn G, Yung GP, Albani S, Kuis W, Wulffraat N, Prakken B. Autologous stem cell transplantation for autoimmunity induces immunologic self-tolerance by reprogramming autoreactive T cells and restoring the CD4+CD25+ immune regulatory network. Blood. 2006;107:1696–1702. doi: 10.1182/blood-2005-07-2800. [DOI] [PubMed] [Google Scholar]
- Dingli D, Michor F. Successful therapy must eradicate cancer stem cells. Stem Cells. 2006;24:2603–2610. doi: 10.1634/stemcells.2006-0136. [DOI] [PubMed] [Google Scholar]
- Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai KL, Karandikar NJ, Casazza JP, Koup RA. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- Dubinsky AN, Burt RK, Martin R, Muraro PA. T-cell clones persisting in the circulation after autologous hematopoietic SCT are undetectable in the peripheral CD34+ selected graft. Bone Marrow Transplant. 2010;45:325–331. doi: 10.1038/bmt.2009.139. [DOI] [PubMed] [Google Scholar]
- Farge D, Labopin M, Tyndall A, Fassas A, Mancardi GL, Van Laar J, Ouyang J, Kozak T, Moore J, Kotter I, et al. Autologous hematopoietic stem cell transplantation for autoimmune diseases: an observational study on 12 years' experience from the European Group for Blood and Marrow Transplantation Working Party on Autoimmune Diseases. Haematologica. 2010;95:284–292. doi: 10.3324/haematol.2009.013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugle T, van Laar JM. Stem cell transplantation for rheumatic autoimmune diseases. Arthritis Res Ther. 2008;10:217. doi: 10.1186/ar2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki KM, Janoff EN. Influenza in immunosuppressed populations: a review of infection frequency, morbidity, mortality, and vaccine responses. Lancet Infect Dis. 2009;9:493–504. doi: 10.1016/S1473-3099(09)70175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamedov IZ, Britanova OV, Chkalina AV, Staroverov DB, Amosova AL, Mishin AS, Kurnikova MA, Zvyagin IV, Mutovina ZY, Gordeev AV, et al. Individual characterization of stably expanded T cell clones in ankylosing spondylitis patients. Autoimmunity. 2009;42:525–536. doi: 10.1080/08916930902960362. [DOI] [PubMed] [Google Scholar]
- Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J, Brooks P, Milliken S, Biggs J, Ma D, Handel M, Cannell P, Will R, Rule S, Joske D, et al. A pilot randomized trial comparing CD34-selected versus unmanipulated hemopoietic stem cell transplantation for severe, refractory rheumatoid arthritis. Arthritis Rheum. 2002;46:2301–2309. doi: 10.1002/art.10495. [DOI] [PubMed] [Google Scholar]
- Muraro PA, Douek DC, Packer A, Chung K, Guenaga FJ, Cassiani-Ingoni R, Campbell C, Memon S, Nagle JW, Hakim FT, et al. Thymic output generates a new and diverse TCR repertoire after autologous stem cell transplantation in multiple sclerosis patients. J Exp Med. 2005;201:805–816. doi: 10.1084/jem.20041679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufer N. Molecular tracking of antigen-specific T-cell clones during immune responses. Curr Opin Immunol. 2005;17:441–447. doi: 10.1016/j.coi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Saccardi R, Di Gioia M, Bosi A. Haematopoietic stem cell transplantation for autoimmune disorders. Curr Opin Hematol. 2008;15:594–600. doi: 10.1097/MOH.0b013e3283136700. [DOI] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Popat U, Hutton G, Zang YC, Krance R, Carrum G, Land GA, Heslop H, Brenner M, Zhang JZ. Characteristics of T-cell receptor repertoire and myelin-reactive T cells reconstituted from autologous haematopoietic stem-cell grafts in multiple sclerosis. Brain. 2004;127:996–1008. doi: 10.1093/brain/awh117. [DOI] [PubMed] [Google Scholar]
- Venturi V, Chin HY, Asher TE, Ladell K, Scheinberg P, Bornstein E, van Bockel D, Kelleher AD, Douek DC, Price DA, et al. TCR beta-chain sharing in human CD8+ T cell responses to cytomegalovirus and EBV. J Immunol. 2008;181:7853–7862. doi: 10.4049/jimmunol.181.11.7853. [DOI] [PubMed] [Google Scholar]
- Wang J, Vock VM, Li S, Olivas OR, Wilkinson MF. A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation. J Biol Chem. 2002;277:18489–18493. doi: 10.1074/jbc.M111781200. [DOI] [PubMed] [Google Scholar]
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.