Abstract
Protein pathways are dynamic and highly coordinated spatially and temporally, capable of performing a diverse range of complex chemistries and enzymatic reactions with precision and at high efficiency. Biotechnology aims to harvest these natural systems to construct more advanced in vitro reactions, capable of new chemistries and operating at high yield. Here, we present an efficient Multiplex Automated Genome Engineering (MAGE) strategy to simultaneously modify and co-purify large protein complexes and pathways from the model organism Escherichia coli to reconstitute functional synthetic proteomes in vitro. By application of over 110 MAGE cycles, we successfully inserted hexa-histidine sequences into 38 essential genes in vivo that encode for the entire translation machinery. Streamlined co-purification and reconstitution of the translation protein complex enabled protein synthesis in vitro. Our approach can be applied to a growing area of applications in in vitro one-pot multi-enzyme catalysis (MEC) to manipulate or enhance in vitro pathways such as natural product or carbohydrate biosynthesis.
Keywords: Genome engineering, MAGE, cell-free protein synthesis, multi-enzyme catalysis, protein purification
INTRODUCTION
Cell-free protein synthesis (CFPS) systems offer the ability to produce a variety of compounds that are otherwise toxic in vivo to a host cell, such as many membrane proteins (1–4). In addition to achieving better yields for hard-to-express proteins, cell-free synthesis can be engineered to incorporate non-standard amino acids into proteins (5). In recent years, a technical renaissance has revolutionized crude extract cell-free protein synthesis systems. Protein yields exceed grams of protein produced per liter reaction volume, batch reactions last for multiple hours, and reaction scale has reached the 100-liter milestone (4). Despite this success, limitations still exist (6). Paramount among these are the presence of proteases, nucleases and tmRNAs in the extract causing truncated protein products (7). Ten years ago, Protein synthesis Using Recombinant Elements (PURE) technology was described for 31 individually purified E. coli translational factors, ribosomes, proper energy supplementary system, and all necessary substrates to reconstitute an in vitro protein translation system (8,9). Because its components are defined, this and related purified translation systems do not contain some detrimental enzymes found in extracts, which can provide advantages for the reconstitution of membrane integration (10), ribosome display for selection or evolution (11,12), and protein synthesis with multiple unnatural amino acids (13–15).
The PURE system is an example of a growing class of multi-enzyme catalysis (MEC) done in a single-pot reaction (16). MEC is an attractive solution for taking a low-cost source molecule through a series of enzymatic reactions to generate high-value compounds (17,18). An elegant example of MEC is the mixing of eight enzymes into a single-pot reaction to convert sucrose to phosphorylated ketoses (19). More recent examples include tour de force synthetic pathway constructions of 28 and 18 enzymes from different species being required for purine (20) and pyrimidine (21) biosynthesis, respectively. In vitro MEC has several advantages over cellular-based systems (17,22). First, rather than attempt to balance the tug-of-war between the cell’s objectives and the engineer’s objectives, in vitro biocatalysis focuses cellular resources toward an exclusive user-defined objectives. Second, cell viability constraints are removed. Third, transport barriers are removed, allowing easy substrate addition, product removal, system monitoring, and rapid sampling. Despite its potential, in vitro multi-enzymatic catalysis has, however, been confined mostly to short reaction cascades due to increased requirements for enzyme isolation and optimization. Therefore, a tool that enables the isolation of all enzymes from a designated pathway from the cell while maintaining the functional state and proper stoichiometries of the components will facilitate both our understanding of that system and our ability to engineer it.
Recently, we reported the Multiplex Automated Genome Engineering (MAGE) method to introduce genetic modifications to the chromosome of Escherichia coli at high efficiency and in a combinatorial manner using short oligonucleotides (23,24). By transfection of 90mer-targeting oligonucleotides into a cell that expresses the λ-Red recombination machinery, strains can be engineered to contain any desired set of genomic changes including insertions, deletions, or mismatches (25). While the efficiency of MAGE can be as high as 35% per iterative cycle, increasing the size of the modification decreases the efficiency of the process (26). Nonetheless, the efficiency of introducing an 18 basepair insertion to the target site is ~2–5% per MAGE cycle, which can be multiplexed across several targets.
Here, we demonstrate the efficient incorporation of poly-histidine purification tags (His-tags) into protein synthesis genes in E. coli using MAGE. We generated a reduced set of strains that can be used to co-purify all components of an in vitro PURE translation system. The incorporation of His-tags into up to 8 different translation factors in a single strain does not dramatically affect the fitness of the cell. Furthermore, we document strategies to apply over 110 MAGE cycles towards engineering genomes in scenarios where individual MAGE cycles may be less efficient (<5%) due to toxicity of the genetic modification or the increased size of the modification. Since our ensembles of PURE (ePURE) strains are grouped functionally by their translation factors, they may serve to facilitate the improvement of in vitro PURE translation reactions. More broadly, this work demonstrates a novel approach towards simultaneous co-purification of assemblies of enzymes associated with metabolic pathways or other complex biochemical reactions such as post-translational modifications.
RESULTS AND DISCUSSION
Nine total strains were constructed for the ensemble PURE (ePURE) system (Table 1). Most of the His-tags in this study were introduced at the terminus specified in the PURE system previously (27), with the exception of glnS, which could only be tagged at the N-terminus by our method. Additionally, pheT was tagged at the C-terminus while pheS was tagged at the N-terminus. The altered genes were grouped by functionality of the translation factors and synthetases (see Table 1). The IEF (initiation and elongation factor) strain is based on a minimal His-tagged translation system (28) and contains His-tags of the three initiation factors (IF1, IF2, and IF3) and three elongation factors (EF-Ts, EF-Tu, EF-G), which included both chromosomal copies of the EF-Tu gene (tufA and tufB). In addition, we included elongation factor 4 (coded by lepA) in the IEF strain, which was not in the original minimal or PURE systems, because EF-4 has only recently been shown to increase the total protein yield in in vitro translation reactions (29). The RF strain contained the release factors (RF1, RF2, and RF3) as well as the ribosomal recycling factor (RRF). Insertion of C-terminal tags into RF1 (prfA) or RF2 (prfB) without refactoring the operon structure was not possible because both genes are in polycistronic operons with the stop codons of prfA and prfB overlapping with the start codons of downstream genes. However, we easily found simultaneous N- and C- terminus tags for RF3 (prfC), which stimulates the release of RF1 and RF2 from the ribosome after peptide chain termination. Interestingly the N-terminal domains of RF1 and RF2 appear to be required to interact with RF3 (30), but they are not affected by N-terminus His-tags. Simultaneous incorporation of His-tags into the N- and C-terminus of RF3 does not appear to affect the cell’s growth rate (Supplementary Figure 1).
Table 1.
The nine strains constructed by MAGE listing the genes, their function, location of hexa-histidine tags, and chromosomal location of each His-tag.
| Strain | Gene | Function | Protein | His-tagged Terminus | Chrom. Location |
|---|---|---|---|---|---|
| IEF | tsf | Translational elongation factor Ts | EF-Ts | C | 191,708 |
| infA | Translational initiation factor 1 | IF1 | N | 925,666 | |
| infC | Translational initiation factor 3 | IF3 | N | 1,798,662 | |
| lepA | Translational elongation factor 4 | EF4 | C | 2,703,347 | |
| infB | Translational initiation factor 2 | IF2 | N | 3,314,036 | |
| tufA | Translational elongation factor Tu | EF-Tu | C | 3,468,167 | |
| fusA | Translational elongation factor G | EF-G | C | 3,469,422 | |
| tufB | Translational elongation factor Tu | EF-Tu | C | 4,175,151 | |
|
| |||||
| RF | frr | Ribosome recycling factor | RRF | C | 193,429 |
| prfA | Translational release factor 1 | RF1 | N | 1,265,317 | |
| prfB | Translational release factor 2 | RF2 | N | 3,033,206 | |
| prfC | Translational release factor 3 | RF3 | N | 4,607,437 | |
| C | 4,609,026 | ||||
|
| |||||
| RS1 | ileS | Ile-tRNA synthetase | Syi | N | 22,391 |
| proS | Pro-tRNA synthetase | Syp | C | 217,057 | |
| cysS | Cys-tRNA synthetase | Syc | C | 555,219 | |
| leuS | Leu-tRNA synthetase | Syl | C | 671,424 | |
| glnS | Gln-tRNA synthetase | Syq | N | 706,980 | |
| serS | Ser-tRNA synthetase | Sys | C | 939,943 | |
|
| |||||
| RS2 | asnS | Asn-tRNA synthetase | Syn | N | 988,208 |
| tyrS | Tyr-tRNA synthetase | Syy | C | 1,713,972 | |
| pheT | Phe-tRNA synthetase B | Syfb | C | 1,793,581 | |
| pheS | Phe-tRNA synthetase A | Syfa | N | 1,796,966 | |
| thrS | Thr-tRNA synthetase | Syt | N | 1,800,594 | |
| aspS | Asp-tRNA synthetase | Syd | C | 1,946,774 | |
|
| |||||
| RS3 | argS | Arg-tRNA synthetase | Syr | N | 1,958,086 |
| metG | Met-tRNA synthetase | Sym | C | 2,194,355 | |
| gltX | Glu-tRNA synthetase | Sye | C | 2,517,279 | |
| hisS | His-tRNA synthetase | Syh | C | 2,637,323 | |
| alaS | Ala-tRNA synthetase | Sya | N | 2,820,033 | |
| lysS | Lys-tRNA synthetase | Syk | C | 3,031,679 | |
|
| |||||
| RS4 | fmt | Met-tRNA formyltransferase | Fmt | C | 3,433,183 |
| trpS | Trp-tRNA synthetase | Syw | C | 3,510,656 | |
| glyS | Gly-tRNA synthetase B | Sygb | C | 3,720,351 | |
| glyQ | Gly-tRNA synthetase A | Syga | C | 3,722,430 | |
| valS | Val-tRNA synthetase | Syv | C | 4,479,005 | |
|
| |||||
| RB1 | rplL | 50S ribosomal protein (L7/L12) | C | 4,178,945 | |
|
| |||||
| RB2 | rpsB | 30S ribosomal protein (S2) | C | 190,599 | |
|
| |||||
| RB3 | rplC | 50S ribosomal protein (L3) | C | 3,450,319 | |
The 23 aminoacyl-tRNA synthetase components (18 single-subunit RSs plus two subunits of both PheRS and GlyRS, and MTF) were split into four separate strains RS1-4, grouped contiguously by their chromosomal positions. In this fashion, RS1-4 can potentially be combined into one strain containing all 23 RSs by merging the contiguous genomic segments together through a conjugation-based genome assembly strategy (25). Finally, we inserted His-tags into the L7/L12, L3, and S2 ribosomal proteins. Previously, the His-tagging of L7/L12 enabled the purification of ribosomes and their subunits (31). An initial set of experiments were done on IEF, RF, and RS4 strains to optimize the MAGE cycling and screening protocol. MAGE was run for 35 cycles with the appropriate set of oligos (8 IEF targeting oligos, 5 RF targeting oligos, or 5 RS4 targeting oligos) for each strain. The three MAGE cycled populations were then plated on LB-agar solid medium, and 96 colonies were isolated for each. Multiplex Allele-Specific Colony PCR (MASC-PCR) was performed on each clone (see Methods and Figure 1b) to query all target sites to identify the clone that contained the most number of insertion tags. This clone was then chosen for the next iteration of MAGE cycling. In the next iteration, oligos targeting only the remaining unmodified sites were used, thereby increasing the efficiency of the process with each stage of this process. Since efficient MAGE required that the oligos target the lagging-strand of the replicating chromosome, two possible sets of oligos existed depending on which replichore the target was located. One oligo set contained sense hexa-histidine sequences (+ oligos), while the other contained anti-sense sequences (− oligos). When the two different sets were placed in the same MAGE reaction, the 18-bp His-tag sequence would anneal to its complement sequence thereby inhibiting the MAGE reaction by generating double stranded DNA. Therefore, we opted to use the two different oligo sets in separate stages of MAGE reaction (i.e. target all +oligo sites, isolate best clone, and then switch to target all −oligo sites next).
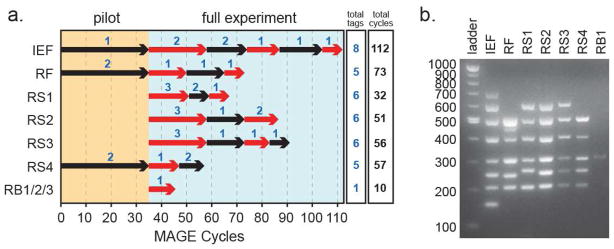
Figure 1. His-tagging of Translation Proteins and Ribosomes by MAGE.
(a). Course of strain construction by MAGE cycling. Each alternating black and red arrow designates one stage of MAGE cycling, ending with multiplex allele-specific PCR (MASC-PCR) of 96 isolates to select the clone with the highest number of His-tag incorporations (each new set of incorporations are designated by the number above the arrows). Total cycles taken to construct each strain and the total targets are detailed in the columns on the right. (b). MASC-PCR verification of the inserted his-tagged sequences in each strain as seen on a 1.5% agarose gel. Amplicons are designed to amplify at 150, 200, 250, 300, 400, 500, 600, 700 bps, corresponding to a maximum of 8 simultaneous His-tags in one strain. RB2 and RB3 tagging verification are not shown.
Figure 1a describes the His-tag progression of all strains. Up to six stages (denoted by alternating black and red arrows) were required to insert all the tags into each strain with periodic MASC-PCR screenings to isolate the best clones. The number of cycles done in any stage varied depending on the number of different oligo species that were involved and was determined by MAGE optimization algorithms previously described (24). The process was generally very efficient, capable of generating 3 of 4 possible insertions in as little as 16 MAGE cycles (strain RS1). Up to 112 MAGE cycles were done on the IEF strain without any discernable negative effect on the cells, which is the highest number of MAGE cycles documented in the literature thus far. Because the cycles were done in a multiplexed fashion, we were able to isolate clones containing a combinatorial set of His-tag inserts. This provided us with the opportunity to potentially identify synthetic lethal or positively synergistic effects associated with the introduction of the non-native poly-histidine tags to the protein-coding region of these essential genes. Furthermore, we could quantify the effect of each His-tag (or combinations of tags) on the fitness of the cell by growth rate measurements.
Insertion of His-tag sequences into all target genes in the ePURE strains was characterized by MASC-PCR (see Figure 1b) and subsequently verified by Sanger sequencing. Phenotypic comparison of the growth rate of most ePURE strains with the ancestral strain show no or modest differences (Supplementary Figure 1), implying that the His-tags proteins are functional since the tagged factors and synthetases are essential genes. Two strains, IEF and RS3, show modest increases in doubling time by 33% and 38%, respectively, from the EcNR2 ancestor. Tracing of the cell lineages and comparison of all other isolated clones showed that the C-terminus argS His-tag was the sole cause of the doubling time increase of RS3, while the cause of IEF growth retardation is unclear.
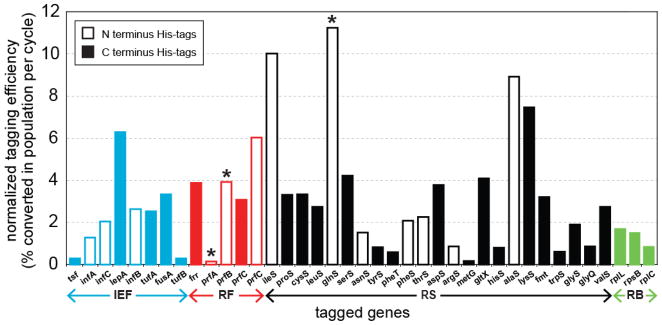
The normalized insertion efficiency per MAGE cycle at each gene is detailed in Figure 2. The normalized insertion efficiency for each gene was determined based on a binomial behavior of the multiplexed process (23) using the formula, k(1 − (1 − f)1/N), where f is the frequency at which inserts are observed in the 96 colonies screened after N MAGE cycles and k is the number of different oligos in the multiplex reaction. The insertion efficiency varied between 0.2% and 11.2% depending on the gene. Fluctuations in the His-tag insertion efficiency between different gene targets were likely due to both technical and biological causes. The efficiency of MAGE reactions can be strongly inhibited by secondary structures (e.g. hairpins) formed by the oligo as previously observed (23). Therefore, all oligos were designed to have a minimized secondary structure (ΔG>-10 kcal/mol) as determined by Mfold (32). However, a few oligos (e.g. tsf, metG, glyQ) could not be further optimized due to sequence constraints at the target site, thus resulting in lower insertion efficiency. On the other hand, the biological causes of lower insertion efficiencies are mostly due to other functional elements within the region of insertion, such as promoters or ribosomal binding sites of downstream gene or in polycistronic operons. Indeed, this was the case for tyrS, pheT, and hisS, which had C-terminal His-tags that might have affected the downstream gene expression.
Figure 2. Normalized efficiency of incorporating His-tags into each gene.
Open bars designate N-terminus tags, solid bards designate C-terminus tags. Functional groups are designated below the sets of colored bars: IEF, RF, RS, RB. Stars above the prfA, prfB and glnS bars indicate that a C-terminus was also attempted but failed to incorporate at adequate efficiency by MASC-PCR screening 96 clones. Efficiencies are calculated from relative efficiency RE, number of cycles N, and number of multiplex sites K using the formula: K*(1−(1−RE)1/N) in accordance to a binomial distribution prediction of the multiplexed cell population.
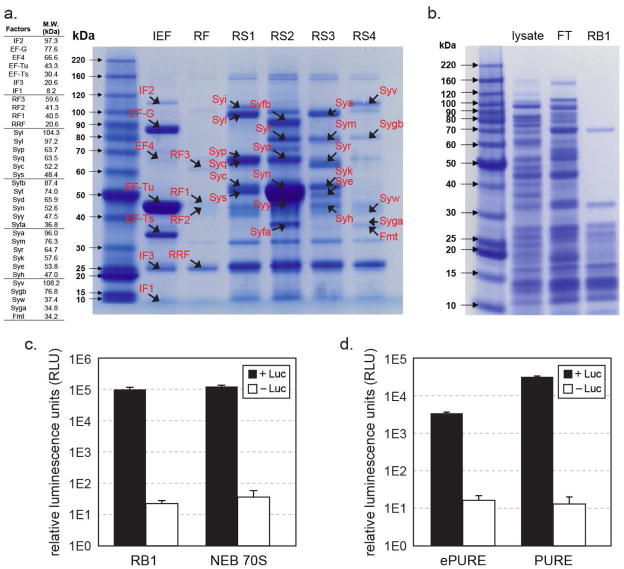
We co-purified the His-tagged translation components from all six ePURE strains (IEF, RF, RS1-4) with Ni-NTA columns. Most components are effectively isolated as shown on protein gels (Figure 3a) and assayed by mass spectrometry (Table 2). The IEF strain showed robust co-purification bands of EF-G, EF-Tu and EF-Ts, while EF-4, IF1, IF2 and IF3 were in low relative concentration. In the RF strain, RF1, RF2 and RRF bound to Ni-NTA weakly as they were washed out at the extended 20 mM imidazole wash step (Supplementary Figure 3). All aaRS were effectively co-purified from strain RS1-4 except ArgRS and GlyRS α subunit, which were in very low concentrations, but still present as confirmed by mass spectrometry (Table 2). Two unintended bands corresponding to EF-Tu and SlyD were present in all pools of purified factors. EF-Tu is the most abundant protein in E. coli cell and SlyD (FKBP-type peptidyl prolyl cis-trans isomerase) is a well-known contaminant in His-tag purification due to its high affinity towards Ni2+ ions (33). These two proteins were also present in the control chromatography with MRE600 lysate.
Figure 3. Assessment of purified His-tagged ribosome and translation factors.
(a). A table of translation factors by protein size is listed on the left panel (not including His-tags). The right panel shows eluted fractions of His-tagged IEF, RF, and RS1-4 factors after Ni-NTA purification on Bis-Tris PAGE gel, stained by Coomassie-blue. The expected migration band of each factor is marked on the gel. Though a few of the visible bands may be non-translation factor, Ni-binding proteins (e.g. at 25 kD), presence of translation factors in the elution fraction was validated by mass spectrum (Table 2). Eluted fractions from RF strains contained very faint amount of proteins. Re-examination of the washed fractions showed that RF1, RF2, RRF and RF3 eluted off the Ni-NTA resin consecutively during the wash step. (b). His-tagged ribosome purified from RB1 cell lysate, FT: flow through. (c). Activity comparison of RB1 ribosome that has His6-tags on L7/L12 ribosomal proteins and was purified over Ni-NTA resin with wild-type E. coli 70S ribosome (New England Biolabs) purified by the traditional sucrose gradient method. Ribosomes were compared using an in vitro translation assay producing firefly luciferase, with chemical luminescence plotted on a log scale. Control reactions were conducted under same conditions but without luciferase gene template. Error bars are ± standard deviations, with n=3. (d). Activity of co-purified ePURE factor mix, supplemented with IF1, IF3, ArgRS, GlyRS, demonstrated by in vitro translation of luciferase and luminescence. PURE factor mix prepared by individual factor purifications is also performed under the same conditions as a positive control. Negative control reactions were conducted in reactions without luciferase gene template. Error bars are ± standard deviations, with n=4.
Table 2.
Mass Spectrum analysis of Ni-NTA purified ePURE factors from each elution fraction. Samples are first alkylated by iodoacetamide, precipitated by trichloroacetic acid, and finally digested by trypsin before subjected to mass spectrum. Spectra count data of detected peptides are presented by their encoded genes and grouped into His-tagged proteins in each factor pools (top of each pool) and top five abundant co-purified contaminants (bottom of each pool, dark gray shade). Light gray shaded texts indicate pools with unexpectedly low amount of detected proteins.
| IEF | RF | ||||||
|---|---|---|---|---|---|---|---|
| Protein | Gene | Spectra Count | MW (kDa) | Protein | Gene | Spectra Count | MW (kDa) |
|
|
|
||||||
| EFG | fusA | 630 | 77.6 | RF3 | prfC | 315 | 59.6 |
| EFTU | tufA, tufB | 589 | 43.3 | RRF | frr | 140 | 20.6 |
| EFTS | tsf | 406 | 30.4 | RF1 | prfA | 98 | 40.5 |
| IF2 | infB | 74 | 97.3 | RF2 | prfB | 83 | 41.3 |
| LEPA | lepA | 17 | 66.6 | ||||
| IF3 | infC | 11 | 20.6 | ||||
| IF1 | infA | 6 | 8.2 | ||||
| SLYD | slyD | 96 | 20.9 | SLYD | slyD | 205 | 20.9 |
| CYOA | cyoA | 68 | 34.9 | GLMS | glmS | 198 | 127.0 |
| IDH | icd | 47 | 45.8 | RPOC | rpoC | 138 | 155.2 |
| RBS2 | rpsB | 45 | 26.7 | CRP | crp | 86 | 23.6 |
| RPOC | rpoC | 43 | 155.2 | RPOB | rpoB | 84 | 150.6 |
| RS1 | RS2 | ||||||
| Protein | Gene | Spectra Count | MW (kDa) | Protein | Gene | Spectra Count | MW (kDa) |
|
|
|
||||||
| SY1 | ileS | 714 | 104.3 | SYN | asnS | 1001 | 52.6 |
| SYS | serS | 467 | 48.4 | SYD | aspS | 239 | 65.9 |
| SYQ | glnS | 385 | 63.5 | SYFB | pheT | 182 | 87.4 |
| SYL | leuS | 310 | 97.2 | SYFA | pheS | 94 | 36.8 |
| SYC | cysS | 283 | 52.2 | SYT | thrS | 81 | 74.0 |
| SYP | proS | 262 | 63.7 | SYY | tyrS | 63 | 47.5 |
| SLYD | slyD | 224 | 20.9 | EFTU | tufA, tufB | 313 | 43.3 |
| EFTU | tufA, tufB | 115 | 43.3 | SLYD | slyD | 227 | 20.9 |
| GLYA | glyA | 112 | 45.3 | SYGB | glyS | 132 | 76.8 |
| FUR | fur | 109 | 16.8 | IDH | icd | 93 | 45.8 |
| CAR8 | carB | 82 | 117.8 | PPSA | ppsA | 76 | 87.4 |
| RS3 | RS4 | ||||||
| Protein | Gene | Spectra Count | MW (kDa) | Protein | Gene | Spectra Count | MW(kDa) |
|
|
|
||||||
| SYE | gltX | 464 | 53.8 | SYV | valS | 538 | 108.2 |
| SYK1 | lysS | 418 | 57.6 | SYGB | glyS | 187 | 76.8 |
| SYA | alaS | 414 | 96.0 | FMT | fmt | 157 | 34.2 |
| SYH | hisS | 161 | 47.0 | SYW | trpS | 82 | 37.4 |
| SYM | metS | 160 | 76.3 | SYGA | glyQ | 19 | 34.8 |
| SYR | argS | 19 | 64.7 | ||||
| EFTU | tufA, tufB | 404 | 43.3 | EFTU | tufA, tufB | 370 | 43.3 |
| SLYD | slyD | 290 | 20.9 | SLYD | slyD | 364 | 20.9 |
| SYGB | glyS | 132 | 76.8 | CARB | carB | 163 | 117.8 |
| SYK2 | lysU | 127 | 57.8 | FUR | fur | 125 | 16.8 |
| CYOA | cyoA | 125 | 34.9 | RBS2 | rpsB | 111 | 26.7 |
|
|
|
||||||
Three strains RB1, RB2, and RB3 that have His-tags on ribosomal proteins L7/L12, S2 and L3 respectively were also subjected to affinity purification. Ribosomes harboring a single His6-tag, RB2 and RB3, bound to Ni-NTA resin with poor and moderate affinities, respectively, as extended washing eluted the tagged ribosomes from the resin. Purification of RB1 ribosomes was much more efficient presumably because the tagged L7/L12 proteins were present in four copies per ribosome (Figure 3b). We tested the activity of purified RB1 ribosomes by a PURE translation assay in comparison to commercial E. coli ribosomes prepared by conventional sucrose gradient methods. The results showed the His-tag on L7/L12 of RB1 ribosome does not affect its activity (Figure 3c), which is consistent with previous literature (31). We also found that RB3 ribosomes, even though not purified as abundantly, were as active as the RB1 ribosomes. Besides activity, it is noteworthy that we also utilized MAGE to remove RNase I in all RB1-3 strains, which otherwise is known to associate with ribosome tightly while purification, leading to degradation of rRNA and mRNA in cell-free translation reactions.
We combined the six pools of translational factors that were purified from ePURE strains at a ratio that mimics their cellular stoichiometry (34) and performed in vitro translation with RB1 ribosomes, phosphoenolypyruvate and pyruvate kinase (35) as an energy regenerating system, and all small molecule substrates needed for a PURE protein synthesis reaction (see Methods). As a control experiment, the 31 individually purified translation factors are also mixed (36) to compare to our ePURE factor mix. The preliminary result showed that the ePURE mix had barely detectable activity, which we hypothesized to be either due to limited concentrations of four factors (IF1, IF3, ArgRS, and GlyRS α subunit) or due to inhibitory contaminants present in the factor mix. To rule out the latter possibility of inhibitory contaminants, we added 2% v/v of each pool of ePURE factors to the commercial PUREXpress translation reaction; these supplements enhanced protein production by 10–49% (Supplementary Figure 2). While our ePURE was designed to co-purify endogenous factors at their physiologically relevant stochiometric ratios (37), this is not completely achieved since some His-tagged factors show unexpectedly low expression and/or binding affinities toward Ni-NTA resin. In contrast with our system and in vivo, the PURE system utilizes different concentrations of factors, such as much higher relative concentrations of three initiation factors (IF1-3). Concentration differences might explain the different activities of the PURE, ePURE and in vivo systems. As predicted, adding extra IF1, IF3, ArgRS and GlyRS to our ePURE mix to amounts reported in literature (36) significantly increased our ePURE activity (Figure 3d). However, the PURE factor mix optimized over the past 15 years was still several-fold more active. Nonetheless, we demonstrated that our ePURE system was active in generating functional protein products.
Here, we applied a highly efficient MAGE method for over 110 cycles to simultaneously modify the protein-coding sequences of many components of an enzyme pathway in vivo. We demonstrated the feasibility of the method by introducing poly-histidine tags into genomic copies of genes associated with all 38 essential components of the translation machinery in E. coli. MAGE-mediated protein modifications can be multiplexed to allow for generation of all combinatorial mutant variants, and they are easily identified by a simple, multiplexed, allele-specific, PCR method. Genomic modifications that cause increased toxicity or reduced fitness (and thus likely to be non-functional in vivo) are selected against in our system, thereby forcing the incorporation of only non-disruptive modifications into the target site. We showed that the genomic modifications of essential translation factors and aminoacyl-tRNA synthetases were well-tolerated by E. coli, and that the co-purification of these factors was possible (38). Additionally, we introduced His-tags in three ribosomal subunit genes to allow for the direct and facile purification of ribosomes using Ni-NTA sepharose. Ribosomes His-tagged in the 50S (rplC, rplL) or 30S (rpsB) ribosomal proteins and purified directly off of nickel resin were equally as active as 70S ribosomes purified using traditional sucrose gradients as tested in an in vitro protein synthesis reaction to make luciferase (a two-domain, 550-amino-acid, eukaryotic protein). The His-tagged strategy for purifying ribosomal parts has the advantage over traditional purification techniques in its ease and convenience. We finally combined all His-tagged components from the first seven strains listed in Table 1 together to produce the ensemble PURE system, which we showed to be 11% active in producing proteins in an in vitro translation reaction comparing to PURE system composed of individually purified components. Based on very recent advances in large scale engineering of the E. coli chromosome (25), it should be possible to combine all 7 strains into one viable strain to further facilitate production of the PURE system, though it would also reduce flexibility in terms of adjusting concentrations of groups of factors. We have made the 7 strains of this ePURE system available to scientists without any restriction, thus already enabling preparation of the PURE system at a much lower cost that currently available by commercial purchase. To address the possibility that certain factors may be more difficult to co-purify, we can use MAGE to further tune the expression level of the individual factors such as by promoter over-expression or ribosomal binding site manipulation (23). In vivo pre-tuning of biological components for downstream in vitro reactions will likely be an attractive approach in many facets where fast optimization and prototyping of constructs are desired.
Our approach of rapid and parallel in vivo protein modification for in vitro reactions highlights the potential utility of co-modification and co-purification of many enzymes simultaneously for multi-enzymatic catalysis through a single-pot reaction. This type of synthetic biology strategy may lead to large-scale production of such single-pot catalytic systems. These endeavors will also likely be useful in many other areas to build in vitro versions of natural product biosynthesis, like polyketides or carbohydrates, and to reconstruct central metabolism with defined components and conditions (39–41) towards minimal cells (42).
METHODS
Media, Chemicals, and Reagents
Unless otherwise specified, all chemicals were obtained from Sigma-Aldrich. Tryptone and yeast extract were obtained from BD Difco. For MAGE cycling, liquid cultures of all strains were grown in LB-min rich media (10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl). For protein purification, liquid cultures of all strains were grown in either 2YTPG media (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl, 5 g/L glucose, 3 g/L KH2PO4, 9 g/L K2HPO4-3H2O), or SB media (24 g/L tryptone, 12 g/L yeast extract, 5 g/L glucose, 2 g/L NaH2PO4, 16.4 g/L K2HPO4-3H2O, 4 mL/L glycerol). Multiplex PCR kits were purchased from Qiagen (Cat #206143). Protein purifications were carried out with ÄKTAprime (GE Healthcare) equipped with 5 mL HisTrap™ HP column (GE Healthcare). Purified protein concentrations were determined by standard Bradford assay (Bio-Rad).
Strains and MAGE Cycling Protocol
The previously described MAGE strain EcNR2 was used throughout this study (23,24). EcNR2 is a derivative of Escherichia coli MG1655 with ΔbioA::λ-Red-bla and ΔmutS::cm. MAGE cycling was initiated by inoculating nine aliquots of 3 mL LB-min liquid media with individual colonies from a freshly-streaked overnight plate. The nine strains were grown in glass tubes stationed in a rotator drum at 300 rpm at 32 °C. When the cultures reached an OD600 of 0.6–0.7, the tubes were moved to a 42 °C shaking water bath for 15 minutes to induce the expression of λ-Red proteins. Cells were then immediately chilled on ice for at least 5 min and subsequent made electrocompetent in 1 mL aliquots by repeated (at least twice) pelleting and re-suspension in cold-sterile dH2O. Cells were concentrated 20-fold into 50 μL reactions containing the appropriate oligonucleotides (typically between 5–25 ng) in dH2O and electroporated with a BioRad GenePulser using a 1-mm gap cuvette (1.8 kV, 200 Ω, 25 μF). The electroporated cells were immediately added to 3 mL of warm LB-min media and recovered for 2 hours, at which point the OD600 reached 0.7 again for the next MAGE cycle. On the last cycle of each day, the cells are allowed to recover into stationary phase overnight. The cycle is restarted the following day by diluting the overnight culture by 1:30 with fresh media. Four MAGE cycles can be manually performed per day, and can be expanded to >12 cycles by automation (23). MAGE cycling can be paused by storing stationary phase cells at 4 °C for up to 2 days prior to reinitiation by dilution. Growth rates of the strains were determined in 96-well microtiter plate format by measuring the OD600 of 200 μL cell cultures grown at 30 °C using a microplate reader (SpectraMax M5, Molecular Devices).
Oligonucleotides (Oligos)
All oligonucleotides and PCR primers were obtained from Integrated DNA Technologies with standard purification. 90mer targeting oligos contained four phosphorothioated bases at the 5′ terminus. All His-tag oligos contained the 18-basepair CACCATCACCATCACCAT insertion that was directed either at the N- or C-terminus of the target gene based on previous literature and crystallographic information (28,36). Standard primers were used for allelic genotyping by PCR or Sanger sequencing. Sanger sequencing was performed by Agencourt Bioscience Corporation. Oligonucleotides used in this study are listed in full in Supplemental Table 1.
Multiplex Allele-Specific Colony PCR (MASC-PCR)
Multiplex allele-specific PCR as previously described (24) was done to simultaneously screen for clones that contained His-tag sequences in up to 8 genes. The querying forward primers contained the first 6 bps of the His-tag insertion sequence at the 3′ terminus. Reverse primers were designed to generate amplicon sizes of 150, 200, 250, 300, 400, 500, 600, or 700 bps, each corresponding to a different genomic locus for allele-specific PCR. Amplified bands reflected the insertion of His-tag sequences at designated loci in each clone. No bands were observed when primer mixes were used in the wild-type control. Primers were designed for a target Tm of 62 °C. Multiple loci are queried in a single PCR reaction using the multiplex PCR kit from Qiagen. In each 20 μL PCR reaction, 1 μL of a 1 in 100 dilution in water of a saturated clonal culture generated the best MASC-PCR signal. PCR cycles were: heat activation and cell lysis for 15 min at 95 °C, denaturing for 30 sec at 94 °C, annealing for 30 sec at experimentally determined optimal Tm of 62 °C, extension for 60 sec at 72 °C, repeated cycling for 26 times, and final extension for 5 min at 72 °C. Gel electrophoresis on a 1.5% agarose gel produced the best separation for a 8-plex MASC-PCR reaction.
Purification of His-tagged Ribosomes
Strains (RB1, RB2, RB3) with His-tagged ribosomal proteins were first inoculated into 5 mL tubes containing LB media, grown for 7 hours at 30 °C and re-inoculated into 200 mL 2YTPG grown for an additional 14 hours. Cultures were then inoculated into 5 L of 2YTPG within a BioFlo 3000 Bioreactor (New Brunswick Scientific, USA) and fermented at 30 °C, pH 7.2 (controlled by 5 M HCl and NaOH), 8 SLPM of air, and 600 rpm of agitation. The cells were harvested at OD600 of 3 and centrifuged at 5000 g for 30 min. The wet cell paste was resuspend in Salt buffer (10 mM Tris-HOAc, pH 7.6, 60 mM NH4Cl, 15 mM MgCl2, and 0.5 mM EDTA) and pass through a French Press, and cell debris was removed by centrifugation twice at 30,000×g for 20 min. The supernatant was then loaded to Ni-NTA columns and washed by Salt buffer with 5mM Imidazole, and eluted by a linear gradient from 5 mM to 150 mM Imidazole. The fractions that contained ribosomes were pooled and concentrated by ultracentrifugation (150,000× g for 4 hr) and resuspended in ribosome storage buffer (20 mM Tris-HOAc, pH 7.6, 30 mM NH4Cl, 15 mM MgCl2, and 150 mM KCl). Ribosome concentration was determined by UV absorbance at 260 nm.
Purification of His-tagged Factors from ePURE Strains
His-tagged ePURE strains (IEF, RF, RS1-4) were grown and harvested in the same manner as His-tagged ribosome strains to yield centrifuged cell pastes. Every 5 g of cell paste was lysed by 40 mL B-PER (Thermo-Fisher), with 100 uL of Halt-protease inhibitor (Thermo-Fisher), 6 mM βME, and 20 mM Imidazole-HOAc (pH 7.4). Cell debris was removed by centrifugation at 150,000× g for 2 hrs. The supernatant was loaded to Ni-NTA columns and washed with wash buffer (20 mM Tris-HOAc, pH 7.6, 30 mM NH4Cl, 150 mM KCl, and 150 mM NaCl) containing 58 mM Imidazole-HOAc (pH 7.4). The His-tagged proteins were eluted with a linear gradient from 58 mM to 400 mM Imidazole-HOAc in wash buffer, pooled, concentrated with Amicon-Ultra-4 concentrator with 3K MWCO for IEF and 10K MWCO for the rest, and dialyzed against 2 L of stock buffer (20 mM Tris-HOAc, pH 7.6, 30 mM NH4Cl, 150 mM KCl, 15 mM Mg(OAc)2, 6 mM βME, and 10 μM GDP) for 3 hrs twice. Each group of factors is added to 20% glycerol and store at −80 °C. RF strain factors were washed with buffer containing 35 mM instead of 58 mM Imidazole to prevent detachment before elution.
Over-expression and Purification of Individual PURE Factors
Plasmids encoding 31 his-tagged translation components (kindly provided by T. Ueda) were transformed to BL21(DE3)pLysS (Agilent) or NEB-Iq (New England Biolab) strain depending on the promoters for expression. Cells were grown in 250 mL SB at 37 °C, induced with 1 mM IPTG when OD600 reached 0.5, and further incubated at 37 °C for 4 hr before harvest. Every 3 g of cell paste was lysed by 15 mL BugBuster Master mix (EMD Chemical), with 100 μL Halt-protease inhibitor (Thermo-Fisher), 6 mM β-ME, and 20 mM Imidazole-HOAc (pH 7.4). Cell debris was removed by centrifugation at 30,000× g twice. The clear lysate with over-expressed PURE factors were then purified according to previously described procedures (36).
In vitro Translation with PURE and ePURE Factors
PURE translation reactions were carried out according to literature conditions (27,43) with minor modifications. A typical 10 μL PURE reaction contained all 31 factors (IF1, IF2, IF3, EF-G, EF-Ts, EF-Tu, RF1, RF2, RF3, RRF, methionyl-tRNA formyl-transferase, and 20 aaRS), 20 μg/mL pyruvate kinase (Sigma), 3.0 μg/mL myokinase (EMD Chemical), 1.1 μg/mL nucleotide diphosphate kinase (Sigma), and 2 U/mL pyrophosphatase (New England Biolab), 10 μg/mL T7 RNA polymerase (Ambion), 0.1 mM of 20 amino acids (Sigma), 54 A260 unit of E. coli total tRNA (Roche), 2 mM ATP, 2 mM GTP, 1 mM CTP, 1mM UTP, 20 mM phosphoenolpyruvate, 10 μg/mL formyl donor, 50mM Hepes-KOH (pH 7.6), 100 mM potassium glutamate, 13 mM Mg(OAc)2, 2 mM spermidine, 1mM DTT, 0.8 U/μL Murine RNase Inhibitor (New England Biolab), 1.2 μM of ribosome, and 10 μg/mL of the pIVEX-Luc plasmid in which the firefly luciferase gene under T7 regulation was cloned into plasmid pIVEX 2.3d (from 5-Prime).
All ePURE translation reactions contained the same components as the PURE reaction except that the 31 factors are replaced by 700 μg/mL IEF (containing IF1, IF2, IF3, EF-G, EFTs, EF-Tu, EF-4), 370 μg/mL RF (containing RF1, RF2, RF3, RRF), 150 μg/mL RS1 (containing CysRS, GlnRS, IleRS, LeuRS, ProRS, SerRS), 550 μg/mL RS2 (containing AsnRS, AspRS, PheRS, ThrRS, TyrRS), 183 μg/mL RS3 (containing AlaRS, ArgRS, GluRS, HisRS, LysRS, MetRS), 79 μg/mL RS4 (containing GlyRS, TrpRS, ValRS, methionyl-tRNA formyl-transferase) and four supplementary, individually prepared factors: 22.3 μg/mL IF1, 30.9 μg/mL IF3, 20.0 μg/mL ArgRS and 96.0 μg/mL GlyRS. The commercial Δ-PURExpress system (New England Biolabs) was setup according to vendor recommendations, except that 0.8 U/μL Murine RNase Inhibitor and 1.2 μM of ribosome were used. All 10 μL translation reactions were run at 37 °C for 1 hr, followed by the addition of 50 μL of luciferase substrate solution (Promega) to each reaction to measure chemical luminescence in relative luminescent units (RLUs).
Mass Spectrometry of ePURE Factors
Ni-NTA co-purified ePURE factors, 100 μg, in 100 μl of 8 M Urea, 25 mM Tris-HCl, and 10 mM DTT were incubated at 56 °C for 30 min to reduce disufide bonds between cysteine residues. After cooling to room temperature (RT), iodoacetmaide was added to the mixture to a concentration of 30 mM and cysteine residues were alkylated at RT for 60 min in the dark. The reaction was quenched with 20 mM of DTT. Proteins were precipitated by adding 1/4 volume of 100% (w/w) trichloroacetic acid (TCA) and re-suspended in 50mM Tris-HCl (pH 7.6), 1M urea. Sequencing grade trypsin, 2 μg, (Promega, WI) was added and proteins were digested overnight at 37°C. Peptide solutions were acidified with 1/2 volume of 5% formic acid (FA), 5% acetonitrile (ACN), and subjected to C18 reversed-phase solid-phase extraction (SPE) using a 100 mg Sep-Pak cartridges (Waters, MA). Peptides were dried, dissolved in 5% FA and 5% ACN, and analyzed by microcapillary liquid chromatography tandem mass spectrometry (LC-MS/MS) on an LTQ Orbitrap Velos (Thermo Scientific, Bremen, Germany) equipped with a Famos autosampler (LC Packings, Sunnyvale, CA) and an Agilent 1100 binary HPLC pump (Agilent Technologies, Santa Clara, CA). Peptides were separated on a 100 μm I.D. microcapillary column in-house packed first with approximately 0.5 cm of Magic C4 resin (5 μm, 100 Å, Michrom Bioresources, Auburn, CA) followed by 20 cm of Maccel C18 AQ resin (3 μm, 200 Å, Nest Group, Southborough, MA) applying a gradient from 9 to 32 % ACN in 0.125 % FA over 75 min at a flow rate of approximately 300 nl/min. The LTQ Orbitrap Velos was operated in a data dependent mode – a survey MS scan over an m/z range of 300–1500 performed with a resolution setting of 6×104 in the Orbitrap was followed by up to 20 ion trap (LTQ) collision induced dissociation (CID) MS/MS spectra on the most intense ions observed in the survey MS scan. The AGC setting was 3×106 for the MS and 2×103 for MS/MS experiments. Maximum ion injection times were set to 1000 and 150 ms for MS and MS/MS experiments, respectively. The precursor ion isolation width was set to 2 m/z, singly charged ions and ions with un-assignable charge state were not selected for MS/MS, and ions within an m/z of −0.52 to 2.52 relative to the m/z of ions selected for MS/MS were excluded from further selection for 30 s. Acquired MS/MS spectra were assigned using the Sequest algorithm (44) by searching them against a database with sequences of proteins encoded by all known E. coli ORFs (NCBI) as well as of known contaminants such as human keratins and porcine trypsin. This forward (target) database component was followed by a decoy component of all the above mentioned protein sequences in reversed order allowing an estimation of the false discovery rate of generated MS2 assignments (45). Searches were performed accepting only sequences for fully tryptic peptides with a precursor ion mass tolerance of 50 p.p.m. Carbamidomethylation of cysteine residues (+57.02146 Da) was used set as static modification and oxidation of methionine residues (+15.99492 Da) was set as variable modification. Assignments of MS2 spectra were filtered essentially as previously described so that the false-discovery rate of identified peptides as well as proteins was smaller than 1 % (46).
Supplementary Material
Acknowledgments
We thank Dr. T. Ueda for kindly supplying plasmids for the PURE system. This work was funded by multiple programs from the National Science Foundation (SynBERC) [Grant #SA5283-11210]; the Department of Energy (Genomes to Life Center) [Grant #DE-FG02-03ER6344]; and the Wyss Institute for Biologically Inspired Engineering. HHW is supported by the Wyss Institute Technology Development Fellowship and the National Institutes of Health Director’s Early Independence Award [Grant Number DP5OD009172]. ACF was funded by the Vanderbilt Institute of Chemical Biology, the National Institutes of Health and the American Cancer Society. MCJ gratefully acknowledges funding from the National Institutes of Health [Grant Number R00GM081450].
Footnotes
AUTHOR CONTRIBUTIONS
HHW, MCJ, AF and GMC designed the study. HHW, GX, AM, and JL performed the MAGE His-tag experiments. PYH performed the ePURE purification and protein translation experiments. WH and SG contributed to the mass-spec data acquisition and analysis. All authors helped draft and edit the final manuscript.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
References
- 1.Katzen F, Fletcher JE, Yang JP, Kang D, Peterson TC, Cappuccio JA, Blanchette CD, Sulchek T, Chromy BA, Hoeprich PD, et al. Insertion of membrane proteins into discoidal membranes using a cell-free protein expression approach. Journal of proteome research. 2008;7:3535–3542. doi: 10.1021/pr800265f. [DOI] [PubMed] [Google Scholar]
- 2.Cappuccio JA, Blanchette CD, Sulchek TA, Arroyo ES, Kralj JM, Hinz AK, Kuhn EA, Chromy BA, Segelke BW, Rothschild KJ, et al. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Mol Cell Proteomics. 2008;7:2246–2253. doi: 10.1074/mcp.M800191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jewett MC, Calhoun KA, Voloshin A, Wuu JJ, Swartz JR. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol Syst Biol. 2008;4:220. doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson ED, Gan R, Hodgman CE, MCJ Cell-free protein synthesis: Applications come of age. Biotechnol Advances. 2011 doi: 10.1016/j.biotechadv.2011.09.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartman MC, Josephson K, Lin CW, Szostak JW. An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS One. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jewett MC, Forster AC. Update on designing and building minimal cells. Curr Opin Biotechnol. 2010;21:697–703. doi: 10.1016/j.copbio.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annual review of biochemistry. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 9.Kung HF, Chu F, Caldwell P, Spears C, Treadwell BV, Eskin B, Brot N, Weissbach H. The mRNA-directed synthesis of the alpha0peptide of beta-galactosidase, ribosomal proteins L12 and L10, and elongation factor Tu, using purified translational factors. Arch Biochem Biophys. 1978;187:457–463. doi: 10.1016/0003-9861(78)90057-7. [DOI] [PubMed] [Google Scholar]
- 10.Nishiyama KI, Maeda M, Abe M, Kanamori T, Shimamoto K, Kusumoto S, Ueda T, Tokuda H. A novel complete reconstitution system for membrane integration of the simplest membrane protein. Biochem Biophys Res Commun. 2010 doi: 10.1016/j.bbrc.2010.03.061. [DOI] [PubMed] [Google Scholar]
- 11.Tan Z, Blacklow SC, Cornish VW, Forster AC. De novo genetic codes and pure translation display. Methods (San Diego, Calif. 2005;36:279–290. doi: 10.1016/j.ymeth.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Forster AC, Cornish VW, Blacklow SC. Pure translation display. Anal Biochem. 2004;333:358–364. doi: 10.1016/j.ab.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 13.Forster AC, Tan Z, Nalam MN, Lin H, Qu H, Cornish VW, Blacklow SC. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc Natl Acad Sci U S A. 2003;100:6353–6357. doi: 10.1073/pnas.1132122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Josephson K, Hartman MC, Szostak JW. Ribosomal synthesis of unnatural peptides. J Am Chem Soc. 2005;127:11727–11735. doi: 10.1021/ja0515809. [DOI] [PubMed] [Google Scholar]
- 15.Murakami H, Ohta A, Ashigai H, Suga H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods. 2006;3:357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Gallego F, Schmidt-Dannert C. Multi-enzymatic synthesis. Curr Opin Chem Biol. 2010;14:174–183. doi: 10.1016/j.cbpa.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 17.Hodgman CE, Jewett MC. Cell-free synthetic biology: thinking outside the cell. Metabolic Engineering. 2011 doi: 10.1016/j.ymben.2011.09.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang PYH, Myung S, You C, Zhu Z, Rollin JA. Toward low-cost biomanufacturing through in vitro synthetic biology: bottom-up design. J Mater Chem. 2011 doi: 10.1039/C1JM12078F. [DOI] [Google Scholar]
- 19.Fessner W-D, WC “Artificial metabolisms” for the asymmetric one-pot synthesis of branched-chain saccharides. Angew Chem Int Ed Engl. 1992;31:614–616. [Google Scholar]
- 20.Schultheisz HL, Szymczyna BR, Scott LG, Williamson JR. Pathway engineered enzymatic de novo purine nucleotide synthesis. ACS Chem Biol. 2008;3:499–511. doi: 10.1021/cb800066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultheisz HL, Szymczyna BR, Scott LG, Williamson JR. Enzymatic De Novo Pyrimidine Nucleotide Synthesis. J Am Chem Soc. 2010 doi: 10.1021/ja1059685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swartz JR. Transforming Biochemical Engineering with Cell-free Biology. AIChE Journal. 2011 in press. [Google Scholar]
- 23.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang HH, Church GM. Multiplexed genome engineering and genotyping methods applications for synthetic biology and metabolic engineering. Methods in enzymology. 2011;498:409–426. doi: 10.1016/B978-0-12-385120-8.00018-8. [DOI] [PubMed] [Google Scholar]
- 25.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang HH, Xu G, Vonner AJ, Church G. Modified bases enable high-efficiency oligonucleotide-mediated allelic replacement via mismatch repair evasion. Nucleic Acids Res. 2011;39:7336–7347. doi: 10.1093/nar/gkr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu Y, Kanamori T, Ueda T. Protein synthesis by pure translation systems. Methods (San Diego, Calif. 2005;36:299–304. doi: 10.1016/j.ymeth.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Forster AC, Weissbach H, Blacklow SC. A simplified reconstitution of mRNA-directed peptide synthesis: activity of the epsilon enhancer and an unnatural amino acid. Anal Biochem. 2001;297:60–70. doi: 10.1006/abio.2001.5329. [DOI] [PubMed] [Google Scholar]
- 29.Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 30.Mora L, Zavialov A, Ehrenberg M, Buckingham RH. Stop codon recognition and interactions with peptide release factor RF3 of truncated and chimeric RF1 and RF2 from Escherichia coli. Mol Microbiol. 2003;50:1467–1476. doi: 10.1046/j.1365-2958.2003.03799.x. [DOI] [PubMed] [Google Scholar]
- 31.Ederth J, Mandava CS, Dasgupta S, Sanyal S. A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli. Nucleic Acids Res. 2009;37:e15. doi: 10.1093/nar/gkn992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolanos-Garcia VM, Davies OR. Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim Biophys Acta. 2006;1760:1304–1313. doi: 10.1016/j.bbagen.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 34.Pingoud A, Gast FU, Block W, Peters F. The elongation factor Tu from Escherichia coli, aminoacyl-tRNA, and guanosine tetraphosphate form a ternary complex which is bound by programmed ribosomes. J Biol Chem. 1983;258:14200–14205. [PubMed] [Google Scholar]
- 35.Kim DM, Swartz JR. Prolonging cell-free protein synthesis with a novel ATP regeneration system. Biotechnol Bioeng. 1999;66:180–188. [PubMed] [Google Scholar]
- 36.Shimizu Y, Ueda T. PURE technology. Methods Mol Biol. 2010;607:11–21. doi: 10.1007/978-1-60327-331-2_2. [DOI] [PubMed] [Google Scholar]
- 37.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 38.Du L, Villarreal S, Forster AC. Multigene expression in vivo: Supremacy of large versus small terminators for T7 RNA polymerase. Biotechnol Bioeng. 2011 doi: 10.1002/bit.24379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meier JL, Burkart MD. The chemical biology of modular biosynthetic enzymes. Chem Soc Rev. 2009;38:2012–2045. doi: 10.1039/b805115c. [DOI] [PubMed] [Google Scholar]
- 40.Kharel MK, Lian H, Rohr J. Characterization of the TDP-D-ravidosamine biosynthetic pathway: one-pot enzymatic synthesis of TDP-D-ravidosamine from thymidine-5-phosphate and glucose-1-phosphate. Org Biomol Chem. 2011;9:1799–1808. doi: 10.1039/c0ob00854k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koeller KM, Wong CH. Complex carbohydrate synthesis tools for glycobiologists: enzyme-based approach and programmable one-pot strategies. Glycobiology. 2000;10:1157–1169. doi: 10.1093/glycob/10.11.1157. [DOI] [PubMed] [Google Scholar]
- 42.Murtas G. Artificial assembly of a minimal cell. Mol Biosyst. 2009;5:1292–1297. doi: 10.1039/b906541e. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu Y, Ueda T. PURE technology. Methods Mol Biol. 607:11–21. doi: 10.1007/978-1-60327-331-2_2. [DOI] [PubMed] [Google Scholar]
- 44.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino-acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 45.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 46.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.