Abstract
Although the adenovirus type 5 (Ad5) E1B 55 kDa protein can bind to RNA in vitro, no UV-light-induced crosslinking of this E1B protein to RNA could be detected in infected cells, under conditions in which RNA binding by a known viral RNA-binding protein (the L4 100kDa protein) was observed readily. Substitution mutations, including substitutions reported to inhibit RNA binding in vitro, did not impair synthesis of viral early or late proteins, or alter significantly the efficiency of viral replication in transformed or normal human cells. However, substitutions of conserved residues in the C-terminal segment of an RNA recognition motif specifically inhibited degradation of Mre11. We conclude that, if the E1B 55 kDa protein binds to RNA in infected cells in the same manner as in in vitro assays, this activity is not required for such well established functions as induction of selective export of viral late mRNAs.
Introduction
During the late phase of infection of permissive cells by species C human adenoviruses, such as adenovirus type 5 (Ad5), large quantities of viral late proteins are made while synthesis of cellular proteins is inhibited (Anderson, Baum, and Gesteland, 1973; Beltz and Flint, 1979). The selective expression of viral late genes, most of which encode structural proteins, is thought to facilitate the self assembly reactions that produce progeny virions, while inhibition of cellular protein synthesis has been reported to be required for efficient release of virus particles (Zhang and Schneider, 1994). Neither transcription of the majority of cellular genes nor processing of primary transcripts are perturbed during the late phase of infection (Babich et al., 1983; Beltz and Flint, 1979; Flint, Beltz, and Linzer, 1983; Granberg et al., 2005; Miller et al., 2007; Yang, Huang, and Flint, 1996; Zhao, Granberg, and Pettersson, 2007). Rather, viral late mRNAs are both selectively translated and selectively exported from the nucleus. Selective translation requires the viral L4 100 kDa protein (Hayes et al., 1990). Translation of major late (ML) mRNAs, which carry the common 202 nucleotide tripartite leader sequence (tpl) (Berget, Moore, and Sharp, 1977; Chow et al., 1977), and of IVa2 mRNA is initiated by an alternative mechanism, cap-independent ribosome shunting (Dolph et al., 1988; Yueh and Schneider, 1996). As this process exhibits a reduced requirement for the cap binding complex elF4F (Huang J. and Schneider, 1991; Zhang and Schneider, 1994), the dephosphorylation and inactivation of the elF4E component of this complex by the L4 100 kDa protein (Cuesta, Xi, and Schneider, 2004) inhibit synthesis of cellular, but not of viral late, proteins. This L4 protein also stimulates initiation of translation of viral ML mRNAs by binding simultaneously to the tpl and the initiation factor elFG to recruit the 40S ribosomal subunit (Xi, Cuesta, and Schneider, 2004).
Studies of the phenotypes exhibited by mutants of Ad5 established that selective export of viral late mRNAs requires the early E1B 55 kDa (Babiss et al., 1983; Pilder et al., 1986; Williams et al., 1986) and E4 Orf6 (Halbert, Cutt, and Shenk, 1985) proteins and the complex (Sarnow et al., 1984) they form in infected cells (Bridge and Ketner, 1990; Cutt, Shenk, and Hearing, 1987). These two viral proteins colocalize at the peripheral zones of viral replication centers (Gonzalez and Flint, 2002; Ornelles and Shenk, 1991), the sites of synthesis and initial processing of viral late transcripts (Aspegren, Rabino, and Bridge, 1998; Bridge and Pettersson, 1995; Pombo et al., 1994; Puvion-Dutilleul et al., 1994). Such colocalization is necessary for efficient export of late mRNAs, as mutations that prevent synthesis of the E4 Orf6 protein or binding of E1B 55 kDa to this E4 protein lead to impaired export of viral late mRNAs and mislocalization of the E1B 55 kDa protein in infected cell nuclei (Gonzalez and Flint, 2002; Ornelles and Shenk, 1991). The E1B and E4 Orf6 proteins associate with not only one another, but also the cellular proteins cullin5, elongins B and C and Rbx1 to form an infected cell-specific E3 ubiquitin ligase (Harada et al., 2002; Querido et al., 2001). The E4 Orf6 protein binds to elongin C and is thought to direct assembly of the ligase, whereas different residues within the E1B 55 kDa protein have been implicated in recognition of different substrates (Blanchette et al., 2004; Schwartz et al., 2008). The substrates identified to date are all cellular proteins, p53 (Harada et al., 2002; Luo et al., 2007; Querido et al., 2001), the MreII and Nbs1 components of the MRN double strand DNA break recognition complex (Carson et al., 2003; Stracker, Carson, and Weitzman, 2002), DNA ligase IV (Baker et al., 2007) and integrin α3 (Dallaire et al., 2009). A substitution in the E4 Orf 6 protein that blocks assembly of the infected cell-specific E3 ubiquitin ligase (the Ad E3 ubiquitin ligase) was observed to impair viral late mRNA export to the same degree of detection of E4 Orf6 (Blanchette et al., 2008). Furthermore, synthesis of a dominant negative derivative of cullin 5 in infected HeLa cells induced decreases in export of viral late mRNAs from the nucleus and in viral late protein synthesis (Woo and Berk, 2007). These 5 observations implicate the activity of the Ad E3 ubiquitin ligase is regulation of mRNA export during the late phase of infection. However the relevant substrate(s) has not been identified, in part because the cellular pathway responsible for export of viral late mRNAs was not known.
The Ad5 E1B 55 kDa protein contains a leucine-rich nuclear export signal (NES) that directs its nuclear export via the exportin1 (Crm-1) export receptor and shuttling between the nucleus and cytoplasm (Dobbelstein et al., 1997; Dosch et al., 2001; Kratzer et al., 2000). In uninfected cells, this receptor is responsible for nuclear export of proteins that contain an NES and of small RNAs bound to them (Gorlich and Kutay, 1999; Stutz and Rosbash, 1998; Weis, 1998). However, exportin-1 is subverted for export of viral unspliced and partially spliced RNAs in human immunodeficiency virus type1-infected cells by the HIV-1 Rev protein, which both contains an NES and binds to a specific sequence in the viral RNAs (Fischer et al., 1995; Fornerod et al., 1997; Malim et al., 1991). Despite the presence of an NES in the E1B 55 kDa protein, viral late mRNAs are not exported via the exportin-1 receptor. Inhibition of this receptor by the drug leptomycin B or an NES-containing, cell-permeable peptide did not impair synthesis of viral late proteins (Carter, Izadpanah, and Bridge, 2003; Rabino et al., 2000) or the export of viral late mRNAs (Flint et al., 2005). Furthermore, substitutions that prevent recognition of the E1B NES by exportin1 had no effect on export of the late fiber mRNA, or viral replication (Kindsmuller et al., 2007). The major receptor for export of mRNAs from the nucleus of all eukaryotes examined is the Nxf1 (Tap) protein, which functions as a heterodimer with Nxt1, see (Carmody and Wente, 2009; Erkmann and Kutay, 2004; Kohler and Hurt, 2007; Reed and Cheng, 2005) for reviews. We have reported recently that inhibition of production of Nxf1 by RNA interference under conditions that do not impair production of the E1B 55 kDa protein reduced the efficiency of viral late mRNA export (Yatherajam, Huang, and Flint, 2011).
Nxf1 was originally identified by virtue of its binding to a specific RNA sequence, the constitutive transport element, which directs export of unspliced RNA of the simple retrovirus, Mason-Pfizer monkey virus (Kang, Bogerd, and Cullen, 2000). Nevertheless, Nxf1 does not interact directly with cellular mRNAs, but rather recognizes its export substrates indirectly by binding to RNA-binding adaptor proteins, such as Ref/Aly, and specific Ser/Arg-rich (SR) proteins (Carmody and Wente, 2009; Huang and Steitz, 2005; Kohler and Hurt, 2007; Reed and Cheng, 2005). The efficient export of viral late mRNAs with concomitant inhibition of cellular mRNA export could therefore be explained by production of a RNA-specific adaptor that sequesters Nxf1 with viral mRNA to block export of cellular mRNAs from the nucleus. The E1B 55 kDa protein interacts with the cellular RNA binding protein E1B-AP5 (Gabler et al., 1998), which has been reported to associate with Nxf1 (Bachi et al., 2000). Overproduction of E1B-AP5 in infected cells led to increases in the cytoplasmic concentration of both viral ML and cellular actin mRNAs (Gabler et al., 1998). However, it was not established that mRNA export was altered, and E1BAP5 was reported subsequently to regulate transcription (Kzhyshkowska et al., 2003). Furthermore, the E1B 55 kDa insertion mutation A143 (Yew, Kao, and Berk, 1990) induces a substantial reduction in the efficiency of viral late mRNA in both normal and transformed human cells (Gonzalez et al., 2006; Gonzalez and Flint, 2002), but does not affect binding of the E1B 55 kDa protein to E1B-Ap5 (Gabler et al., 1998). Another possible candidate adapter for Nxf1-dependent export of viral late mRNAs is the E1B 55 kDa protein itself: this protein has been reported to bind to RNA in vitro and to contain an RNA-recognition motif (RRM) required for such binding (Horridge and Leppard, 1998). We therefore examined RNA-binding by the E1B protein in infected cells and the functional role of the RNA recognition motif.
Results
Binding of the E1B 55 kDa protein to RNA in Ad5-infected cells
In the one previous study of the RNA binding properties of the E1B 55 kDa protein, non-specific binding of the viral protein synthesized in E. coli was detected by electrophoretic mobility shift and filter-binding assays (Horridge and Leppard, 1998). Substitutions in an RNP1 consensus sequence of an RNA recognition motif (RRM, also known as the RNP motif) identified in the E1B protein inhibited such in vitro RNA-binding activity (Horridge and Leppard, 1998). Neither RNA binding by the E1B 55 kDa protein in Ad5-infected cells nor the importance of the RNP1 motif has been investigated. We therefore attempted to examine binding of the E1B 55 kDa protein to RNA in infected cells by using UV lightinduced cross-linking of protein to RNA.
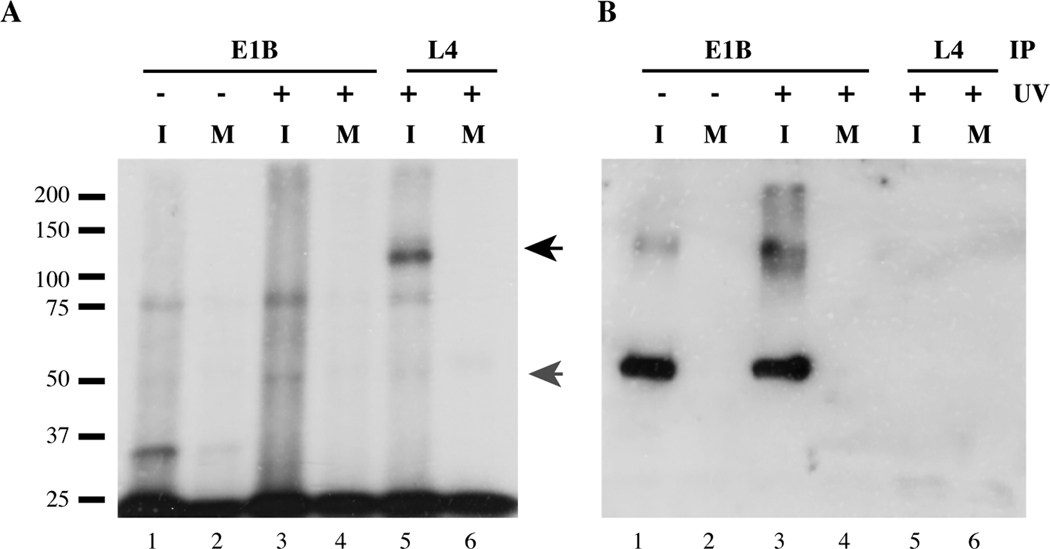
HeLa cells infected with 20 pfu/cell Ad5, typically for 18 hrs, were exposed on ice to 254 nm UV light for various periods, and nuclear lysates prepared and treated with RNase T1 as described in Materials and Methods. The E1B 55 kDa protein was then immunoprecipitated using monoclonal antibody 2A6 (Sarnow, Sullivan, and Levine, 1982) IgG and protein A/G agarose beads. Any RNA fragments that were covalently attached to the immunoprecipitated protein were indirectly labeled with [32P], by ligation of their 3′ ends to [32P]-labeled pCp, as described in Materials and Methods. After extensive washing of the bead-bound material, proteins were eluted as described in Materials and Methods, prior to SDS-polyacrylamide gel electrophoresis, transfer of proteins to nitrocellulose membranes and autoradiography followed by immunoblotting to detect the E1B 55 kDa protein. To provide a positive control, the same assay was applied in parallel to the viral L4 100 kDa protein, which can be readily crosslinked to RNA in Ad5-infected cells (Adam and Dreyfuss, 1989). One example of the results obtained in numerous attempts to crosslink the E1B 55 kDa protein to RNA, in this case by exposure of infected cells to UV light for 4 minutes, is shown in Figure 1.
Figure 1. The E1B 55 kDa protein cannot be cross-linked to RNA in Ad5-infected cells.
HeLa cells were infected with 20 pfu/cell Ad5 for 18 hrs (I) or mock-infected (M) and either exposed (+) or not exposed (−) to 254nm light for 4 minutes. The E1B 55kDa or He L4 100kDa protein was immunoprecipitated (IP) from nuclear lysates, and any covalently attached RNA fragments indirectly labeled with [32P], as described in Materials and Methods. Following electrophoresis in an 8% SDS-polyacrylamide gel, proteins were transferred to nitrocellulose and the membrane subjected sequentially to autoradiography (A) and immunoblotting to detect the E1B 55 kDa protein (B). The positions of molecular mass markers are listed at the left of panel A. The black and gray arrows at the right of panel A indicate the positions of the L4 100 kDa protein and a protein of some 50 kDa discussed in the text, respectively.
Labeled RNA attached to the L4 100 kDa protein was observed specifically in Ad5-infected cells (Fig 1A, lines 5 and 6, black arrow), as anticipated from the results of a previous study (Adam and Dreyfuss, 1989). A protein labeled at low efficiency by RNA transfer that migrated slightly more slowly than the 50 kDa molecular mass marker was also evident in infected cell samples (Fig 1A, lanes 1, 3 and 5, gray arrow). However, this species was present regardless of whether infected cells had been exposed to UV light (Fig 1A, lanes 1 and 3), and in both E1B 55 kDa and L4 100 kDa immunoprecipitates (Fig 1A, lanes 3 and 5). Furthermore, it migrated slightly more rapidly than the E1B 55 kDa protein detected by immunoblotting of the same gel after autoradiography (Fig 1B, lanes 1 and 3). Similar results were obtained when Ad5-infected cells were exposed to UV light for longer periods, or were treated later in the infectious cycle, when greater quantities of viral RNAs are synthesized (data not shown).
There are several possible explanations for the failure to observe binding of the E1B 55 kDa protein to RNA in infected cells in these experiments. The efficiency with which UV light induces formation of photo-adducts between proteins and nucleic acids depends on several parameters, including the nucleotide composition of the nucleic acids, and the proximity and geometry of appropriate amino acids and bases in the protein-nucleic acid complex (see (Chodosh, 2001); (Zhang et al., 2004)). The atomic structure of any interface between the E1B 55 kDa protein and RNA might therefore preclude efficient UV light-induced crosslinking. It is also possible that association of the E1B 55 kDa protein with RNA in infected cells is dynamic and transient, properties that would decrease the likelihood of formation of photo-adducts between the protein and RNA. For such reasons, it is not possible to conclude from results like those shown in Figure 1 that the E1B protein does not bind to RNA in vivo.
Mutation of the E1B 55 kDa protein RNA binding motif does not impair viral late gene expression or replication
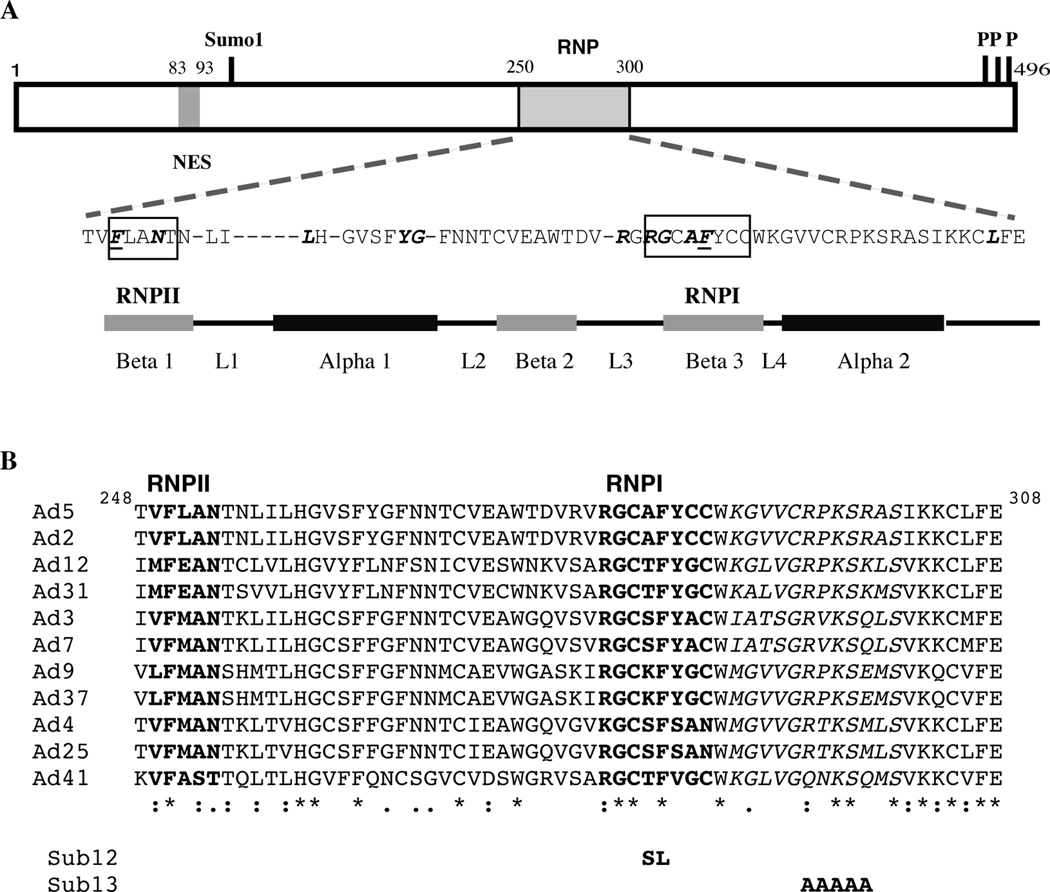
The RRM typically comprises some 90 amino acids containing a highly conserved RNP1 consensus sequence and a less well conserved RNPII sequence (Adam et al., 1986; Kenan, Query, and Keene, 1991; Swanson et al., 1987). The E1B 55 kDa RRM identified by Horridge and Leppard (Horridge and Leppard, 1998) and the conserved secondary structure of the RRM, an α β sandwich (Maris, Dominguez, and Allain, 2005), are shown in Figure 2A. The viral sequence contains many of the residues typically conserved among cellular RRMs (indicated by bold Italic face in Figure 2A), and residues within the Ad5 E1B 55 kDa protein RNPI and RNPII motifs are conserved among serotypes representative of the 6 human adenovirus species (Fig 2B). Structural studies have established that amino acids that form the RNP1 beta 3 strand make direct contacts with bases or the phosphate backbone of single-stranded nucleic acids (Maris, Dominguez, and Allain, 2005), and substitutions within the Ad5 E1B 55 kDa RNP1 sequence reduced or eliminated in vitro RNA binding activity (Horridge and Leppard, 1998). To investigate the role of RNA binding in viral replication, mutations resulting in two of these substitutions, A284S and F285L (E1BSub12, Fig 2B), were introduced simultaneously into the E1B 55 kDa protein coding sequence in the viral genome. It has been recognized more recently that RRMs can also participate in protein-protein interactions, to mediate interaction of two such domains within a single protein or association of two different proteins (Maris, Dominguez, and Allain, 2005). In several such cases, binding of RRMs to one another is via residues located within the C-terminal α2 helix (Maris, Dominguez, and Allain, 2005). The corresponding sequence of the Ad5 E1B 55 kDa protein contains several residues that are completely or highly conserved among human adenoviruses (Fig 2B). To assess the functional importance of this conserved sequence, we designed mutations that result in substitution of five residues within it by Ala (E1BSub13, Fig 2B).
Figure 2. The RNA recognition motif of the E1B 55 kDa protein.
A. The Ad5 E1B 55 kDa protein is represented by the box at the top, with the positions of the nuclear export signal (NES) (Dobbelstein et al., 1997; Kratzer et al., 2000), the RRM (Horridge and Leppard, 1998), and sites of sumoylation (Sumo-1) (Teodoro and Branton, 1997) and phosphorylation (P) (Teodoro and Branton, 1997) indicated. The sequence of the RRM is shown below, with the RNPI and RNPII motifs boxed, and amino acids conserved among RRMs indicated in bold, italic face. The underlined amino acids are aromatic residues important for RNA binding (Maris, Dominguez, and Allain, 2005). The conserved secondary structure of the RRM (Maris, Dominguez, and Allain, 2005) is shown below. B. The sequences of the E1B 55 kDa proteins of human adenoviruses of species C (Ad5 and Ad2), species A (Ad9 and Ad37), species B (Ad3 and Ad7), species D (Ad9 and Ad37), species E (Ad4 and Ad25) and species F (Ad41) corresponding to residues 248 to 308 of the Ad5 protein were aligned using Clustal W (1.83) (Larkin et al., 2007; Thompson, Higgins, and Gibson, 1994). The sequences of the RRM are shown, with the RNPI and RNPII motifs in bold face and the alpha2 helix in italic face. Below are indicated amino acids that are identical in all sequences (*) or represent conservative (:) or semi-conservative (.) substitutions. The amino acid substitutions introduced by the E1BSub12 and E1BSub13 mutations are listed below.
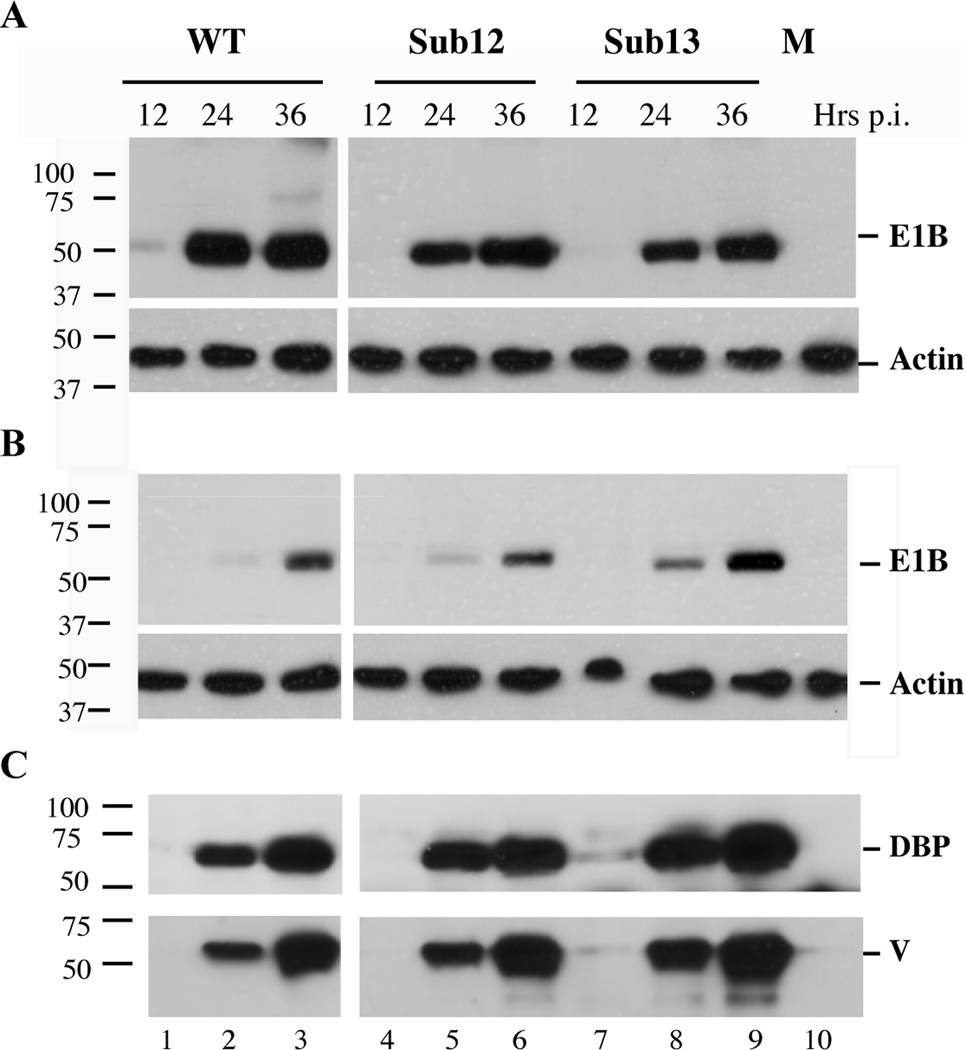
The E1B 55 kDa substitution mutations were introduced into the viral genome and mutant viruses, designated AdEasyE1BSub12 and AdEasyE1BSub13, isolated in complementing 293 cells (Aiello et al., 1979; Graham et al., 1977), as described in Materials and Methods. The concentrations of the E1B 55 kDa protein accumulating in HeLa cells and normal human fibroblasts infected by the mutants and the phenotypically wild-type parent were compared by immunoblotting. No significant differences in the steady state concentration of this viral protein, relative to those of an internal β-actin control, were observed in either HeLa cells or HFFs infected by the mutant and the wild type viruses (Figs 3A and 3B). These observations indicate that neither set of substitutions reduces the stability of the E1B 55 kDa protein.
Figure 3. Steady State concentrations of viral proteins in wild type- and mutant virus- infected cells.
A. HeLa cells were infected with 10 pfu/cell AdEasyE1 (WT), AdEasyE1Sub12 or AdEasyE1Sub13 for the periods indicated at the top, or mock infected (M). Total protein extracts were prepared and the E1B 55 kDa protein and β-actin examined by immunoblotting, as described in Materials and Methods. B. As panel A, except that HFFs were infected with 30 pfu/cell of the viruses listed at the top of panel A. C. The viral E2 DNA binding protein (DBP) and late protein V were examined by immunoblotting of the samples used in the experiment shown in panel B. In all panels, the positions of molecular mass markers are indicated at the left.
To begin to assess the effects of these mutations on viral replication, the concentrations of additional viral proteins were compared in transformed and normal human cells infected by the wild-type and mutant viruses. Neither of the substitutions resulted in impaired production of the early E2 DNA-binding protein (DBP) or of protein V, a late structural protein, in HFFs (Fig 3C) or HeLa cells (data not shown). The former result is not surprising, as no effects on synthesis of other early proteins have been observed in transformed or normal human cells infected by E1B 55 kDa-null mutant viruses (Harada and Berk, 1999; O'Shea et al., 2004; Pilder et al., 1986; Williams et al., 1986). As discussed previously, viral late mRNAs are not exported efficiently from the nucleus in cells infected by such mutants (see Introduction). This defect results in a significantly more severe inhibition of late protein synthesis (Goodrum and Ornelles, 1999; Harada and Berk, 1999; Pilder et al., 1986; Williams et al., 1986), in part because specific late proteins are required for late phase-specific stimulation of transcription from the major late (ML) promoter (Ali et al., 2007; Tribouley et al., 1994), to establish the late pattern of alternative processing of ML pre-mRNAs (Farley, Brown, and Leppard, 2004; Tormanen et al., 2006) or for efficient translation of viral late mRNAs (Cuesta, Xi, and Schneider, 2000; Hayes et al., 1990). Towards the end of the late phase of infection, the activity of the Ad E3 ubiquitin ligase also prevents activation of the RNA-dependent protein kinase Pkr, phosphorylation of elF2α and reduced translation of viral late mRNAs (Spurgeon and Ornelles, 2009). The observation that the concentration of protein V is not reduced in cells infected by the mutants therefore indicates that the E1BSub12 and E1BSub13 mutations impair neither viral late mRNA export nor the ability of the Ad E3 ligase to block activation of Pkr.
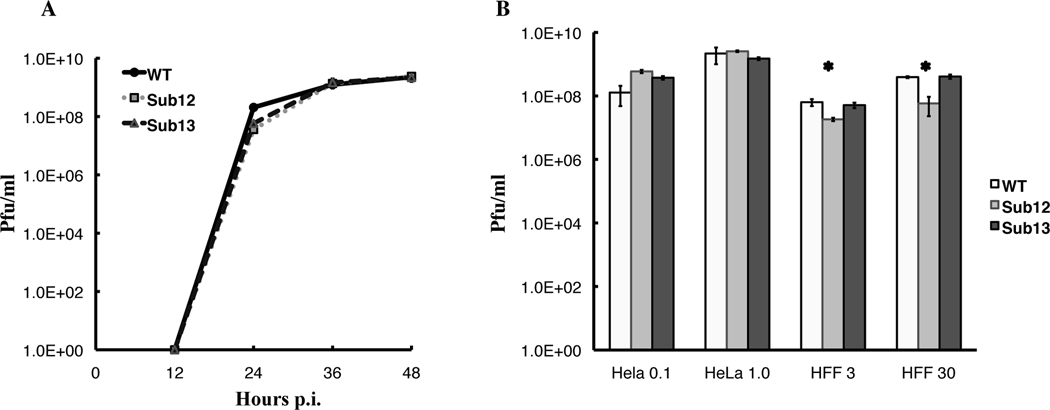
The efficiency of replication of the mutant viruses was next compared to that of the wild type, initially by measuring production of infectious particles as a function of time after infection at typical multiplicities. The kinetics of virus production were virtually identical in HeLa cells infected with 10 pfu/cell wild type and the AdEasyE1BSub12 and AdEasyE1BSub13 mutants (Fig 4A) and in HFFs infected with 30 pfu/cell (data not shown). In separate experiments, the yield of virus 44 hours after low or high multiplicity infection of the two cell types were also compared with no significant differences detected in HeLa cells (Fig 4B). However, in HFFs infected at low or high multiplicity, the yields of AdEasy E1B Sub12 were reduced 4 to 6-fold. These decreases were statistically significant, but whether they have biological relevance is not clear: in these cells, replication of the E1B 55kDa null mutant Hr6 is reduced by three orders of magnitude (Gonzalez et al., 2006).
Figure 4. The RRM mutations do not impair viral replication.
A. HeLa cells were infected with 10 pfu/cell AdEasyE1 (WT), AdEasyE1Sub12 or AdEasyE1Sub13, and samples harvested after increasing periods. The yield of intracellular infectious particles was determined as described in Materials and Methods. In both panels, the values represent the means of at least three technical replicates, with the standard deviations shown by the error bars. B. Yields of intracellular infectious particles were determined 44 hrs after infection of HeLa cells with 0.1 or 1.0 pfu/cell, and of HFFs with 3 or 30 pfu/cell wild type or mutant viruses. The asterisks indicate difference between wild-type and mutant virus replication determined to be significant (P=0.005 and 8 × 10−6 for 3 and 30 Pfu/cell, respectively) by application of the Students t test.
Contacts between RRM domains and single-stranded nucleic acids can be made via residues only in the β-strand formed by the RNP1 motif, by amino acids in both RNP motifs or by only RNPII residues (Maris, Dominguez, and Allain, 2005). We therefore wished to address the question of whether the RNPII motif contributed to the functions or the E1B 55 kDa protein. Mutations that result in substitution of RNPII amino acids conserved among the E1B 55 kDa proteins of human adenovirus (Fig 2B) were therefore introduced into the E1BSub12 background to create a double RNP mutant. However, this approach was not informative, as the steady state concentration of the E1B 55 kDa protein was reduced some 10-fold in cells infected by the double mutant virus (data not shown).
Effects of E1B RNP mutations on degradation of substrates of the infected cell-specific E3 ubiquitin ligase
The results of previous studies have suggested that the E1B 55 kDa protein is the substrate recognition subunit of the Ad E3 ubiquitin ligase (see the Introduction). As RNA recognition motifs can also participate in protein-protein interactions (Maris, Dominguez, and Allain, 2005), it was of interest to determine whether the substitutions in the RNP1 motif or adjacent α2 segment of the E1B protein (Fig. 2A) impaired degradation of proteins modified by this ligase. These proteins include the cellular tumor suppressor p53 (Harada et al., 2002; Luo et al., 2007; Querido et al., 2001), which is absent from, or altered in, many lines of human tumor cells, reviewed in (Hainaut and Hollstein, 2000; Hollstein et al., 1991). Consequently, these experiments were performed in normal human fibroblasts. Initial examination of the concentrations of the Ad E3 ubiquitin ligase substrate Mre11 (Stracker, Carson, and Weitzman, 2002) in the infected HFF samples used in the analyses shown in Figure 3B suggested that this protein was not degraded as efficiently in cells infected by AdEasyE1BSub13 as following AdEasyE1BSub12 (data not shown). To investigate this effect more rigorously by comparison to a positive control, we introduced a deletion of base pair 2347 in the viral genome into the same genetic background to create the mutant AdEasyE1Δ2347, as described in Materials and Methods. This deletion, which is present in the genome of Hr6 (Williams et al., 1986), alters only the coding sequence of the E1B 55 kDa protein and results in premature termination of translation a short distance downstream. Consequently, no E1B 55 kDa protein (nor the predicted N-terminally truncated derivative) can be detected in Hr6-infected cells (Gonzalez et al., 2006; Lassam, Bayley, and Graham, 1979; Williams et al., 1986). Sequencing of plaque-purified isolates of AdEasy E1Δ2347 confirmed the presence of this single base pair deletion, but no other mutations, in the E1B or E1A genes.
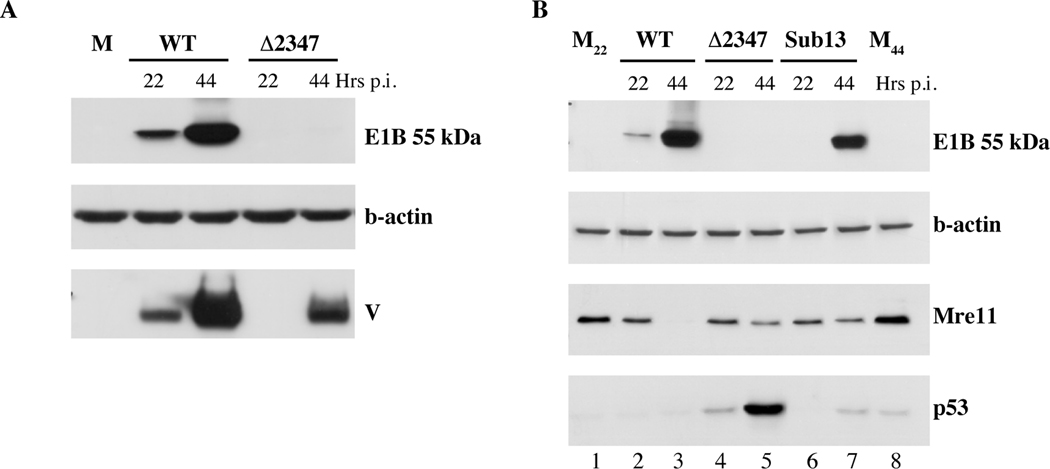
To establish whether this deletion reproduces phenotypes exhibited by Hr6, we first compared accumulation of the E1B 55 KDa protein in cells infected by AdEasyE1 or AdEasy E1Δ2347 by immunoblotting after increasing periods of infection. The concentrations of the internal control β-actin were very similar in all samples (Fig. 5A). However, the E1B 55kDa protein could not be detected in mutant-infected cells, even at late times when high concentrations were present in cells infected by the parental wild-type virus (Fig. 5A). These observations indicate that, as predicted from the properties of Hr6 described above, deletion of the bp2347 from the AdEasyE1 genome prevents production of the E1B 55kDa protein in infected cells. When this E1B protein cannot be made in infected cells, synthesis of viral late proteins is impaired (Goodrum and Ornelles, 1999; Harada and Berk, 1999; Pilder et al., 1986; Williams et al., 1986), a phenotype that is particularly dramatic in HFFs (Gonzalez et al., 2006). The effects of the deletion on accumulation of a late structural protein, protein V, were therefore also examined by immunoblotting. This late protein was readily detected by 22 hr. after infection with AdEasyE1, and increased considerably in concentration by 44 hrs (Fig. 5A). In contrast, protein V was not observed 22 hrs. after infection with AdEasyE1Δ2347 and attained significantly lower concentrations than in wild-type infected cells by 44 hrs. (Fig. 5A). We therefore conclude that deletion of base pair 2347 from the AdEasyE1 genome resulted in failure to produce the E1B 55kDa protein, as predicted, and reproduced the defect in viral late gene previously observed in transformed and normal human cells infected by Hr6 and other E1B 55 kDa-null mutants (Gonzalez et al., 2006; Goodrum and Ornelles, 1999; Harada and Berk, 1999; Pilder et al., 1986; Williams et al., 1986).
Figure 5. Effects of the RRM mutations on degradation of Mre11 and p53.
A. HFFs were infected with AdEasyE1 (WT) or AdEasyE1Δ2347 (Δ2347) for the periods indicated, or mock infected (M). Total cell extracts were prepared and the proteins listed at the right examined by immunoblotting. B. HFFs were infected with the same wild-type and E1B 55 kDa-null mutant or AdEasyE1Sub13 (Sub13) for the periods indicated, or mock infected (M22, M44). The proteins listed at the right were examined by immunoblotting of total cell extracts.
The effects of this E1B 55 kDa null mutation and the E1BSub13 substitutions on degradation of substrates of the AdE3 ubiquitin ligase were next compared by immunoblotting. Although the concentration of Mre11 was observed to be reduced in AdEasyE1Δ2347-infected cells at 22 and 44 hrs. p.i., this decrease was considerably less than in cells infected in parallel by AdEasyE1 (Fig. 5B, lanes 1–5), as expected (Stracker, Carson, and Weitzman, 2002). The degree and rate of decline in Mre11 concentration were virtually identical in cells infected by AdEasyE1Sub13 and the E1B-null mutant, even though the E1B 55 kDa protein was produced efficiently in the former case (Fig. 5B, compare lanes 4 and 5 with lanes 6 and 7). This phenotype indicated the E1BSub13 substitutions might impair interaction of the E1B with the E4 Orf6 protein, and hence assembly of the Ad E3 ubiquitin ligase and targeting of all known substrates for proteasomal degradation. Alternatively, the substitutions could disrupt specifically the binding of the E1B substrate recognition-subunit of the ligase to Mre11. To distinguish between these possibilities, we examined the effects of the mutations on concentrations of a second substrate, p53. Despite accumulation to much increased concentrations by 44 hrs. after infection with AdEasyE1Δ2347, p53 was detected at only very low concentrations in cells infected by either AdEasyE1Sub13 or its wild-type parent (Fig. 5B), indicating that the E1BSub13 substitutions do not preclude targeting of p53 for degradation by the action of the Ad E3 ubiquitin ligase.
Discussion
The E1B 55 kDa protein RRM is one of the few sequence motifs associated with specific functions reported or suggested to be present in this protein (Blackford and Grand, 2009; Flint and Gonzalez, 2003; Horridge and Leppard, 1998). Although the integrity of the RNP1 sequence within this motif was reported to be required for in vitro binding of the E1B protein to RNA (Horridge and Leppard, 1998), no interaction of the E1B protein with RNA could be detected in Ad5-infected cells by UV light-induced crosslinking (Fig. 1). This negative result can be explained by either the fact that E1B 55 kDa protein does not bind to RNA during the infectious cycle or properties of the protein-RNA interaction that preclude efficient crosslinking, such as a very short lifetime or an atomic structure of the protein-RNA interface that precludes efficient crosslinking. An analysis of the phenotypes exhibited by substitutions within the RRM, including RNPI substitutions reported to inhibit severely RNA binding In vitro (Horridge and Leppard, 1998), was undertaken to help distinguish between these possibilities. The failure of these mutations to impair significantly viral late gene expression (Fig. 3) or replication (Fig. 4) indicates that, if the E1B 55 kDa protein interacts with RNA in infected cells in the same way as it does in vitro, this activity is not required for efficient progression through the infectious cycle. Furthermore, the absence of any defect in viral late gene expression (Fig. 3) provides strong evidence that such RNA-binding activity does not contribute to the E1B 55 kDa protein-dependent selective export of viral late mRNA (see Introduction) or inactivation of Pkr (Spurgeon and Ornelles, 2009).
The substitutions introduced into the E1B 55kDa protein targeted amino acids of the RNPI sequence conserved among representatives of all 6 human adenovirus species (E1BSub12), or included such completely conserved residues in the C-terminal segment of the RRM (E1BSub13)(Fig 2B). There are several possible explanations for our finding that replacement of these highly conserved residues had no significant impact on viral replication. It may be that there is redundancy between the function fulfilled by the E1B RRM and one or more additional viral proteins. An excellent example of this phenomenon in Ad5-infected cells is provided by E1B 55kDa and E4 Orf6 protein-containing Ad E3 ubiquitin ligase and the E4 Orf3 protein: either the ligase or the E4 Orf3 protein alone can inactivate components of the MRN double strand break repair complex to a degree sufficient to protect against inhibition of viral DNA synthesis (Araujo et al., 2005; Evans and Hearing, 2005; Liu, Shevchenko, and Berk, 2005; Stracker, Carson, and Weitzman, 2002). Consequently, viral genome replication is impaired only when both the Ad E3 ubiquitin ligase and the E4 Orf3 protein are not present (Bridge and Ketner, 1989; Evans and Hearing, 2005; Jayaram and Bridge, 2005; Mathew and Bridge, 2007). In fact, such functional redundancy was observed in these experiments: viral late gene expression and replication were as efficient in AdEasySub13-infected HFFs as in cells infected by the wild-type parent (Figs. 3 and 4), even though these mutations impaired degradation of Mre11 to the same degree as the absence of the E1B 55 kDa protein (Fig. 5B).
The E1B 55 kDa protein has been proposed to function as the substrate-recognition subunit of the Ad E3 ubiquitin ligase (Blanchette et al., 2004): it can associate with the substrates p53 (Cathomen and Weitzman, 2000; Kao, Yew, and Berk, 1990; Liu and Green, 1994; Yew, Liu, and Berk, 1994), components of the MRN complex (Cathomen and Weitzman, 2000) and DNA ligase IV (Baker et al., 2007) in the absence of the E4 Orf6 protein. A previous analysis of the effects of insertions and substitutions in the E1B 55 kDa protein on binding to p53 and Mre11 and their subsequent degradation indicated that the viral protein recognizes these substrates via different sequences (Schwartz et al., 2008). Our observation that the E1BSub13 substitutions within the C-terminal α-helix of the putative RRM impair degradation of Mre11 but not of p53 (Fig. 5B) is consistent with this conclusion: as p53 is degraded efficiently in cells infected by this mutant (Fig. 5B), we can conclude that the E1BSub13 substitutions do not interfere with assembly of the Ad E3 ubiquitin ligase. These substitutions, which replace amino acids 295 to 299 with alanine, are more N-terminal than mutations previously reported to impair degradation of Mre11, such as insertions at residues 354, 373 and 443 (Schwartz et al., 2008). These observations suggest that amino acids that are well separated in the primary sequence lie in close proximity in the native E1B 55 kDa protein to form a discrete Mre11 recognition pocket, or surface.
It also remains possible that the E1B 55 kDa protein RNPI motif and the sequence immediately C-terminal to it targeted in these studies mediate functions of the protein that could not be detected using the assays described here. One such function is repression of expression of specific cellular genes in infected HFFs (Miller et al., 2009). It has been known for some time that the E1B 55kDa protein can repress transcription, including p53-dependent transcription, in in vitro and transient co-expression assays (Martin and Berk, 1998; Teodoro and Branton, 1997; Yew and Berk, 1992; Yew, Liu, and Berk, 1994). Such repression has been correlated with the ability of the E1B protein to cooperate with viral E1A proteins in transformation of rodent cells (Endter et al., 2005; Teodoro and Branton, 1997; Yew and Berk, 1992). When HFFs and other types of normal human cells are infected by E1B 55 kDa-null mutants in which the Ad E3 ubiquitin ligase is not present, p53 accumulates to high concentrations (Cardoso et al., 2008; O'Shea et al., 2004). Nevertheless, the p53 transcriptional program is reversed as efficiently in the absence of the E1B 55 kDa protein as it is in Ad5-infected cells (Miller et al., 2007; Miller et al., 2009). On the other hand, expression of a set of cellular genes highly enriched for those associated with innate anti-viral and adaptive immune responses was increased significantly in Hr6-infected cells (Miller et al., 2009). We have observed that viral replication is more sensitive to inhibition by exogenous type I interferon in the absence of the E1B 55 kDa protein (J. Qi, J. Chahal and S.J.F. unpublished observations), suggesting that repression of expression of host immune defense genes by the E1B protein serves as a countermeasure to cellular anti-viral defense mechanisms. It will therefore be of interest to investigate whether the RRM substitutions impact this function of the E1B 55 kDa protein.
Materials and Methods
Cells and Virus
HeLa cells and primary human foreskin fibroblasts (HFFs) were maintained in monolayer cultures in DMEM (Gibco-BRL) containing 5% (v/v) calf serum (Gibco-BRL) plus 5% (v/v) Hyclone bovine growth serum (Thermo Science – Fisher) and 10% (v/v) bovine growth serum, respectively. Wild-type and mutant viruses were propagated in 293 cells (Graham et al., 1977) and concentrations of infectious particles determined by plaque assay on these same cells (Williams, 1973).
A phenotypically wild-type derivative of the AdEasy genome (He et al., 1998) that contains the complete E1A and E1B genes was constructed by homologous recombination between pShuttle E1, which has been described previously (Miller et al., 2009), and pAdEasy-1 (He et al., 1998) in E. coli BJ 5183 cells. The E1BSub12 and E1BSub13 mutations that result in substitutions of amino acids within the putative RNA recognition motif of the E1B 55 kDa protein (Fig 2B) were introduced into the plasmid pShuttle E1 (Miller et al., 2009) by using the QuikChangeII site-directed mutagenesis kit (Stratagene-Agilent Technologies) as recommended by the manufacturer. Plasmids carrying the E1BSub13 mutations were identified by the presence of a new restriction endonuclease cleavage site, whereas recovery of the E1BSub12 mutations was assayed by sequencing of the relevant region of the E1B gene. The deletion of base pair 2347 present in the E1B gene of the Ad5 mutant Hr6 was introduced into pShuttleE1 by the same method, and plasmids carrying this mutation identified by loss of an MwoI cleavage site. Recovery of the mutations into the viral genome, isolation, plaque-purification and amplification of mutant viruses in 293 cells and confirmation of the presence of the mutations in viral DNA were as described previously (Miller et al., 2009).
To examine viral replication, HeLa cells or HFFs in 6 well dishes were infected with wild type AdEasyE1, Ad5E1BSub12 or Ad5E1B Sub13 for increasing periods, or with different multiplicities for 44 hrs. Cells were harvested and washed once in PBS, and resuspended in 0.125ml/well of 0.01M Tris-HCl, pH7.4, containing 0.15 NaCl, 0.005M KCl, 10mM MgCl2 and 0.1 (w/v) dextrose. After 5 cycles of freeze-thawing, and centrifugation at 500×g for 10 mins at 4°C, concentrations of infectious particles were determined by plaque assay on 293 cells.
Crosslinking of proteins to RNA
HeLa cells were infected with 20 pfu/cell Ad5 for 18 to 24 hrs, or mock infected, and washed twice with 5ml/10 cm dish ice-cold phosphate-buffered saline (PBS, Gibco-BRL), prior to addition of 2 ml/plate PBS. Cells on ice were then irradiated with ~4000 µwatts/cm2 254 nm UV light for 2–8 minutes using a Stratalinker UV crosslinker 2400 (Stratagene). They were washed twice with 5 ml PBS and lysed by incubation for 5 mins at 4°C in 0.05M Tris-HCl, pH 7.4 containing 0.15M NaCl, 1.5mM MgCl2, 0.6% (v/v) nonidetP40 (NP-40), 1mm PMSF, 1 µg/ml antipain, 2 µg/ml leupeptin and 100 units/ml RNasin (Promega). Nuclei were collected by centrifugation and suspended in 0. 05M Tris-HCl, pH 7.4 containing 0.15M NaCl, 3mM MgCl2, 0.4% NP-40 and the protease and RNase inhibitors listed above, and sonicated on ice in 30 sec bursts until sample viscosity was reduced. RNase-free DNase I (Roche Applied Science) was added to a final concentration of 0.1 units/ml, and samples incubated for 30 mins at room temperature. RNase T1 (Ambion Inc.) was added to a final concentration of 0.04 units/ml prior to incubation for a further 30 mins at room temperature. Samples were centrifuged at 16,000 ×g for 10 mins at 4°C. Supernatants were removed and pre-cleared by incubation for 1 hr at 4°C with protein A/G agarose beads (Santa Cruz Biotechnology) that had been washed twice with 0.05M Tris-HCl, pH7.4 containing 0.15 M NaCl, 1 mM EDTA, 1% (v/v) NP-40, 1mM PMSF, 2µg/ml leupeptin, 1 µg/ml antipain and 100 units/ml RNasin (buffer A), and resuspended in this buffer. The beads were removed by centrifugation at 800 ×g for 1 min at 4°C. The E1B 55kDa or L4 100kDa proteins were then immunoprecipitated by sequential incubation of supernatants with monoclonal antibodies 2A6 IgG (Sarnow, Sullivan, and Levine, 1982) or 2100–1, respectively, (Cepko and Sharp, 1982) (overnight at 4°C with rotation), and protein A/G agarose (4 hrs at 4°C with rotation) prepared as described above. The beads were collected by centrifugation at 800 ×g for 1 min at 4°C and washed twice with buffer A, twice with the same buffer containing 0.5M rather than 0.15M NaCl, and twice with 0.05M Tris-HCl, pH7.4, containing 10 mM MgCl2 and 0.5% (v/v) NP-40 (buffer B). They were then resuspended in 0.05M Tris-HCl, pH8.5 containing 1mM EDTA and incubated with 0.75 units/ml calf intestinal alkaline phosphatase (Roche) for 30 mins at 37°C. After centrifugation as described above, beads were washed twice with 0.05M Tris-HCl, pH7.4, containing 20 mM EGTA and 0.5% (v/v) NP-40, and twice with buffer B. Following resuspension in 0.05M Tris-HCl, pH7.8, containing 10 mM MgCl2, 1mM ATP, 1.6 µg bovine serum albumen and 1 mM dithiothreitol, samples were incubated overnight at 4°C with 100 µCi [32p]-pcp (300 Ci/mmol, Perkin Elmer) and 0.5 units/ml T4 RNA ligase 1 (New England Biolabs). Following ligation, the beads were collected by centrifugation as described above and washed four times with buffer B. They were then resuspended in 0.12M Tris-HCl, pH6.8 containing 4% (w/v) SDS, 20% (v/v) glycerol and 0.004% (w/v) bromphenol blue. Proteins were released by incubation at 100°C for 10 mins followed by centrifugation at 15,000×g for 10 mins at room temperature to remove protein A/G agarose. Proteins were subjected to electrophoresis in 8% polyacrylamide gels and transferred to 0.2 µm nitrocellulose membranes (Millipore). The membranes were rinsed in PBS and exposed to HyBlot Cl™ autoradiography film (Denville Scientific Inc.) at −80°C in the presence of intensifying screen. Subsequently, the E1B 55 kDa protein was detected by immunoblotting with the 2A6 monoclonal antibody as described (Gonzalez and Flint, 2002).
Immunoblotting
HeLa cells and HFFs were infected with 10 pfu/cell and 30 pfu/cell, respectively of wild-type AdEasyE1 or the AdEasyE1BSub12, AdEasy5E1BSub13 or AdEasyE1Δ2347 mutants, for increasing periods, or mock-infected. Cells were lysed by addition of 0.2 ml/well of a 6 well dish of 0.06M Tris – HCl, pH6.8, containing 2% (w/v) SDS, 10% (v/v) glycerol, 5% (v/v) β-mercaptoethanol and 0.002% (w/v) bromphenol blue. Lysates were collected and stored at −20°C. Prior to electrophoresis in 10% polyacrylamide gels cast and run in 0.1M Tris, 0.192 M Glycine, pH 8.8, containing 0.1% (w/v) SDS, lysates were sonicated as described above and incubated at 100°C for 10 mins. The viral E1B 55 kDa protein, the E2 single-stranded DNA-binding protein and protein V were detected by immunoblotting with monoclonal antibodies 2A6 (Sarnow, Sullivan, and Levine, 1982), B6 (Reich and Levine, 1984) and F58#1 (Lunt et al., 1988), respectively. The cellular Mre11 and p53 proteins were examined using rabbit anti-Mre11 polyclonal antibodies (Novus Biologicals) and anti-p53 mouse monoclonal antibody DO1 directly conjugated to horse radish peroxidase (Santa Cruz). To provide internal controls, the concentrations of β-actin were examined in parallel by using an anti-human β-actin monoclonal antibody conjugated to horse radish peroxidase (Abcam). Primary antibodies were visualized by enhanced chemiluminescence (Amersham Bioscience) following binding by anti-mouse or anti-rabbit IgG conjugated to horse radish peroxidase (Millipore Corporation).
Acknowledgements
We thank Ellen Brindle-Clark for assistance with preparation of the manuscript. This work was supported by a grant (RO1 AI050809) from the National Institute of Allergy and Infectious Disease, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam S, Dreyfuss G. Adenovirus proteins associated with mRNA and hnRNA in infected HeLa cells. J. Virol. 1989;61:3276–3283. doi: 10.1128/jvi.61.10.3276-3283.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam S, Nakagawa T, Swanson MS, Woodruff TK, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and identification of a ribonucleoprotein consensus sequence. Mol. Cell. Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello L, Guilfoyle R, Huebner K, Weinmann R. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293) Virology. 1979;94(2):460–469. doi: 10.1016/0042-6822(79)90476-8. [DOI] [PubMed] [Google Scholar]
- Ali H, LeRoy G, Bridge G, Flint SJ. The adenovirus L4 33-kilodalton protein binds to intragenic sequences of the major late promoter required for late phase-specific stimulation of transcription. J Virol. 2007;81(3):1327–1338. doi: 10.1128/JVI.01584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CW, Baum PR, Gesteland RF. Processing of adenovirus 2-induced proteins. J Virol. 1973;12(2):241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo FD, Stracker TH, Carson CT, Lee DV, Weitzman MD. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J Virol. 2005;79(17):11382–11391. doi: 10.1128/JVI.79.17.11382-11391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspegren A, Rabino C, Bridge E. Organization of splicing factors in adenovirus-infected cells reflects changes in gene expression during the early to late phase transition. Exp. Cell Res. 1998;245(1):203–213. doi: 10.1006/excr.1998.4264. [DOI] [PubMed] [Google Scholar]
- Babich A, Feldman LT, Nevins JR, Darnell JE, Jr, Weinberger C. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translational discrimination. Mol Cell Biol. 1983;3(7):1212–1221. doi: 10.1128/mcb.3.7.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiss LE, Young CSH, Fisher PP, Ginsberg HS. Expression of adenovirus E1A and E1B gene products and the Escheriria coli XPRT gene in KB cells. J. Virol. 1983;46:454–465. doi: 10.1128/jvi.46.2.454-465.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6(1):136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A, Rohleder KJ, Hanakahi LA, Ketner G. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J Virol. 2007;81(13):7034–7040. doi: 10.1128/JVI.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz GA, Flint SJ. Inhibition of HeLa cell protein synthesis during adenovirus infection: restriction of cellular messenger RNA sequences to the nucleus. J. Mol. Biol. 1979;131:353–373. doi: 10.1016/0022-2836(79)90081-0. [DOI] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford AN, Grand RJ. Adenovirus E1B 55-kilodalton protein: multiple roles in viral infection and cell transformation. J Virol. 2009;83(9):4000–4012. doi: 10.1128/JVI.02417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette P, Cheng CY, Yan Q, Ketner G, Ornelles DA, Dobner T, Conaway RC, Conaway JW, Branton PE. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol Cell Biol. 2004;24(21):9619–9629. doi: 10.1128/MCB.24.21.9619-9629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette P, Kindsmuller K, Groitl P, Dallaire F, Speiseder T, Branton PE, Dobner T. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J Virol. 2008;82(6):2642–2651. doi: 10.1128/JVI.02309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge E, Ketner G. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 1989;63:631–638. doi: 10.1128/jvi.63.2.631-638.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge E, Ketner G. Interaction of adenoviral E4 and E1b products in late gene expression. Virology. 1990;174:345–343. doi: 10.1016/0042-6822(90)90088-9. [DOI] [PubMed] [Google Scholar]
- Bridge E, Pettersson U. Nuclear Organization of Replication and Gene Expression in Adenovirus-Infected Cells. In: Doerfler W, Böhm P, editors. The Molecular Repertoire of Adenoviruses I. Vol. 199/I. Germany: Springer-Verlag; 1995. pp. 99–117. [DOI] [PubMed] [Google Scholar]
- Cardoso FM, Kato SE, Huang W, Flint SJ, Gonzalez RA. An early function of the adenoviral E1B 55 kDa protein is required for the nuclear relocalization of the cellular p53 protein in adenovirus-infected normal human cells. Virology. 2008;378(2):339–346. doi: 10.1016/j.virol.2008.06.016. [DOI] [PubMed] [Google Scholar]
- Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122(Pt 12):1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22(24):6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CC, Izadpanah R, Bridge E. Evaluating the role of CRM1-mediated export for adenovirus gene expression. Virology. 2003;315(1):224–233. doi: 10.1016/s0042-6822(03)00526-9. [DOI] [PubMed] [Google Scholar]
- Cathomen T, Weitzman MD. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J Virol. 2000;74(23):11407–11412. doi: 10.1128/jvi.74.23.11407-11412.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko CL, Sharp PA. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell. 1982;31:407–415. doi: 10.1016/0092-8674(82)90134-9. [DOI] [PubMed] [Google Scholar]
- Chodosh LA. UV crosslinking of proteins to nucleic acids. Curr Protoc Mol Biol. 2001;Chapter 12(Unit 12):5. doi: 10.1002/0471142727.mb1205s36. [DOI] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Cuesta R, Xi Q, Schneider RJ. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 2000;19(13):3465–3474. doi: 10.1093/emboj/19.13.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Xi Q, Schneider RJ. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J Virol. 2004;78(14):7707–7716. doi: 10.1128/JVI.78.14.7707-7716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutt JR, Shenk T, Hearing P. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J. Virol. 1987;61:543–552. doi: 10.1128/jvi.61.2.543-552.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallaire F, Blanchette P, Groitl P, Dobner T, Branton PE. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J Virol. 2009;83(11):5329–5338. doi: 10.1128/JVI.00089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M, Roth J, Kimberly WT, Levine AJ, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolph PJ, Racaniello V, Villamarin A, Pallodino F, Schneider RJ. The adenovirus tripartite leader may eliminate the requiremnt of cap-binding protein complex during translation initiation. J. Virol. 1988;62:2059–2066. doi: 10.1128/jvi.62.6.2059-2066.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch T, Horn F, Schneider G, Kratzer F, Dobner T, Hauber J, Stauber RH. The adenovirus type 5 E1B-55k oncoprotein actively shuttles in virus- infected cells, whereas transport of E4Orf6 is mediated by a CRM1- independent mechanism. J. Virol. 2001;75(12):5677–5683. doi: 10.1128/JVI.75.12.5677-5683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endter C, Hartl B, Spruss T, Hauber J, Dobner T. Blockage of CRM1-dependent nuclear export of the adenovirus type 5 early region 1B 55-kDa protein augments oncogenic transformation of primary rat cells. Oncogene. 2005;24(1):55–64. doi: 10.1038/sj.onc.1208170. [DOI] [PubMed] [Google Scholar]
- Erkmann JA, Kutay U. Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp Cell Res. 2004;296(1):12–20. doi: 10.1016/j.yexcr.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Evans JD, Hearing P. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J Virol. 2005;79(10):6207–6215. doi: 10.1128/JVI.79.10.6207-6215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley DC, Brown JL, Leppard KN. Activation of the early-late switch in adenovirus type 5 major late transcription unit expression by L4 gene products. J Virol. 2004;78(4):1782–1791. doi: 10.1128/JVI.78.4.1782-1791.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Huber J, Boulens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Flint SJ, Beltz GA, Linzer D. Synthesis and processing of SV40-specific RNA in adenovirus-infected, SV40-transformed human cells. J. Mol. Biol. 1983;167:335–359. doi: 10.1016/s0022-2836(83)80339-8. [DOI] [PubMed] [Google Scholar]
- Flint SJ, Gonzalez RA. Regulation of mRNA production by the adenoviral E1B 55kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 2003;272:287–330. doi: 10.1007/978-3-662-05597-7_10. [DOI] [PubMed] [Google Scholar]
- Flint SJ, Huang W, Goodhouse J, Kyin S. A peptide inhibitor of exportin1 blocks shuttling of the adenoviral E1B 55 kDa protein but not export of viral late mRNAs. . Virology. 2005;337:7–17. doi: 10.1016/j.virol.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Gabler S, Schutt H, Groitl P, Wolf H, Shenk T, Dobner T. E1B 55-kilodalton-associated protein: a cellular protein with RNA- binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 1998;72(10):7960–7971. doi: 10.1128/jvi.72.10.7960-7971.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R, Huang W, Finnen R, Bragg C, Flint SJ. Adenovirus E1B 55-kilodalton protein is required for both regulation of mRNA export and efficient entry into the late phase of infection in normal human fibroblasts. J Virol. 2006;80(2):964–974. doi: 10.1128/JVI.80.2.964-974.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez RA, Flint SJ. Effects of mutations in the adenoviral E1B 55 kDa protein coding sequence on viral late mRNA metabolism. J. Virol. 2002;76:4507–4519. doi: 10.1128/JVI.76.9.4507-4519.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrum FD, Ornelles DA. Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication. J. Virol. 1999;73(9):7474–7488. doi: 10.1128/jvi.73.9.7474-7488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Granberg F, Svensson C, Pettersson U, Zhao H. Modulation of host cell gene expression during onset of the late phase of an adenovirus infection is focused on growth inhibition and cell architecture. Virology. 2005;343(2):236–245. doi: 10.1016/j.virol.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- Halbert DN, Cutt JR, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada JN, Berk AJ. p53-Independent and -dependent requirements for E1B-55K in adenovirus type 5 replication. J. Virol. 1999;73(7):5333–5344. doi: 10.1128/jvi.73.7.5333-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada JN, Shevchenko A, Pallas DC, Berk AJ. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 2002;76(18):9194–9206. doi: 10.1128/JVI.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BW, Telling GC, Myat MM, Williams JF, Flint SJ. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 1990;64:2732–2742. doi: 10.1128/jvi.64.6.2732-2742.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95(5):2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Horridge JJ, Leppard KN. RNA-binding activity of the E1B 55-kilodalton protein from human adenovirus type 5. J. Virol. 1998;(11):9374–9379. doi: 10.1128/jvi.72.11.9374-9379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Schneider RJ. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell. 1991;65:271–280. doi: 10.1016/0092-8674(91)90161-q. [DOI] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. SRprises along a messenger's journey. Mol Cell. 2005;17(5):613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Jayaram S, Bridge E. Genome concatenation contributes to the late gene expression defect of an adenovirus E4 mutant. Virology. 2005;342(2):286–296. doi: 10.1016/j.virol.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Kang Y, Bogerd HP, Cullen BR. Analysis of cellular factors that mediate nuclear export of RNAs bearing the Mason-Pfizer monkey virus constitutive transport element. J. Virol. 2000;74(13):5863–5871. doi: 10.1128/jvi.74.13.5863-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CC, Yew PR, Berk AJ. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179(2):806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- Kenan DJ, Query CC, Keene JD. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16(6):214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- Kindsmuller K, Groitl P, Hartl B, Blanchette P, Hauber J, Dobner T. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc Natl Acad Sci U S A. 2007;104(16):6684–6689. doi: 10.1073/pnas.0702158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8(10):761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Kratzer F, Rosorius O, Heger P, Hirschmann N, Dobner T, Hauber J, Stauber RH. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene. 2000;19(7):850–857. doi: 10.1038/sj.onc.1203395. [DOI] [PubMed] [Google Scholar]
- Kzhyshkowska J, Rusch A, Wolf H, Dobner T. Regulation of transcription by the heterogeneous nuclear ribonucleoprotein E1B-AP5 is mediated by complex formation with the novel bromodomain-containing protein BRD7. Biochem J. 2003;371(Pt 2):385–393. doi: 10.1042/BJ20021281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lassam NJ, Bayley ST, Graham FL. Tumor antigens of human Ad5 in transformed cells and in cells infected with transformation defective host range mutants. Cell. 1979;18:781–791. doi: 10.1016/0092-8674(79)90131-4. [DOI] [PubMed] [Google Scholar]
- Liu F, Green MR. Promoter targeting by an adenovirus E1A through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shevchenko A, Berk AJ. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J Virol. 2005;79(22):14004–14016. doi: 10.1128/JVI.79.22.14004-14016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt R, Vayda ME, Young M, Flint SJ. Isolation and characterization of monoclonal antibodies against the adenovirus core proteins. Virology. 1988;164:275–279. doi: 10.1016/0042-6822(88)90645-9. [DOI] [PubMed] [Google Scholar]
- Luo K, Ehrlich E, Xiao Z, Zhang W, Ketner G, Yu XF. Adenovirus E4orf6 assembles with Cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. FASEB J. 2007;21(8):1742–1750. doi: 10.1096/fj.06-7241com. [DOI] [PubMed] [Google Scholar]
- Malim MH, McCarn DF, Tiley LS, Cullen BR. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65(8):4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272(9):2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Martin ME, Berk AJ. Adenovirus E1B 55K represses p53 activation in vitro. J Virol. 1998;72(4):3146–3154. doi: 10.1128/jvi.72.4.3146-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew SS, Bridge E. The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology. 2007;365(2):346–355. doi: 10.1016/j.virol.2007.03.049. [DOI] [PubMed] [Google Scholar]
- Miller DL, Myers CL, Rickards B, Coller HA, Flint SJ. Adenovirus type 5 exerts genome-wide control over cellular programs governing proliferation, quiescence, and survival. Genome Biol. 2007;8(4):R58. doi: 10.1186/gb-2007-8-4-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Rickards B, Mashiba M, Huang W, Flint SJ. The adenoviral E1B 55-kilodalton protein controls expression of immune response genes but not p53-dependent transcription. J Virol. 2009;83(8):3591–3603. doi: 10.1128/JVI.02269-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea CC, Johnson L, Bagus B, Choi S, Nicholas C, Shen A, Boyle L, Pandey K, Soria C, Kunich J, Shen Y, Habets G, Ginzinger D, McCormick F. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell. 2004;6(6):611–623. doi: 10.1016/j.ccr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Ornelles D, Shenk T. Location of the adenovirus early region 1B 55 kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34 kilodalton protein. J. Virol. 1991;65:424–439. doi: 10.1128/jvi.65.1.424-429.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55kd transforming polypeptide modulates transport or cytoplasmic stablization of viral and host cell mRNAs. Mol. Cell. Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 1994;13(21):5075–5085. doi: 10.1002/j.1460-2075.1994.tb06837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Bachellerie JP, Visa N, Puvion E. Rearrangements of intranuclear structures involved in RNA processing in response to adenovirus infection. J Cell Sci. 1994;107(Pt 6):1457–1468. doi: 10.1242/jcs.107.6.1457. [DOI] [PubMed] [Google Scholar]
- Querido E, Morrison MR, Chu-Pham-Dang H, Thirlwell SW, Boivin D, Branton PE. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J Virol. 2001;75(2):699–709. doi: 10.1128/JVI.75.2.699-709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabino C, Aspegren A, Corbin-Lickfett K, Bridge E. Adenovirus late gene expression does not require a Rev-like nuclear RNA export pathway. J. Virol. 2000;74(14):6684–6688. doi: 10.1128/jvi.74.14.6684-6688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Cheng H. TREX, SR proteins and export of mRNA. Curr Opin Cell Biol. 2005;17(3):269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Reich NC, Levine AJ. Growth regulation of a cellular tumor antigen, p53, in non-transformed cells. Nature. 1984;308:199–201. doi: 10.1038/308199a0. [DOI] [PubMed] [Google Scholar]
- Sarnow P, Hearing P, Anderson CW, Halbert DN, Shenk T, Levine AJ. Adenovirus early region 1B 58,000 dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J. Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P, Sullivan CA, Levine AJ. A monoclonal antibody detecting the Ad5 E1B-58K tumor antigen in adenovirus-infected and transformed cells. Virology. 1982;120:387–394. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- Schwartz RA, Lakdawala SS, Eshleman HD, Russell MR, Carson CT, Weitzman MD. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J Virol. 2008;82(18):9043–9055. doi: 10.1128/JVI.00925-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgeon ME, Ornelles DA. The adenovirus E1B 55-kilodalton and E4 open reading frame 6 proteins limit phosphorylation of eIF2alpha during the late phase of infection. J Virol. 2009;83(19):9970–9982. doi: 10.1128/JVI.01113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418(6895):348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12(21):3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Nakagawa TY, LeVan K, Dreyfuss G. Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-rRNA-binding proteins. Mol Cell Biol. 1987;7(5):1731–1739. doi: 10.1128/mcb.7.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro JG, Branton PE. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 1997;71(5):3620–3627. doi: 10.1128/jvi.71.5.3620-3627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormanen H, Backstrom E, Carlsson A, Akusjarvi G. L4-33K, an adenovirus-encoded alternative RNA splicing factor. J Biol Chem. 2006;281(48):36510–36517. doi: 10.1074/jbc.M607601200. [DOI] [PubMed] [Google Scholar]
- Tribouley C, Lutz P, Staub A, Kedinger C. The product of the adenovirus intermediate gene IVa2 is a transcription activator of the major late promoter. J. Virol. 1994;68:4450–4457. doi: 10.1128/jvi.68.7.4450-4457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23(5):185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- Williams J, Karger BD, Ho YS, Castiglia CL, Mann T, Flint SJ. The adenovirus E1B 495R protein plays a role in regulating the transport and stability of the viral late messages. Cancer Cells. 1986;4:275–284. [Google Scholar]
- Williams JF. Oncogenic transformation of hamster embryo cells in vitro by adenovirus type 5. Nature. 1973;243:162–163. doi: 10.1038/243162a0. [DOI] [PubMed] [Google Scholar]
- Woo JL, Berk AJ. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J Virol. 2007;81(2):575–587. doi: 10.1128/JVI.01725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Cuesta R, Schneider RJ. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 2004;18(16):1997–2009. doi: 10.1101/gad.1212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang U-C, Huang W, Flint SJ. mRNA export correlates with activation of transcription in human subgroup C adenovirus-infected cells. J. Virol. 1996;70:4071–4080. doi: 10.1128/jvi.70.6.4071-4080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatherajam G, Huang W, Flint SJ. Export of adenoviral late mRNA from the nucleus requires the Nxf1/Tap export receptor. J Virol. 2011;85(4):1429–1438. doi: 10.1128/JVI.02108-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew PR, Berk AJ. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357(6373):82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- Yew PR, Kao CC, Berk AJ. Dissection of functional domains in the adenovirus 2 early 1B 55k polypeptide by suppressor-linker-insertional mutagenesis. Virol. 1990;179:795–805. doi: 10.1016/0042-6822(90)90147-j. [DOI] [PubMed] [Google Scholar]
- Yew PR, Liu X, Berk AJ. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8(2):190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10(12):1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang K, Prandl R, Schoffl F. Detecting DNA-binding of proteins in vivo by UV-crosslinking and immunoprecipitation. Biochem Biophys Res Commun. 2004;322(3):705–711. doi: 10.1016/j.bbrc.2004.07.202. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Schneider RJ. Adenovirus inhibition of cell translation facilitates release of virus particles and enhances degradation of the cytokeratin network. J Virol. 1994;68(4):2544–2555. doi: 10.1128/jvi.68.4.2544-2555.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Granberg F, Pettersson U. How adenovirus strives to control cellular gene expression. Virology. 2007;363(2):357–375. doi: 10.1016/j.virol.2007.02.013. [DOI] [PubMed] [Google Scholar]