Abstract
Epigenetic regulation is important for stable maintenance of cell identity. For continued function of organs and tissues, illegitimate changes in cell identity must be avoided. Failure to do so can trigger tumour development and disease. How epigenetic patterns are established during cell differentiation has been explored by studying model systems such as X inactivation. Mammals balance the X-linked gene dosage between the sexes by silencing of one of the two X chromosomes in females. This is initiated by expression of the non-coding X-inactive specific transcript (Xist) RNA and depends on specific cellular contexts, in which essential silencing factors are expressed. Normally X inactivation is initiated in early embryogenesis, but recent reports identified instances where Xist is expressed and can initiate gene repression. Here we describe the features that characterize the cellular permissivity to initiation of X inactivation and note that these can also occur in cancer cells and in specific haematopoietic progenitors. We propose that embryonic pathways for epigenetic regulation are re-established in adult progenitor cells and tumour cells. Understanding their reactivation will deepen our understanding of tumourigenesis and may be exploited for cancer therapy.
Keywords: X inactivation, Xist RNA, chromatin, cancer, epigenetics
Introduction
Current research into epigenetic mechanisms aims at an understanding how the genome is regulated in development and disease. Applying findings to a clinical setting is not straightforward and will require the development of new conceptual frameworks. Even for basic questions the investigation of the establishment of epigenetic patterns in development has been difficult. If epigenetic pathways are blocked, often many developmental aberrations arise resulting in a complex phenotype. The difficulty in interpreting such experiments hinders the development of a coherent view of epigenetic regulation in cell differentiation. Model systems in which epigenetic regulation can be studied independently of developmental anomalies provide an alternative and facilitate the understanding of some basic questions. Such a system is X chromosome inactivation. Here we focus on recent findings in X inactivation and present a view of their relevance for development and disease.
How is epigenetic regulation relevant for disease?
Deoxyribonucleic acid (DNA) methyltransferases, Polycomb group (PcG) proteins and chromatin remodelling complexes have all been linked to human disease in a number of studies (Table I). DNA cytosine methylation has been examined in different types of cancer (Laird & Jaenisch, 1996). Promoters of tumour suppressor genes can become methylated when they are transcriptionally repressed (Gal-Yam et al, 2008; Laird & Jaenisch, 1996). In addition, the global DNA cytosine methylation level of tumour cell genomes is often reduced when compared to normal cells and hypomethylation has been associated with genomic instability (Eden et al, 2003). Frequently, loss of imprinted methylation and expression is correlated with tumourigenesis (reviewed in Iacobuzio-Donahue, 2009). Defects in the DNA methylation system have also been linked to developmental diseases. Mutations in the DNA de novo methyltransferase DNMT3B gene are associated with immunodeficiency, centromeric region instability, facial anomalies (ICF) syndrome (Hansen et al, 1999). A mutation in the methyl-cytosine DNA binding protein methyl CpG binding protein 2 (MeCP2) has been linked to RETT syndrome (Amir et al, 1999). Interestingly, restoring MeCP2 function in a mouse model of RETT syndrome can ameliorate the symptoms of the disease (Guy et al, 2007). While technically challenging, restoring MeCP2 expression could therefore be considered for therapeutic approaches.
Table I.
Epigenetics related to diseases
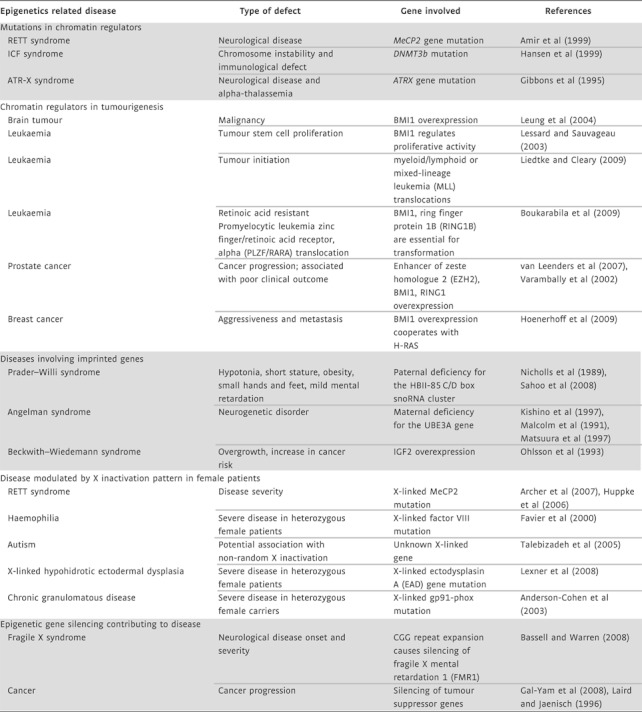 |
Polycomb group proteins form complexes that modify chromatin and regulate gene expression (reviewed in Ringrose & Paro, 2007; Schuettengruber et al, 2007; Schwartz & Pirrotta, 2008). PcG proteins are also important players in human diseases. Examples are provided by the observations that elevated expression levels of PcG proteins are associated with brain tumour development (Leung et al, 2004) and prostate cancer progression (van Leenders et al, 2007; Varambally et al, 2002). Significantly, the PcG protein B lymphoma Mo-MLV insertion region 1 oncogene (BMI1) has been shown to collaborate with Harvey rat sarcoma oncogene (H-RAS) to induce an aggressive and metastatic phenotype in a breast cancer model (Hoenerhoff et al, 2009). Recent work also shows that PcG regulation in leukaemia could have important implications for treatment outcomes (Boukarabila et al, 2009). In addition to PcG genes, other components of chromatin remodelling complexes have been associated with diseases such as X-linked alpha-thalassemia with mental retardation (ATR-X syndrome) which is caused by mutations in the SWI/SNF (Switch/Sucrose NonFermentable) homologue 2 (SNF2) chromatin remodelling protein Alpha thalassemia/mental retardation syndrome X-linked (ATRX) (Gibbons et al, 1995). These examples illustrate that understanding the basic principles of chromatin biology is of immediate relevance for a variety of human diseases.
Glossary
Chromatin
The genomic DNA as well as the structural and regulatory proteins of the chromosomes form the chromatin.
DNA methyltransferase
DNA methyltransferase catalyses the methylation of DNA by converting cytosine into 5-methyl-cytosine. Three catalytically active DNA methyltransferase genes (DNMT1, DNMT3A, and DNMT3B) are known in mammals.
Dosage compensation
The process used for adjusting the dosage differences arising from heteromorphic sex chromosomes. In mammals males have an XY and females have an XX sex chromosome pair and one of the X chromosomes is inactivated in female cells leaving a single X to be transcribed in both sexes.
Embryonic stem (ES) cells
ES cells are derived from blastocysts stage embryos. They maintain the ability to differentiate into all cell types of the embryo in culture.
Epigenetic regulation
Regulation of the genome mainly by chromatin modifying activity for control of gene expression in a developmental manner.
Imprinted methylation
DNA methylation that is present depending on the parental inheritance of a gene or regulatory region.
Incontinentia pigmenti
Incontinentia pigmenti is a rare disease caused by mutations of the X-linked NEMO gene. In female patients patches of defective cells can be observed in characteristic patterns in skin pigmentation as a result of X inactivation.
Polycomb group proteins
Polycomb group proteins act as repressors for developmental control genes and have a role in the regulation of cell proliferation in mammals. Some of these proteins form complexes that have catalytic activity such as the posttranslational modification of histones.
RETT syndrome
RETT syndrome is a neurodevelopmental disorder caused by mutations in the X-linked MeCP2 gene; it is characterized by autism, dementia and ataxia.
Stem cells
Stem cells can self-renew but also produce more than one cell type in the process of differentiation.
X chromosome inactivation
The process leading to the chromosome-wide silencing of one the two X chromosome in females to balance X-linked gene dosage between the XY male and XX female karyotype.
Epigenetic processes also influence human diseases through their normal function in development. Such processes include, but are not limited to, X inactivation and genomic imprinting (Table I). One of the two X chromosomes in female cells becomes inactivated in a random manner to compensate the gene doses for X-linked genes between the sexes. X inactivation leads to a genetic mosaic of cells expressing either the maternal or the paternal X chromosome. Normally about half the cells are expected to express either one of the X chromosomes but deviations from this ratio can be observed. Non-random patterns of X inactivation lead to phenotypic variability in a number of human diseases associated with mutations on one of the two X chromosomes in female patients. The severity of symptoms arising from mutations of the X-linked MeCP2 gene in female RETT patients has been observed to correlate with the pattern of X inactivation (reviewed in Chahrour & Zoghbi, 2007). In Incontinentia pigmenti, which is caused by mutations in the X-linked NF kappa B essential modulator (NEMO) gene, the patches of X inactivation result in a characteristic whorled pattern of skin hyperpigmentation (reviewed in Sun & Tsao, 2008). Similarly, mutations in the X-linked ectodysplasin A gene lead to anhidrotic ectodermal dysplasia with an absence of sweat glands in affected tissue patches (Lexner et al, 2008). Non-random patterns of X inactivation have also been linked to the development of female-predominant autoimmune disease (reviewed in Invernizzi et al, 2008). Non-randomness in X inactivation is often associated with mutations or structural changes of one of the X chromosomes, but its exact cause is not known (Morleo & Franco, 2008). It is likely that mutations on the X chromosome can influence the X inactivation pattern either by cell selection during development or via direct influence on the choice of which X chromosome to inactivate. In general, mutations that affect non-randomness do not appear to necessarily overlap with the disease causing mutation.
Genomic imprinting is a related mechanism that may also contribute to disease development (reviewed in Charalambous et al, 2007). A small number of genes are expressed in a mono-allelic fashion dependent upon from which parent they were inherited. As a result, mutation of the expressed copy of an imprinted gene has phenotypic consequences. Prader–Willi syndrome is caused by maternal inheritance of deletions on the chromosome 11 that affect the HBII-85 C/D box small nucleolar ribonucleic acid (RNA) cluster and is associated with hypotonia, short stature, obesity, small hands and feet, hypogonadism and mild mental retardation (Sahoo et al, 2008). Other examples are Angelman syndrome, a neurogenetic disease caused by maternal deficiency of the UBE3A gene (Kishino et al, 1997; Matsuura et al, 1997), and Beckwith–Wiedemann syndrome, an overgrowth syndrome with elevated cancer predisposition associated with over expression of insulin like growth factor 2 (Ohlsson et al, 1993). These examples illustrate how epigenetic processes contribute to human diseases and modulate the severity of disease symptoms.
A glimpse at the establishment of epigenetic patterns through X inactivation
Understanding X inactivation is also important to address basic questions in epigenetic regulation in mammalian cells. Epigenetic modifications of the genome are thought to prevent illegitimate changes in cell identity and protect an organism from cancer and other diseases. In mammals, the genetic information within each cell's nucleus contains the information required to specify over 200 distinct cell types. Cells perform specific functions within tissues and must ensure a stable phenotype and maintain their identity, in some cases over lifetime, to support organ function. Many of the cells are replenished by differentiation of stem cells and immature cell types (Fig 1). To allow a phenotypic change from a stem cell to a distinct differentiated cell, cell identity must become unstable at some point in differentiation. This point is of central importance to epigenetic regulation. How epigenetic patterns are established for stable maintenance of cell identity is poorly understood at present.
Figure 1. A schematic view of stem cell differentiation.
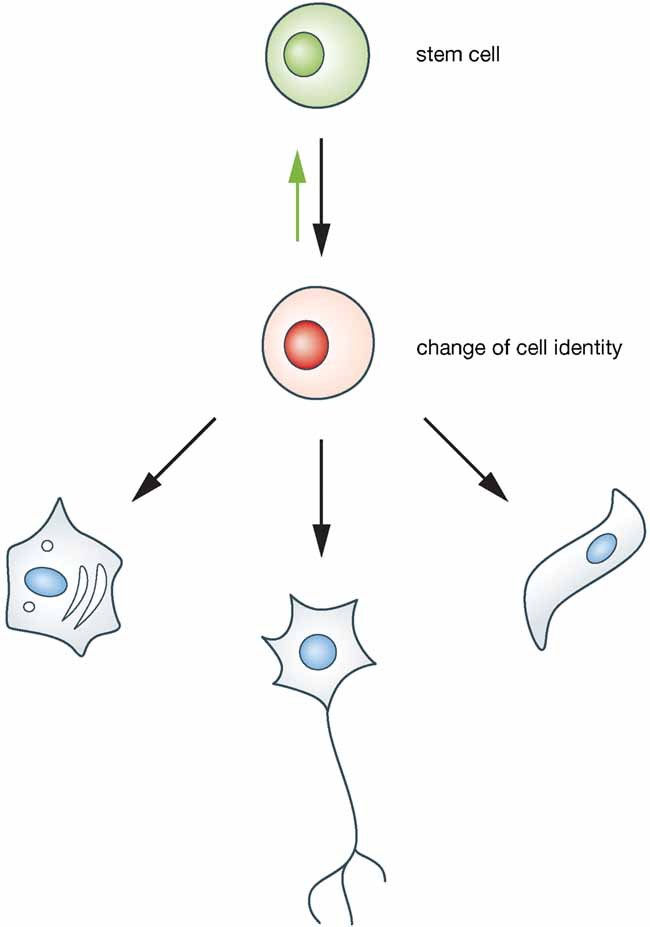
Differentiated cells that constitute organs and tissues are generated during development. Their characteristics and identity are faithfully maintained to ensure proper organ function. Throughout life many of these cells are replenished by the differentiation of stem cells. Therefore, adult stem cells have to be maintained stably. Yet, the differentiation of stem cells requires the cell identity to be changed from a stem cell to a differentiated cell with a specialized function. This transition passes through a point when cell identity is not stable (red). Notably, during embryogenesis adult stem cells, themselves, are generated from progenitors (green arrow).
Dosage compensation systems have been useful to dissect epigenetic mechanisms in flies, worms and mammals (Lucchesi et al, 2005). Chromosome-wide epigenetic changes are applied to balance the different sex chromosome constitution between the sexes (Fig 2). In the case of X inactivation in mammals, an unusual non-coding RNA—X-inactive-specific transcript (Xist) for inactive X specific transcript (Borsani et al, 1991; Brockdorff et al, 1991; Brown et al, 1991)—triggers the silencing of one of the two X chromosomes in female cells. Xist is transcribed from the X inactivation centre (Xic) locus on the X chromosome and then spreads over the entire chromosome. During the differentiation of mouse embryonic stem (ES) cells, the transition from an active X chromosome to an inactive X chromosome (Xi) one can be followed (Rastan & Robertson, 1985). Studying this process continuously during development has shown that X inactivation is separated into an INITIATION and a MAINTENANCE phase (Fig 2B). The accumulation of Xist initiates the repression of genes along the entire X chromosome. Initially, Xist is required for repression (Marahrens et al, 1997; Penny et al, 1996). Using genetic manipulation of Xist expression it has been shown that gene silencing is reversible and that genes will be reactivated once Xist expression is lost (Wutz & Jaenisch, 2000). Upon differentiation of the cell the Xi becomes subject to further modifications (Blewitt et al, 2008; Chaumeil et al, 2002; Keohane et al, 1996). Heritable changes including DNA methylation and histone hypoacetylation keep genes repressed on the Xi without requiring constant expression of Xist (Brown & Willard, 1994; Csankovszki et al, 1999; Wutz & Jaenisch, 2000). The initial trigger—Xist—is not required for maintenance of repression in differentiated cells, and the shift from Xist dependent initiation to stabilization of the inactive state could be regarded as a change of epigenetic context.
Figure 2. In mammals X inactivation compensates the difference in X-linked gene dosage between males and females.
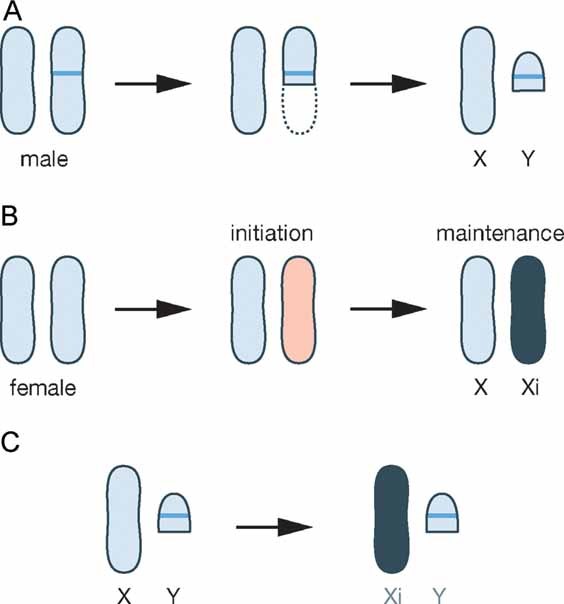
- It is thought that the mammalian sex chromosomes X and Y originated from an autosome pair, when a spontaneous mutation arose specifying male sex (blue line). Evolution of the sex chromosomes led to the loss of sequences from the Y chromosome leading to a gene dosage difference between XY males and XX females.
- For balancing gene dosage between the sexes random inactivation of one of the two X chromosomes in female cells is initiated by Xist (pink) and maintained by other mechanisms in differentiated cells (black).
- Experimental expression of Xist in male cells can be used for probing the epigenetic context. In cells expressing initiation factors, Xist mediated gene silencing is initiated and loss of X-linked gene expression then causes the death of the cells. Therefore, a phenotype can be observed depending on the epigenetic context of the cell.
Studies of the mechanism of X inactivation have progressed to a point where it is now possible to apply Xist for probing the epigenetic context of cells (Wutz, 2007). Importantly, Xist can initiate gene silencing only in very specific cell types. Naturally, X inactivation is initiated in the cells of the early embryo (Mak et al, 2004; Okamoto et al, 2004; Takagi et al, 1982) but experimental manipulation of Xist expression has shown that also in the adult mouse, cells exist in which Xist can initiate gene silencing. These cells include lineage-restricted progenitors within the haematopoietic system (Savarese et al, 2006). The haematopoietic stem cell (HSC) and mature blood cells, however, do not provide the appropriate cellular context for Xist-mediated silencing (Fig 2C). It therefore appears that the function of Xist in initiating chromosome-wide silencing is dependent on specific pathways that exist in the early embryo and are reactivated transiently during the differentiation of adult stem cells of the blood system. Recently, it has been observed that Xist is able to trigger gene silencing also in certain tumour cells (Agrelo et al, 2009). This was shown using a mouse thymic lymphoma model triggered by an oncogenic nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) fusion kinase within the T-cell compartment (Chiarle et al, 2008). In this tumour model, experimental Xist induction did cause ectopic X inactivation and could block tumour development (Agrelo et al, 2009). Hence, lymphoma cells can also display the cellular context that allows initiation of Xist-mediated gene silencing. The special AT-rich binding protein (SATB1) has been identified as an initiation factor for Xist-mediated silencing by comparing expression profiles from Xist-responsive and Xist-resistant tumours (Agrelo et al, 2009). Expression of SATB1 is restricted in development and has been proposed to characterize the cellular context for Xist-mediated silencing in ES cell differentiation and in T lymphocyte development (Agrelo et al, 2009).
SATB1 is a DNA binding protein that interacts with AT-rich DNA sequences and facilitates the organization of chromatin structure (de Belle et al, 1998; Dickinson et al, 1992). In T-cells, SATB1 forms a network which overlaps the base of chromatin loops, suggesting a topological function in gene regulation (Fig 3) (Cai et al, 2006). SATB1 has also been shown to interact with chromatin modifying complexes and to regulate histone modification and nucleosomal positioning over large regions (Yasui et al, 2002). Furthermore, it is subject to posttranslational modifications that could direct its activity (Pavan Kumar et al, 2006; Purbey et al, 2009; Tan et al, 2008). A role for SATB1 in regulating gene expression in a coordinate manner was first observed in T-cell development (Cai et al, 2006). In X inactivation SATB1 could function to organize the chromatin of genes to enable silencing by Xist (Brockdorff, 2009). Such a model is consistent with the observation that Xist initially localizes to the repetitive core of the X chromosome territory (Chaumeil et al, 2006). Genes are positioned in the periphery of the chromosome territory (Chaumeil et al, 2006), where also SATB1 is observed (Agrelo et al, 2009). These observations suggest that SATB1 is a node for signal integration on chromatin consistent with the view that it has a central role in the establishment of epigenetic patterns. Its precise expression control in development and differentiation make it an interesting marker for certain cellular contexts that are associated with epigenetic transitions and potentially, changes in cell identity.
Figure 3. SATB1 organizes the genome for coordinate gene regulation.
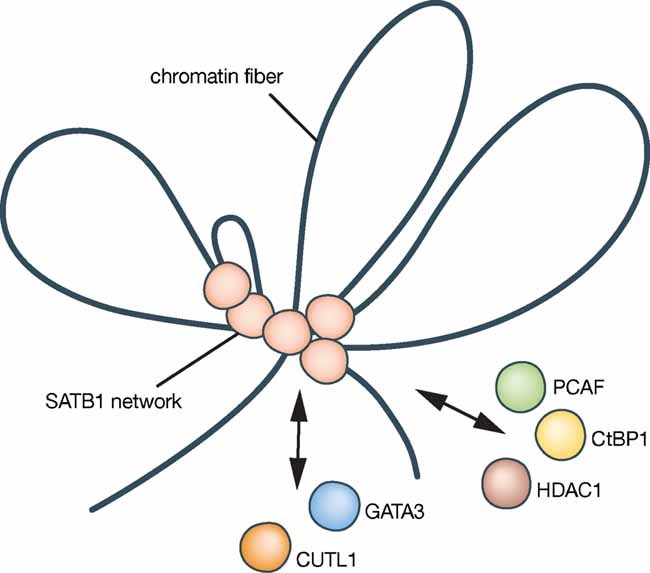
SATB1 forms a network that overlaps the base of chromatin loops and has been shown to recruit chromatin modifying complexes. SATB1 interacts with C-terminal binding protein 1 (CtBP1), histone deacetylase 1 (HDAC1), p300/CBP-associated factor (PCAF) and cut-like 1 (CUTL1) and colocalizes with GATA binding protein 3 (GATA3). The current view is that spatial and molecular organization of the genome is modulated by SATB1, which might be important for establishing epigenetic patterns and for coordinating gene expression.
An epigenetic context of progenitors?
The ability of Xist to initiate silencing is restricted to very specific cell types indicating that the underlying silencing pathways are under developmental control. In the haematopoietic system the silencing pathway becomes re-established in all haematopoietic lineages at the stage of lineage committed progenitor or precursor cells (Savarese et al, 2006). This has been explicitly shown for pre-B cells and CD4+ CD8+ double positive T-cells. Presently, it is not clear if Xist responsive cell types are exclusive to the embryo and the haematopoietic system or if other adult stem cell niches such as the gut or skin are also characterized by re-establishing an appropriate epigenetic context. However, there are indications that the Xist silencing pathway is active in a variety of tumour cells. An ectopic human XIST transgene has been found to induce chromosome inactivation in human HT-1080 fibrosarcoma cells (Hall et al, 2002). Further, XIST expression has been observed in human testicular germ cell tumours, (Kawakami et al, 2003, 2004a; Looijenga et al, 1997). This is unusual as XIST is normally not expressed in male cells. However, in these tumours multiple inactive X chromosomes seem to be present suggesting that the context for initiation of X inactivation is associated with the development of certain testicular tumours (Kawakami et al, 2003, 2004a; Looijenga et al, 1997). A recent study by Han and colleagues has further linked SATB1 expression to an aggressive and metastatic phenotype in breast cancer (Han et al, 2008). SATB1 expression conferred metastatic activity to non-aggressive breast cells suggesting that SATB1 reorganizes the genome of cancer cells to become metastatic (Han et al, 2008; Richon, 2008). These observations suggest that the context for Xist-mediated silencing might be found in a wider range of tissues and could define the biology of a class of cancer cells. This could provide an opportunity to distinguish certain tumour cells from normal stem cells and differentiated cells and be amenable to the development of new therapies (Fig 4A and B). Consistent with this, links between tumour cells and embryonic cells have been suggested previously. Similarities between gene expression patterns of ES cells and poorly differentiated aggressive forms of cancer have been noted by bioinformatic analysis of expression data for a large set of tumours (Ben-Porath et al, 2008). Although ES cell specific transcription factors do not appear to play a major role in tumourigenesis, certain gene expression signatures are shared between tumour cells and ES cells. Similarities in chromatin composition and organization between tumour cells and ES cells have also been found (Ohm et al, 2007; Tiwari et al, 2008). This supports the idea that certain pathways for chromatin regulation are shared between the epigenetic context of embryonic cells, progenitor cells and tumour cells.
Figure 4. Epigenetic context in tumourigenesis.
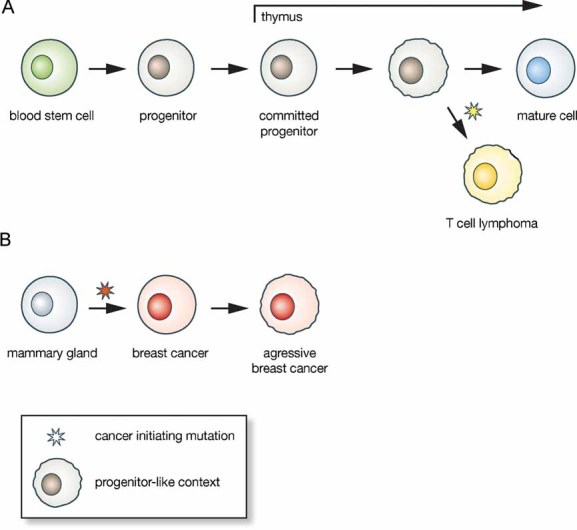
- Tumours can develop from progenitors that provide an epigenetic context for the establishment of epigenetic patterns, which can be identified by an active gene silencing pathway. Aggressive T-cell lymphomas provide an example, as they maintain properties of their T-cell progenitor origin such as SATB1 expression.
- Other tumours are thought to establish a similar cellular context when they progress to malignant disease. Breast cancer cells provide an example for this type of tumour. When breast cancers progress to a more malignant and highly metastatic phenotype, SATB1 expression can be elevated. This might go along with a reprogramming of the tumour cell transcriptome, and albeit direct evidence is still awaited, with the re-establishment of pathways for gene silencing. In breast cancer SATB1 expression is unrelated to the cellular context from which the tumour originated and is acquired at a late stage in tumourigenesis. Establishing a progenitor-like context might endow tumour cells with developmental pathways that contribute to metastasis.
X inactivation and tumourigenesis
A wider link between X inactivation and tumourigenesis has been made in a number of studies observing a gain or loss of X chromosomes in tumour cells (Table II). Testicular germ cell tumours frequently display an increase in the number of X chromosomes as well as of chromosome 12 and 17 (reviewed in Lind et al, 2007). Although evidence for inactivation of the additional X chromosomes has been obtained, more analysis is needed to fully address this point (Looijenga et al, 1997). Of note, the presence of an active unmethylated XIST promoter has been suggested as a marker for tumour diagnosis (Kawakami et al, 2004a). The presence of an undermethylated XIST promoter is also reported in prostate cancer, albeit XIST was not found expressed in this type of cancer (Laner et al, 2005). In addition, a gain of an X chromosome is also a frequent karyotypic alteration of malignant lymphomas (McDonald et al, 2000).
Table II.
Gain and loss of X chromosomes in human tumours
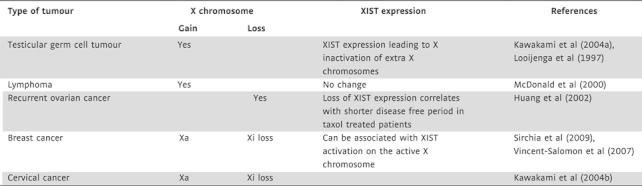 |
A correlation between the response to chemotherapy and the level of XIST expression has been observed in ovarian cancer (Huang et al, 2002). In this case, XIST was found down-regulated in recurrent tumours and the level of XIST expression was correlated with the disease-free period of cancer patients with Taxol in the therapeutic regimen (Huang et al, 2002). A likely explanation for the absence of XIST expression in recurrent ovarian cancer could be a loss of the inactive X chromosome in the tumour cells. This is consistent with a frequent Xi loss and amplification of the active X chromosome reported in a range of female tumours such as ovarian, breast and cervical cancer (Kawakami et al, 2004b). Loss of the inactive X chromosome could be a consequence of the genomic instability of cancer cells. Hence, a reduction of XIST expression might be a useful marker for disease progression in certain female tumours.
The gain of active X chromosomes is observed among all classes of breast cancer and is accompanied by a considerable heterogeneity of XIST expression within a tumour sample (Sirchia et al, 2009; Vincent-Salomon et al, 2007). Activation of XIST expression from the active X chromosome has also been observed (Sirchia et al, 2009). Xi also appears to be absent in a number of breast cancer 1 (BRCA1)-deficient breast cancer cells, which has led to the proposal that the breast and ovarian tumour suppressor BRCA1 contributes to XIST localization and X inactivation (Ganesan et al, 2002). However, more detailed analysis has shown that BRCA1 status does not correlate with XIST expression or localization (Pageau et al, 2007; Vincent-Salomon et al, 2007). X chromosome abnormalities have further been linked to the pathogenesis of sporadic basal-like breast cancers, a distinct class of breast cancer with phenotypic similarities to BRCA1-deficient breast tumours (Richardson et al, 2006). Taken together, analyses of XIST expression and X chromosome number might provide opportunities for developing diagnostic markers with strictly tumour type-specific interpretation.
Conclusions
X inactivation is important to human disease in a variety of ways. Non-random patterns of X inactivation modulate disease onset or the severity of symptoms. The gain and loss of X chromosomes and deregulation of XIST expression has been associated with certain human tumours and holds potential for development of diagnostic markers. There are further aspects of X inactivation that are relevant to human disease without direct clinical application. X inactivation is a model system for addressing basic questions in epigenetic regulation. Recent results suggest that X inactivation studies can facilitate the identification of pathways that are activated in mammals for setting up epigenetic patterns in narrow developmental windows. Detailing the mechanisms behind Xist mediated repression could help defining the cellular context permissive for epigenetic modifications and highlight differences between tumour and normal cells. While understanding the Xist-mediated gene silencing mechanism provides a potential handle on some of the key mammalian pathways for epigenetic regulation, only a restricted subset of mammalian nuclear pathways can be probed with Xist. X inactivation is specific to placental mammals (Duret et al, 2006). Its recent evolution makes it likely that other components of mammalian dosage compensation already existed before the advent of Xist. These components, which include the Polycomb system or SATB1, are evolutionary older and can be expected to be involved in a wider epigenetic regulation of development. Importantly, the pathways for Xist-mediated gene silencing are limited to very specific cellular contexts. A potential reason for restricting the activity of this silencing pathway might be to safeguard against illegitimate changes of cell identity which could lead to malignant transformation. Studies into this pathway will further our understanding of certain tumour cell contexts and, thus, could help developing new therapies.
Pending issues
Defining the existence or not of a ‘context for Xist gene silencing’ within adult stem cell systems other than the haematopoietic system.
Analysing Xist function in a wider range of tumour models to understand how general the silencing context is in tumour formation. Can tumours be classified into Xist-responsive tumours and non-responsive? Does this correlate with specific tumour properties such as metastasis?
A better understanding of the Xi chromatin structure is needed so that strategies for efficient and selective reactivation of Xi genes can be devised. Note the promising results obtained by reactivating the RETT syndrome associated MeCP2 gene.
A better understanding of the complex mechanism(s) that regulate(s) random X inactivation, e.g. how do non-random patterns of X inactivation arise?
Acknowledgments
We thank Carrie Cowan and David Keays for critically reading the manuscript. This work was supported by a grant from the Vienna Science and Technology Fund (WWTF), and by the IMP through Boehringer Ingelheim.
The authors declare that they have no conflict of interest.
References
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, et al. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell. 2009;16:507–516. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. RETT syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Anderson-Cohen M, Holland SM, Kuhns DB, Fleisher TA, Ding L, Brenner S, Malech HL, Roesler J. Severe phenotype of chronic granulomatous disease presenting in a female with a de novo mutation in gp91-phox and a non familial, extremely skewed X chromosome inactivation. Clin Immunol (Orlando, FL) 2003;109:308–317. doi: 10.1016/j.clim.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Archer H, Evans J, Leonard H, Colvin L, Ravine D, Christodoulou J, Williamson S, Charman T, Bailey ME, Sampson J, et al. Correlation between clinical severity in patients with RETT syndrome with a p.R168X or p.T158M MECP2 mutation, and the direction and degree of skewing of X-chromosome inactivation. J Med Genet. 2007;44:148–152. doi: 10.1136/jmg.2006.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- Boukarabila H, Saurin AJ, Batsche E, Mossadegh N, van Lohuizen M, Otte AP, Pradel J, Muchardt C, Sieweke M, Duprez E. The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 2009;23:1195–1206. doi: 10.1101/gad.512009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. SAT in silence. Dev Cell. 2009;16:483–484. doi: 10.1016/j.devcel.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The story of RETT syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diab Obes. 2007;14:3–12. doi: 10.1097/MED.0b013e328013daa2. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Okamoto I, Guggiari M, Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res. 2002;99:75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
- Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- de Belle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J Cell Biol. 1998;141:335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science (New York, NY) 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science (New York, NY) 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- Favier R, Lavergne JM, Costa JM, Caron C, Mazurier C, Viemont M, Delpech M, Valleix S. Unbalanced X-chromosome inactivation with a novel FVIII gene mutation resulting in severe hemophilia A in a female. Blood. 2000;96:4373–4375. [PubMed] [Google Scholar]
- Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, Mok SC, Randrianarison V, Brodie S, Salstrom J, et al. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111:393–405. doi: 10.1016/s0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Picketts DJ, Villard L, Higgs DR. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of RETT syndrome. Science (New York, NY) 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci USA. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenerhoff MJ, Chu I, Barkan D, Liu ZY, Datta S, Dimri GP, Green JE. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009;28:3022–3032. doi: 10.1038/onc.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Rao PH, Lau CC, Heard E, Ng SK, Brown C, Mok SC, Berkowitz RS, Ng SW. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol Cancer Ther. 2002;1:769–776. [PubMed] [Google Scholar]
- Huppke P, Maier EM, Warnke A, Brendel C, Laccone F, Gartner J. Very mild cases of RETT syndrome with skewed X inactivation. J Med Genet. 2006;43:814–816. doi: 10.1136/jmg.2006.042077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA. Epigenetic changes in cancer. Annu Rev Pathol. 2009;4:229–249. doi: 10.1146/annurev.pathol.3.121806.151442. [DOI] [PubMed] [Google Scholar]
- Invernizzi P, Pasini S, Selmi C, Miozzo M, Podda M. Skewing of X chromosome inactivation in autoimmunity. Autoimmunity. 2008;41:272–277. doi: 10.1080/08916930802024574. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Okamoto K, Ogawa O, Okada Y. XIST unmethylated DNA fragments in male-derived plasma as a tumour marker for testicular cancer. Lancet. 2004a;363:40–42. doi: 10.1016/S0140-6736(03)15170-7. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Okamoto K, Sugihara H, Hattori T, Reeve AE, Ogawa O, Okada Y. The roles of supernumerical X chromosomes and XIST expression in testicular germ cell tumors. J Urol. 2003;169:1546–1552. doi: 10.1097/01.ju.0000044927.23323.5a. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Zhang C, Taniguchi T, Kim CJ, Okada Y, Sugihara H, Hattori T, Reeve AE, Ogawa O, Okamoto K. Characterization of loss-of-inactive X in Klinefelter syndrome and female-derived cancer cells. Oncogene. 2004b;23:6163–6169. doi: 10.1038/sj.onc.1207808. [DOI] [PubMed] [Google Scholar]
- Keohane AM, O'Neill LP, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Laird PW, Jaenisch R. The role of DNA methylation in cancer genetic and epigenetics. Annu Rev Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- Laner T, Schulz WA, Engers R, Muller M, Florl AR. Hypomethylation of the XIST gene promoter in prostate cancer. Oncol Res. 2005;15:257–264. doi: 10.3727/096504005776404607. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Leung C, Lingbeek M, Shakhova O, Liu J, Tanger E, Saremaslani P, Van Lohuizen M, Marino S. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- Lexner MO, Bardow A, Juncker I, Jensen LG, Almer L, Kreiborg S, Hertz JM. X-linked hypohidrotic ectodermal dysplasia. Genetic and dental findings in 67 Danish patients from 19 families. Clin Genet. 2008;74:252–259. doi: 10.1111/j.1399-0004.2008.01037.x. [DOI] [PubMed] [Google Scholar]
- Liedtke M, Cleary ML. Therapeutic targeting of MLL. Blood. 2009;113:6061–6068. doi: 10.1182/blood-2008-12-197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind GE, Skotheim RI, Lothe RA. The epigenome of testicular germ cell tumors. APMIS. 2007;115:1147–1160. doi: 10.1111/j.1600-0463.2007.apm_660.xml.x. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, van Gurp RJ, Verkerk AJ, Oosterhuis JW. X inactivation in human testicular tumors. XIST expression and androgen receptor methylation status. Am J Pathol. 1997;151:581–590. [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science (New York, NY) 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- Malcolm S, Clayton-Smith J, Nichols M, Robb S, Webb T, Armour JA, Jeffreys AJ, Pembrey ME. Uniparental paternal disomy in Angelman's syndrome. Lancet. 1991;337:694–697. doi: 10.1016/0140-6736(91)90278-w. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- McDonald HL, Gascoyne RD, Horsman D, Brown CJ. Involvement of the X chromosome in non-Hodgkin lymphoma. Genes Chromosomes Cancer. 2000;28:246–257. [PubMed] [Google Scholar]
- Morleo M, Franco B. Dosage compensation of the mammalian X chromosome influences the phenotypic variability of X-linked dominant male-lethal disorders. J Med Genet. 2008;45:401–408. doi: 10.1136/jmg.2008.058305. [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knoll JH, Butler MG, Karam S, Lalande M. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader–Willi syndrome. Nature. 1989;342:281–285. doi: 10.1038/342281a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Nystrom A, Pfeifer-Ohlsson S, Tohonen V, Hedborg F, Schofield P, Flam F, Ekstrom TJ. IGF2 is parentally imprinted during human embryogenesis and in the Beckwith–Wiedemann syndrome. Nat Genet. 1993;4:94–97. doi: 10.1038/ng0593-94. [DOI] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science (New York, NY) 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Pageau GJ, Hall LL, Lawrence JB. BRCA1 does not paint the inactive X to localize XIST RNA but may contribute to broad changes in cancer that impact XIST and Xi heterochromatin. J Cell Biochem. 2007;100:835–850. doi: 10.1002/jcb.21188. [DOI] [PubMed] [Google Scholar]
- Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Purbey PK, Singh S, Notani D, Kumar PP, Limaye AS, Galande S. Acetylation-dependent interaction of SATB1 and CtBP1 mediates transcriptional repression by SATB1. Mol Cell Biol. 2009;29:1321–1337. doi: 10.1128/MCB.00822-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastan S, Robertson EJ. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol. 1985;90:379–388. [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Richon VM. A new path to the cancer epigenome. Nat Biotechnol. 2008;26:655–656. doi: 10.1038/nbt0608-655. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development (Cambridge, England) 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader–Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Sirchia SM, Tabano S, Monti L, Recalcati MP, Gariboldi M, Grati FR, Porta G, Finelli P, Radice P, Miozzo M. Misbehaviour of XIST RNA in breast cancer cells. PLoS One. 2009;4:e5559. doi: 10.1371/journal.pone.0005559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Tsao H. X-chromosome inactivation and skin disease. J Invest Dermatol. 2008;128:2753–2759. doi: 10.1038/jid.2008.145. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sugawara O, Sasaki M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- Talebizadeh Z, Bittel DC, Veatch OJ, Kibiryeva N, Butler MG. Brief report: non-random X chromosome inactivation in females with autism. J Autism Dev Disord. 2005;35:675–681. doi: 10.1007/s10803-005-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JA, Sun Y, Song J, Chen Y, Krontiris TG, Durrin LK. SUMO conjugation to the matrix attachment region-binding protein, special AT-rich sequence-binding protein-1 (SATB1), targets SATB1 to promyelocytic nuclear bodies where it undergoes caspase cleavage. J Biol Chem. 2008;283:18124–18134. doi: 10.1074/jbc.M800512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leenders GJ, Dukers D, Hessels D, van den Kieboom SW, Hulsbergen CA, Witjes JA, Otte AP, Meijer CJ, Raaphorst FM. Polycomb-group oncogenes EZH2, BMI1, and RING1 are overexpressed in prostate cancer with adverse pathologic and clinical features. Eur Urol. 2007;52:455–463. doi: 10.1016/j.eururo.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Vincent-Salomon A, Ganem-Elbaz C, Manie E, Raynal V, Sastre-Garau X, Stoppa-Lyonnet D, Stern MH, Heard E. X inactive-specific transcript RNA coating and genetic instability of the X chromosome in BRCA1 breast tumors. Cancer Res. 2007;67:5134–5140. doi: 10.1158/0008-5472.CAN-07-0465. [DOI] [PubMed] [Google Scholar]
- Wutz A. Xist function: bridging chromatin and stem cells. Trends Genet. 2007;23:457–464. doi: 10.1016/j.tig.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]


