In the early eighties a third tRNA binding site, the E site, was found in addition to the classical A and P sites; the E site seems to be a universal feature of ribosomes (for review see ref. 1). Two models exist that describe different aspects of the three sites. The hybrid-site model is based on protection experiments (2) introducing hybrid states of tRNA binding: The tRNAs move alternately on one or the other subunits through the tRNA binding sites in the course of elongation. An analysis of the functional interplay of the three ribosomal sites led to the second model, the allosteric three-site model. The key features are that the deacylated tRNA is stably bound to the E site in a codon-dependent manner and that the A site and the E site are reciprocally linked (3). Both models highlight different features of the elongation cycle and do not necessarily exclude each other.
Recently, the demise of the allosteric three-site model was proclaimed by attempting to demonstrate that (i) the tRNA binds to the E site in a labile fashion and (ii) no reciprocal linkage exists between the A and E sites (4). Concerning the first statement, the old finding was reproduced that the stability of E-site binding depends on the buffer conditions: A tRNA is stably bound to the E site and is not released upon translocation in our polyamine/Mg2+ buffer, in contrast to the buffer system without polyamines and low Mg2+ (4). Therefore, the argument concerning labile E-site binding can be reduced to the question of which buffer conditions might reflect the in vivo situation.
A thorough analysis of native polysomes derived from Escherichia coli cells demonstrated that at least 75% of the posttranslocational (POST) ribosomes carry a deacylated tRNA confirming stable binding to the E site in vivo (5). E site-specific protection of 23S rRNA is found in polysomes arguing in favor of an occupied E site (6). Also Wintermeyer and colleagues (7) reported an occupied E site in native disomes after an incubation with EF-G for 10 min at 37°C. Furthermore, codon-anticodon interaction in the E site before and after a (−1) frameshift at a so-called slippery sequence of seven nucleotides (8) requires stable E site binding in vivo. Altogether, stable binding of deacylated tRNA to the E site is a valid feature in vivo. The polyamine/Mg2+ buffer matches the corresponding ion concentrations in vivo (see ref. 9 for discussion).
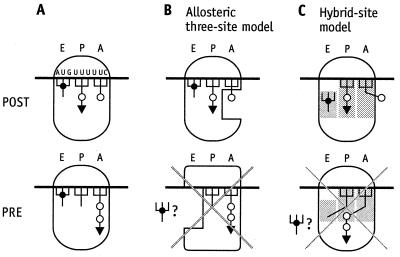
The rejection of reciprocal linkage between the A and E sites rests on figure 4 in ref. 4. A POST complex was constructed under polyamine/Mg2+ buffer conditions carrying a dipeptidyl tRNA at the P site and a deacylated tRNA at the E site, where it remained stably bound for more than 30 min and during some manipulations that removed EF-G from the assay system. At the final step a further ternary complex Phe-tRNA⋅EF-Tu⋅GTP was quantitatively bound to the A site, whereas the binding of most of the deacylated tRNA at the E site was not affected (Fig. 1A). The simultaneous binding of three tRNAs was thought to reject the allosteric three-site model.
Figure 1.
Interpretation of the experiment reported in figure 4 of ref. 4. (A) A POST complex was constructed carrying a dipeptidyl tRNA at the P site and a deacylated tRNA at the E site. At the final step a further ternary complex Phe-tRNA⋅EF-Tu⋅GTP was quantitatively bound to the A site, while most of the deacylated tRNA remains at the E site. Because the final complex has not been characterized, the following products are possible: (i) no peptide-bond formation (Upper), and (ii) peptide-bond formation leading to a tripeptidyl tRNA (Lower). (B) The POST complex carrying three tRNAs agrees with the allosteric three-site model, whereas the PRE complex does not. (C) Similarly, the corresponding POST complex is consistent with the hybrid-site model, whereas the PRE complex is not.
According to the allosteric three-site model, three tRNAs can be bound to a ribosome in the POST state during the decoding process, where a deacylated tRNA resides at the E site, a peptidyl tRNA at the P site and an aminoacyl tRNA to the low-affinity A site (Fig. 1B, Upper). Similarly, three tRNAs can be accommodated by a POST ribosome in the frame of the hybrid-site model (Fig. 1C, Upper). This complex is related (although not identical) to a ribosome that binds three deacylated tRNAs simultaneously. The first tRNA binds to the P site, the second to the E site thus establishing a POST state. Filling the A site requires the presence of a large excess of tRNA (10), indicating a low-affinity A site (9). However, only two tRNAs can be bound to pretranslocational (PRE) ribosomes according to the allosteric three-site model (Fig. 1B, Lower). The same is true for the hybrid-site model, because a peptidyl tRNA is located at the A/P site after peptide-bond formation and the deacylated tRNA at the P/E site, leaving no site for a further deacylated tRNA (Fig. 1C, Lower). A demonstration of programmed ribosomes in the PRE state carrying three tRNAs would be incompatible with both models.
The decisive question concerning whether the ribosome (Fig. 1A) carrying three tRNAs is in the PRE or POST state cannot be answered. Neither a peptide analysis (quantitative tripeptide formation would indicate a PRE state) nor a puromycin reaction after the addition of the third tRNA were performed (a negative puromycin reaction would also indicate a PRE state; ref. 4).
In vivo evidence for a reciprocal linkage between A and E sites has been presented. (i) If the tRNA is not released from the E site upon A site occupation but at a later step, e.g., the translocation reaction, three tRNAs would be present at the PRE and two at the POST ribosome. Statistically 2.4 tRNAs per ribosome would be expected in disagreement with the observed 1.9 in native polysomes (5). (ii) A disturbance of the tRNA E-site interaction caused by a tRNA mutation provokes an increase in error of the selection process at the A site in vivo (11). Such an influence of the E site on accuracy has been inferred from the reciprocal coupling of the A and E sites and demonstrated in vitro (12).
In summary, the interpretation that led to the statement that “the allosteric three-site model is untenable” (4) would refute the hybrid site model as well. However, the corresponding data cannot be interpreted conclusively.
References
- 1.Nierhaus K H. Mol Microbiol. 1993;9:661–669. doi: 10.1111/j.1365-2958.1993.tb01726.x. [DOI] [PubMed] [Google Scholar]
- 2.Moazed D, Noller F. Nature (London) 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 3.Nierhaus K H. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- 4.Semenkov Y P, Rodnina M V, Wintermeyer W. Proc Natl Acad Sci USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remme J, Margus T, Villems R, Nierhaus K H. Eur J Biochem. 1989;183:281–284. doi: 10.1111/j.1432-1033.1989.tb14925.x. [DOI] [PubMed] [Google Scholar]
- 6.Moazed D, Noller H F. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 7.Stark H, Orlova E V, Rinke-Appel J, Jünke N, Mueller F, Rodnina M, Wintermeyer W, Brimacombe R, van Heel M. Cell. 1997;88:19–28. doi: 10.1016/s0092-8674(00)81854-1. [DOI] [PubMed] [Google Scholar]
- 8.Horsfield J A, Wilson D N, Mannering S A, Adamski F M, Tate W P. Nucleic Acids Res. 1995;23:1487–1494. doi: 10.1093/nar/23.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schilling-Bartetzko S, Franceschi F, Sternbach H, Nierhaus K H. J Biol Chem. 1992;267:4693–4702. [PubMed] [Google Scholar]
- 10.Rheinberger H J, Sternbach H, Nierhaus K H. Proc Natl Acad Sci USA. 1981;78:5310–5314. doi: 10.1073/pnas.78.9.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connor M, Willis N M, Bossi L, Gesteland R F, Atkins J F. EMBO J. 1993;12:2559–2566. doi: 10.1002/j.1460-2075.1993.tb05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geigenmüller U, Nierhaus K H. EMBO J. 1990;9:4527–4533. doi: 10.1002/j.1460-2075.1990.tb07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]