Abstract
The article by McConoughey et al in the current issue of EMBO Molecular Medicine examines the contribution of transglutaminase 2 (TG2) to Huntington's disease (HD) pathogenesis. The authors find that TG2 inhibition can ameliorate HD neurodegeneration, and thereby elevate the status of transglutaminases (TGs) to a major therapeutic target—not because of their well-known activity in mutant protein aggregation, but instead based upon their ability to epigenetically modulate transcription and energy production. While the reintroduction of TG inhibition as a therapy for HD may evoke feelings of déjà vu, the outcome this time around could go in a dramatically different direction.
Keywords: epigenetics, Huntington's disease, PGC-1-alpha, transcription, transglutaminase
See related article in EMBO Mol Med (Stephen J. McConoughey et al. (2010) EMBO Mol Med 2: 349–370)
Ever since the discovery of abnormally expanded polyglutamine repeats as the molecular basis of a novel class of hereditary, late-onset neurodegenerative disorders, neurogenetics researchers have been searching for the factors that mediate the toxicity of polyglutamine (polyQ) tracts. Later called the ‘CAG-polyglutamine repeat diseases’, this class of neurological disorders includes nine diseases that are mostly very rare—with the notable exception of Huntington's disease (HD). Unfortunately, discovery of the affected genes did not immediately reveal a causative mechanism for the neuronal degeneration. In fact, most of the proteins, encoded by the mutated disease genes, lacked obvious similarity to other known proteins. Thus, the most obvious feature of all of them, the expanded polyQ repeat tract, became the subject of investigation as to the normal and pathogenic function of these proteins. In the early uncertain days of this field, transglutaminases (TGs) emerged as a logical candidate for a key role in polyQ toxicity, as TGs cross-link the γ-glutamyl with inter- or intra-molecular ε-lysine residues in adjacent polypeptides, or polyaminate glutamine residues (Cooper et al, 2000).
»…the rapid ascension of TGs as mediators of polyQ toxicity soon met with serious skepticism.«
But the rapid ascension of TGs as mediators of polyQ toxicity soon met with serious skepticism. In the mid-1990s, many TG experts concluded that expanded polyglutamines were unlikely to be good substrates for TGs, as their catalytic pocket had strict amino acid sequence requirements surrounding the modified glutamine residue (Etoh et al, 1986; Simon & Green, 1988). Nonetheless, with scant attractive alternative hypotheses on hand, in vitro experiments to investigate expanded polyglutamines as substrates for TGs advanced in earnest. And defiance and persistence paid off—as expanded polyglutamines turned out to be excellent substrates for certain TGs, given that they enhanced the reactivity of authentic substrates such as involucrin (Kahlem et al, 1996). The relationship between TGs and expanded polyglutamines soon suggested therapeutic approaches with inhibitors of TGs (e.g. cystamine), and this therapeutic approach ameliorated the toxic effects of polyQ disease proteins in cell culture and in mice (Karpuj et al, 2002). The involvement of TGs in the polyQ disorders was corroborated by evidence from patient samples, where total TG activity was elevated in brain extracts from HD patients (Karpuj et al, 1999), and an isodipeptide, a biomarker of TG activity, was elevated over threefold in HD cerebrospinal fluid compared to control (Jeitner et al, 2001). Interestingly, and relevant to the work of McConoughey et al (2010), the increased TG activity was more prominent in the cortical nuclear extracts of HD as compared to cytosolic extracts (Karpuj et al, 1999). Furthermore, initial hints that TGs are involved in both bioenergetics and transcriptional dysfunction due to expanded polyQ proteins came from two sets of studies from the same group. First, glyceraldehyde 3-phosphate dehydrogenase and the α-ketoglutarate dehydrogenase complex were inactivated when cross-linked in vitro to a heterologous protein with expanded polyQ, hence possibly contributing to disruption of cerebral energy homeostasis (Cooper et al, 1997). Second, H1 histone was identified as a suitable substrate for tissue TG (Cooper et al, 2000), suggesting that perhaps TGs participate in chromatin remodelling and gene expression regulation.
»…reshuffling and rewriting the mechanistic basis of TG involvement by implicating transglutaminase 2 (TG2) in transcription dysregulation and defective energy homeostasis in HD pathogenesis«
With the historical narrative in mind, readers of the McConoughey et al (2010) paper may thus initially experience a sense of déjà vu at the return of TGs to the HD stage; however, the authors are actually reshuffling and rewriting the mechanistic basis of TG involvement by implicating transglutaminase 2 (TG2) in transcription dysregulation and defective energy homeostasis in HD pathogenesis. First, they show that TG2, which cross-links the mutant huntingtin (htt) protein, and is elevated in HD cortex and striatum (Zainelli et al, 2005), polyaminates the N-terminal tail of histone 3 (H3). Polyamination of the H3 N-terminal tail increases its positive charge and its propensity to more tightly interact with negatively charged deoxyribonucleic acid (DNA), and hence participate in facultative heterochromatin formation. Thus, TG2 hyperactivity could dysregulate transcription. In support of this thesis, when the authors chemically inhibited TG2 in ST-Hdh Q111/Q111 striatal-like neurons derived from knock-in HD mice expressing full-length mutant htt protein, they found that 196 out of 461 dysregulated genes (i.e. 42%) shifted back toward normal levels by at least 25%. Second, using a chromatin immunoprecipitation (ChIP) assay, they showed that in ST-Hdh Q111/Q111 striatal-like neurons, TG2 excessively occupies the promoter/enhancer regions of two genes essential for energy production: peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) and cytochrome c. Based upon transcription from a cytochrome c promoter–reporter construct, small interfering RNA against TG2, mouse embryonic fibroblast TG2 knock-out cell line and a host of other corroborating experiments, the promoter occupation was found to be associated with transcriptional repression of these two genes. The importance of PGC-1α to HD pathogenesis was established by a set of studies four years ago (Cui et al, 2006; Weydt et al, 2006), and remains an important therapeutic target for this disease. In the current work, McConoughey et al (2010) thus throw another twist into the mechanism of bioenergetics deficiency mediated by transcriptional dysregulation of PGC-1α and its downstream target cytochrome c by connecting pathologically elevated TG2 cross-linking activity, epigenetic transcriptional dysregulation of bioenergetics genes and mitochondrial biosynthetic deficiency (Fig 1). Equally remarkable, in the process, they introduce us to a TG2 peptide inhibitor Z-DON-Gln-Ile-Val-OMe (ZDON) that could be a promising lead compound for development of central nervous system (CNS) drugs that would reduce the deleterious effects of TG2 hyperactivity in HD. Their findings shed light on the basis of metabolic and bioenergetics abnormalities in HD, reemphasizing the need to direct translational efforts to these questions. Indeed, as the authors point out, future studies aimed at delineating the relative contribution of PGC-1α vis-à-vis TG2 inhibition should occur in parallel to determine how best to strategize therapy development for HD going forward.
Figure 1. Model for action of transglutamine 2 in Huntington's disease.
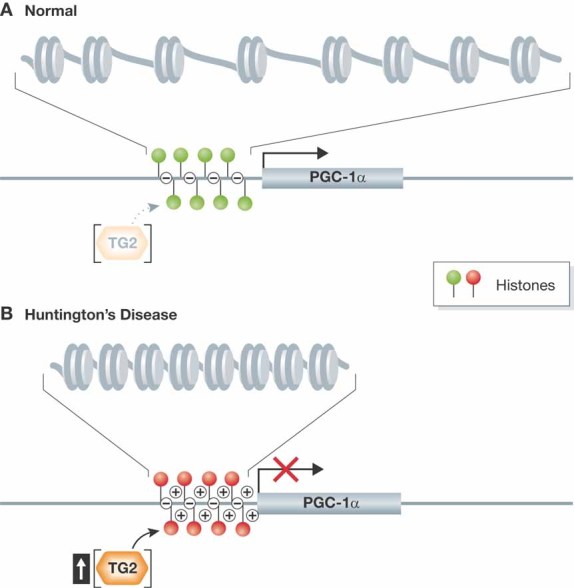
- Under normal conditions, transglutaminase 2 (TG2) activity is at baseline and does not interfere with transcription of key genes involved in the regulation of mitochondrial and metabolic functions, such as PGC-1α.
- In Huntington's disease, TG2 activity increases, resulting in polyamination of histones yielding an increased net positive charge that promotes tighter packing of DNA with histones. This alteration of chromatin structure can repress the transcription of target genes. Reduced expression from PGC-1α and related genes thus contributes to the mitochondrial and metabolic dysfunction characteristic of Huntington's disease.
»…introduce us to a TG2 peptide inhibitor (ZDON) that could be a promising lead compound for development of CNS drugs«
Acknowledgments
The authors' research on Huntington's disease is supported by R01 NS65874 and the CHDI.
The authors declare that they have no conflict of interest.
References
- Cooper AJ, et al. Proc Natl Acad Sci USA. 1997;94:12604–12609. doi: 10.1073/pnas.94.23.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AJ, et al. Dev Neurosci. 2000;22:404–417. doi: 10.1159/000017470. [DOI] [PubMed] [Google Scholar]
- Cui L, et al. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Etoh Y, et al. Biochem Biophys Res Commun. 1986;136:51–56. doi: 10.1016/0006-291x(86)90875-2. [DOI] [PubMed] [Google Scholar]
- Jeitner TM, et al. J Neurochem. 2001;79:1109–1112. doi: 10.1046/j.1471-4159.2001.00673.x. [DOI] [PubMed] [Google Scholar]
- Kahlem P, et al. Proc Natl Acad Sci USA. 1996;93:14580–14585. doi: 10.1073/pnas.93.25.14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpuj MV, et al. Proc Natl Acad Sci USA. 1999;96:7388–7393. doi: 10.1073/pnas.96.13.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpuj MV, et al. Nat Med. 2002;8:143–149. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- McConoughey SJ, et al. Embo Mol Med. 2010;2 doi: 10.1002/emmm.201000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, et al. J Biol Chem. 1988;263:18093–18098. [PubMed] [Google Scholar]
- Weydt P, et al. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Zainelli GM, et al. J Neuropathol Exp Neurol. 2005;64:58–65. doi: 10.1093/jnen/64.1.58. [DOI] [PubMed] [Google Scholar]


