Abstract
The epithelial apical membrane Na+/H+ exchangers (NHE2, NHE3) and Cl−/HCO3− exchangers (DRA and PAT-1) are key luminal membrane transporters involved in electroneutral NaCl absorption in the mammalian intestine. During the past decade, there has been a surge of studies focusing on short-term regulation of these electrolyte transporters particularly for NHE3 regulation. However, the long-term regulation of the electrolyte transporters involving transcriptional mechanisms and transcription factors that govern their basal regulation or dysregulation in diseased states has only now started to unfold with the cloning and characterization of their gene promoters. This review updates the readers with a detailed analysis of the core promoters of NHE2, NHE3, DRA and PAT-1 and outlines the transcription factors involved in their basal regulation as well as in response to both physiological (butyrate, protein kinases, probiotics) and pathophysioloical (cytokines and high levels of serotonin) stimuli. The available information on the transcriptional regulation of the recently identified NHE8 isoform is also highlighted. This review, therefore, bridges a gap in our knowledge of the transcriptional mechanisms underlying the alterations in the gene expression of intestinal epithelial luminal membrane Na+ and Cl− transporters involved in electroneutral NaCl absorption. An understanding of the mechanisms of modulation of gene expression of these transporters is important for a better assessment of the pathophysiology of diarrhea associated with inflammatory and infectious diseases and may aid in designing better management protocols.
Keywords: Na+/H+ exchangers, Cl−/HCO3− exchangers, NHE2, NHE3, NHE8, DRA, PAT1, Gene expression, Transcription factors
INTRODUCTION
Electroneutral NaCl absorption is the predominant mechanism underlying Na+, Cl− and fluid absorption in the ileum and colon of the mammalian gastrointestinal tract and involves coupling of Na+-H+ and Cl−-HCO3− exchangers [1–3]. The molecular identity of the proteins involved in this process has only recently become apparent. It is now well established that the luminal membrane sodium hydrogen exchange is mediated via SLC9A2 (NHE2) and SLC9A3 (NHE3) of the SLC9 gene family while the luminal membrane Cl−-HCO3− exchange is mediated by SLC26A3 (also termed as DRA, Down Regulated in Adenoma) and SLC26A6 (also termed as PAT1, Putative Anion Transporter 1) of the SLC26 gene family [3–6]. Recent studies have also identified the presence of NHE8 as another luminal membrane NHE of intestinal epithelial cells suggested to be important during early life [7]. These proteins specifically NHE2, NHE3, PAT1 and DRA have been well characterized for their functional roles as well as their short-term regulation utilizing in vitro studies [8–13] and knockout mouse models [14–19]. Since the normal function of these transporters is vital to the intestinal fluid absorption and disturbances in their function and expression have been associated with pathophysiology of diarrheal diseases, it is important to understand the molecular basis of the regulation of their gene expression in health and disease. Understanding the long-term regulation of the gene expression of these transporters is also necessary to gain insights into the mechanisms underlying tissue specific and the intestinal region and vertical axis (crypt/villus) expression of these genes. In this regard, the knowledge about the regulation of the gene expression of these important transporters has started to emerge only very recently. To date, however, no comprehensive review detailing the mechanisms of transcriptional regulation of these transporters is available. This review, therefore, focuses on advances in the promoter characterization as well as the mechanisms underlying transcriptional regulation of these transporters under basal conditions and their modulation by physiological and pathophysiological agents.
NA+/H+ EXCHANGERS (NHES)
Members of the Na+/H+ exchange (NHE) gene family are the key transporters involved in the predominant Na+ absorptive pathway in the ileum and proximal colon by mediating electroneutral transport of an extracellular Na+ for a cytosolic H+. Of the nine members of the NHE gene family identified to date, NHE1, NHE2, NHE3, and NHE8 are expressed in the mammalian intestinal tract [20, 21]. NHE1 is localized to the basolateral membrane of intestinal epithelial cells (IECs) where it serves housekeeping functions such as regulation of pHi and cell volume homeostasis [22]. NHE2 and NHE3 are localized to the apical membrane and play important roles in intestinal Na+ absorption with varying activities in different segments of the GI tract [23]. For example, NHE2 is predominantly expressed in the colon while NHE3 is in the ileum [14, 20, 23]. The importance of NHE3 in Na+ absorption and water homeostasis in the intestine became evident from NHE3 knockout mice, which developed mild diarrhea, low blood pressure and mild metabolic acidosis [24]. Surprisingly, NHE2 knockout mice exhibited no apparent abnormality in intestinal Na+ absorption [16, 25]. NHE8, the recently cloned isoform [21, 26] has been localized to the brush border membranes in both the kidney and IECs [21, 27]. NHE8 is differentially expressed along the human gastrointestinal tract from stomach to colon [28]. However, its functional role in Na+ absorption in adult intestine is not clear at the present.
Studies during the past 2 decades have implicated dysregulation of Na+ absorption and NHE3 down-regulation in various diarrheal disorders including inflammatory bowel diseases (IBD) [29, 30]. The majority of the studies on NHE regulation have focused on the short-term regulation generating a wealth of information on post-translational events of NHE3 regulation involving protein phosphorylation, trafficking, protein turnover, signal transduction pathways, and interaction with other regulatory proteins [8, 9]. However, the long–term regulation afforded by transcriptional mechanisms provides basis for adaptation, and is relevant to disease states or disease intervention as well as understanding the basis for differential expression of NHE isoforms along the length of the intestine. This section highlights our current understanding of the transcriptional mechanisms that impact the expression of NHE2, NHE3 and NHE8, which are expressed on the apical membrane of intestinal epithelial cells. The cloning and characterization of NHE2, 3, and 8 promoters have advanced our knowledge of transcriptional regulation and have provided critical insights into NHE gene regulation.
NHE2
NHE2 Gene
The human NHE2 (hNHE2), encoded by a single copy gene located on chromosome 2q11.2, is composed of 11 introns and 12 exons and occupies ~90 kb of the genome [31]. NHE2 mRNA is expressed at high levels in skeletal muscle, colon, and kidney and at low levels in the testis, prostate, ovary and small intestine, suggesting its tissue-specific expression [32]. The protein product encoded by hNHE2 gene is 812 amino acids long and when expressed in an in vitro cell-free system shows a band of ~75 kDa [32], representing the unglycosylated immature form, while the mature form expressed in human intestinal C2BBE1 cells is of ~85 kDa [33].
NHE2 Promoter
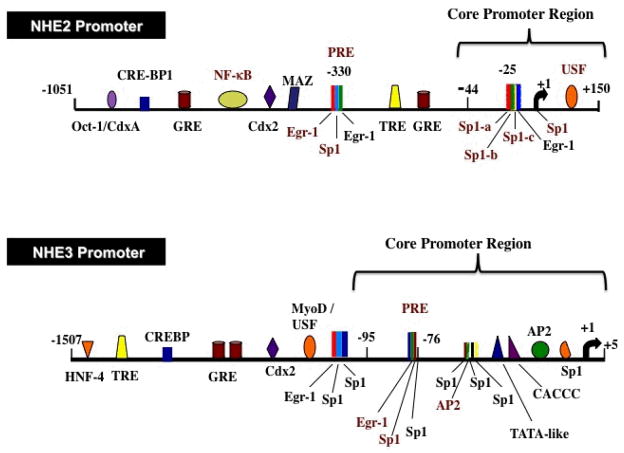
To date human and rat NHE2 promoters have been cloned [31, 34]. Transcription initiation site (TIS) of hNHE2 was localized to the adenine residue 316 nucleotides upstream from the ATG translation start codon. Comparison of the nucleotide sequences of the cloned region with the promoter databases revealed the presence of numerous potential transcription factor-binding sites including Sp1, Egr-1, AP-2, GATA, NFκB, Oct-1, Cdx-2 and a number of half-sites for glucocorticoid and thyroid hormone receptors, suggesting that regulation of this gene involves a complex array of regulatory factors [31] (Figure 1). The human and rat NHE2 promoter sequences exhibited an overall 59% sequence homology with several conserved binding sites for transcription factors AP-2, Sp1, Egr-1, CACCC, NFκB and Oct-1 [31]. There were also unique motifs for other transcription factor binding sites suggesting that distinct mechanisms may also be involved in regulation of these orthologs [31]. The human and rat NHE2 promoters both lack TATA- and CAAT-boxes. As typical of all other TATA-less and CAAT-less promoters, the NHE2 promoter is highly GC-rich and contains multiple GC-boxes. These motifs are targeted by Sp1 transcription factor family members as well as other transcription factors that show high affinity for GC-rich sequences, such as Egr-1, Krüpple-like factors, ZBP-89 and MAZ [35, 36].
Figure 1. Schematics of the human NHE2 and NHE3 promoters.
Predicted protein binding sites are indicated. Experimentally verified functional protein binding sites are shown in red text.
NHE2 Core Promoter Region
Functional characterization of 5′-deletion constructs of the hNHE2 promoter localized the core promoter region to nucleotides −40 to +150 bp (Figure 1), suggesting that cis-elements required for maximal basal transcriptional activity are located in this region. Further 5′-deletions, which caused 70% reduction in promoter activity, suggested the critical role of GC-elements contained in the deleted region (−40 to −20 bp) in promoter regulation. EMSA and ChIP assays demonstrated Sp1 and Sp3 transcription factors to bind to GC-element centered at −25 bp region [37, 38]. Competition and mutational analyses identified the Sp1-b and Sp1-c binding sites to be essential for interaction with Sp1 and Sp3 and also suggested a cooperative role between Sp1-b/Sp1-c and downstream sequences for maximal NHE2 promoter activity [38] (Figure 1). The Initiator (Inr) element, a functional analog of TATA-box, in conjunction with or without a TATA-box can promote transcription initiation [39]. The sequences surrounding NHE2 transcription initiation site exhibit a weak homology to Inr consensus sequence [40] and were found to interact with transcription factors Sp1 and Sp3 [38]. In addition to Sp1 [14], other proteins such as USF1 [41], YY1 [42] may also interact with the Inr element and enhance transcription. A USF binding site is present in the 5′-UTR of the hNHE2 promoter. Deletion of this site diminished basal NHE2 promoter activity [38].
Regulation of NHE2 by Sp1 and Sp3
Sp1 binds with high affinity to consensus GC-box elements 5′-G/TGGGCGGG/AG/AC/T-3′ [43, 44]. Consistent with the involvement of Sp1 and Sp3 in the regulation of basal NHE2 transcription, multiple binding sites for Sp1 have been identified in both the human and rat NHE2 promoters [31, 34, 38, 45, 46]. The importance of Sp1 and Sp3 in promoter activity of NHE2 was also shown in cotransfection assays in Drosophila SL2 cells, which are devoid of endogenous Sp1 related factors [47]. Both Sp1 and Sp3 were capable of trans-activating NHE2 promoter and Sp3 stimulated NHE2 to a greater extent than Sp1 [38].
In contrast to the stimulatory effects of Sp1 or Sp3 on hNHE2 promoter in intestinal epithelial cells, Sp3 inhibited while Sp1 stimulated rNHE2 transcription in renal cells [45]. While the rNHE2 minimal promoter (bp −36/+116) was actively expressed in the mouse renal cell line mIMCD-3, it was insufficient to drive transcription in either the rat intestinal epithelial cells (RIE) or the human intestinal cell line (Caco-2) [46]. A larger rNHE2 promoter fragment (−69/+116) was required for basal promoter activity in RIE cells. Similar to hNHE2 promoter, both Sp1 and Sp3 displayed positive effects on rNHE2 promoter activity in intestinal epithelial cells, whereby, Sp3 exhibited higher transactivation potential than Sp1. The lack of promoter activity of the rNHE2 −36/+116 bp region in intestinal epithelial cells may stem from cell-specific expression (mIMCD-3 vs. human intestinal C2BBE1 cells), as species differences did not appear to be involved. Regulation of the human and rat NHE genes by Sp1/Sp3 appears to be mediated through distinct response elements [38, 46]. This is evident by functional diversity of Sp1 binding sites in the human −40 to +150 bp and rat −36 to +116 bp promoter region. Furthermore, the rat Sp1 elements at −69 to −36 bp, which are responsible for its expression in the intestinal epithelial cells are not conserved in the human NHE2 promoter sequence [31]. Based on the current data, the basal regulation of NHE2 appears to involve various Sp1 binding sites.
NHE3
NHE3 Gene
The human NHE3 gene, located on chromosome 5p15.3 [48], is expressed in a tissue-specific manner. Northern blot analysis detected hNHE3 mRNA in decreasing amounts in following order: kidney > small intestine > testes > ovary > colon ≥ prostate > thymus > peripheral leukocyte ≥ brain > spleen > placenta. The cDNA encodes for a protein of 834 amino acids with calculated molecular mass of 92,906 [49]. A single transcription initiation site (TIS) was identified 116 nucleotides upstream from the hNHE3 translation start site [50]. The genomic organization of the hNHE3 gene is not fully characterized. However, in silico analysis of hNHE3 cDNA sequence revealed that the hNHE3 gene spans >50 kb of the chromosome 5 and contains 17 exons and 16 introns (Malakooti, unpublished data). The organization of exons and introns of the hNHE3 gene is similar to the reported rNHE3 gene [51, 52]. The genomic organization of NHE8 has not been defined.
NHE3 Promoter
hNHE3 promoter was characterized by cloning a ~3.2 kb human genomic DNA spanning from −3117 to +131 bp [50]. The nucleotide sequence of 1.5 kb of the 5′-flanking region from (TIS), which contained an atypical TATA-box and multiple GC-elements that contained the overlapping binding sites for Sp1 as well as Egr-1, AP-2 and CACCC [50] Further upstream, putative binding sites with relevance to the regulation of intestinal genes or tissue-specific and developmentally controlled genes, such as Cdx-2, Sp1, AP-2, USF, MyoD, cAMP-response element binding site (CREB), VDR, T3R, half-sites for GRE, and CACCC were identified [50] (Figure 1).
The rNHE3 promoter was cloned by two different groups and potential transcription factor binding sites were reported [51, 52]. A comparison of the nucleotide sequences of the human and rat NHE3 promoters revealed an overall sequence homology of 38%, whereas, significant conservation of the nucleotide sequences (79%) and transcription factor binding sites were found within the proximal promoter region [50]. Both the human and rat NHE3 promoter regions lack an authentic TATA-box and CAAT-box in close proximity of the TIS. Furthermore, this region is highly GC-rich in both promoters, which is typical of many developmentally and differentially regulated gene promoters [39]. The consensus Sp1, AP-2, CACCC, and the atypical TATA-like sequences appeared at the same positions in both species in this region.
NHE3 Core Promoter
Deletion analyses of the hNHE3 promoter-reporter constructs and expression in C2BBE1 cells identified the core promoter region that extends from −88/+5 bp and confers maximal promoter activity [50, 53]. The promoter activity was reduced by 60% with further deletion to −76 bp. The sequences downstream from −76 bp represents the minimal promoter, which confers ~40% of the maximal basal promoter activity [50, 53] (Figure 1).
Regulation of NHE3 by Sp1 and Sp3
The basal activity of the hNHE3 promoter is driven by the functional overlapping Egr-1/Sp1 motif contained within the −89/−69 bp region and overlapping Sp1/AP-2/Sp1 sequences at −72/−49 bp. Deletion or disruption of these motifs significantly decreased the promoter activity [50, 53]. DNA-binding assays demonstrated that the Egr-1/Sp1 element at −89/−69 bp binds to Sp1 and Sp3 at basal growth conditions. This motif in conjunction with downstream cis-elements confers maximal transcriptional activity to the hNHE3 promoter. This site is of central importance to the basal as well as regulated expression of NHE3 promoter [50, 53–56].
The Sp1/AP-2/Sp1 element at −72/−49 bp of the hNHE3 promoter was shown to bind AP-2 [50]. The critical role of Sp1 and Sp3 in the basal expression of hNHE3 was demonstrated by siRNA knockdown of these transcription factors in C2BBE1 cells that caused dramatic decrease in the basal hNHE3 promoter activity [54]. Further, transactivation of hNHE3 promoter by Sp1 and Sp3 was confirmed by exogenous overexpression of Sp1 and Sp3 in SL2 cells [53]. Kiela et al. [57] investigated the mechanism of rNHE3 gene regulation in transiently transfected Caco-2, IEC-6, QT6, and Drosophila SL2 cells. Deletion and mutational analysis showed that the atypical TATA element in the proximal promoter region did not play a role in the transcriptional activation of rNHE3 promoter [57]. Three potential Sp1 motifs in promoter region downstream from −81 bp were shown to be critical for rNHE3 promoter activity. It was also shown that Sp1 and Sp3 interact synergistically with a GATA-5 site in the exon-1 leading to enhanced promoter activity, which may be important for a gradient of intestinal NHE3 expression along the crypt-villus axis [57]. The hNHE3 promoter lacks the GATA motif in the 5′-untranslated region (5′-UTR). However, exon-1 sequence harbors two sets of direct repeat elements [50] and a 7-amino acid long minicistron [49]. These elements could be involved in downregulation of NHE3 gene expression, as deletion of this 5′-UTR resulted in a moderate increase in hNHE3 promoter activity [50].
NHE3 expression in cultured cells has been shown to be increased during cell confluence in a STAT-3-dependent manner, which appeared to be critical for dome formation in polarized epithelial cells [58]. Furthermore, a cooperative interaction between STAT-3 and Sp1/Sp3 has been shown in this effect of STAT3 on NHE3 expression [58]. This further emphasizes the importance of Sp1/Sp3 in NHE3 expression during cell confluence in polarized epithelial cells.
NHE8
NHE8, the newly identified apical membrane Na+/H+ exchanger, which was initially cloned from mouse kidney cDNA library [26]. The rat NHE8 cDNA encodes for a protein of 575 amino acids residues. The protein product of this gene was detected at 65 kDa in BBM preparations from the rat jujenal mucosa by Western blot analysis [21, 59, 60]. The genomic organization of NHE8 gene has not been characterized.
NHE8 is present intracellularly in most cells [61], but also in the apical membrane of epithelial cells of intestine [21, 59, 60] and kidney [26], serving as Na+/H+ exchanger [28]. Contrary to the expression levels of NHE2 and NHE3 during development, NHE8 mRNA expression was higher in 2–3 -week old rats and lower in adult rats [21, 59, 60]. This suggests an important role of NHE8 in the early stages of development.
NHE8 Promoter
The human NHE8 promoter was recently cloned [62]. The transcription initiation site was mapped to a Guanine residue 95 bp upstream from the translation initiation codon by 5′-RACE analysis. Characterization of the promoter region localized the minimal promoter to a fragment spanning −32 /+17 bp with respect to TIS, which interacts with Sp3 [62].
MODULATION OF NHE GENE EXPRESSION
The following sections highlight the mechanisms underlying the modulation of NHE gene expression by physiological and pathophysiological agents (Table 1).
Table 1.
Modulation of NHE2, 3, 8, DRA and PAT-1 Expression by Various Stimuli.
| Stimuli | NHE2 | NHE3 | NHE8 | DRA | PAT-1 |
|---|---|---|---|---|---|
| Butyrate | NE (67) | ↑ (55,67,68,69) | ND | ↑ (103) | ND |
| EGF | ↑ (33,75) | NE (75) | ↓ (76) | ND | ND |
| Glucocorticoids | NE (70) | ↑ (51,52,70) | ↓ (74) | ND | ND |
| PMA | ↑ (37) | ↑ (53) | ND | ND | ND |
| TNF | ↓ (33) | ↓ (54) | ↓ (62) | ND | ND |
| IFN-γ | ↓ (81) | ↓ (54,81) | ND | ↓ (100) | ↓ (101) |
| IL1-β | ND | ND | ND | ↓ (96) | ND |
| Serotonin | ND | ↓ (56) | ND | ND | ND |
| L. acidophilus | ND | ND | ND | ↑ (102) | NE |
ND: Not determined, NE: No effect. Arrows indicate the direction of change. Numbers in parentheses indicate the reference numbers.
Physiological Stimuli
Butyrate
Butyrate, a short chain fatty acid, is produced in the colon by microbial fermentation of dietary fiber. Butyrate exerts diverse effects on cellular functions and has been shown to regulate cell proliferation, differentiation, and apoptosis [63]. Butyrate is also implicated in suppressing inflammation and carcinogenesis [64]. It enhances the colonic NaCl absorption [65, 66], and butyrate-dependent increase in Na+ absorption was demonstrated to be mediated by both NHE2 and NHE3 in rat distal colon [66]. Musch et al. [67], demonstrated that butyrate enhanced NHE3 activity in C2BBE1 cells via elevated expression of NHE3 mRNA and protein. Further, in vivo studies with fiber-supplemented rats for 2-days displayed increased 22Na+-uptake in colon secondary to enhanced NHE3 mRNA and protein levels [67]. These findings indicated transcriptional activation of NHE3 by butyrate as the likely mechanism for NHE3 response to butyrate. Amin et al. [55], demonstrated that butyrate-induced enhancement of hNHE3 mRNA expression and promoter activity in C2BBE1 cells required Sp1 and Sp3 binding to the NHE3 core promoter region (−89/−69 bp). siRNA silencing and studies utilizing pharmacological inhibitors of specific protein phosphatases further suggested that dephosphorylation of Sp1/Sp3 in response to butyrate may play a role in their enhanced binding affinity to the hNHE3 promoter and stimulation of NHE3 by sodium butyrate.
Sodium butyrate also activates rNHE3 gene promoter via a ser/thr kinase-dependent mechanism [68] and involved response elements within −320/−34 bp of the rNHE3 promoter. Subsequent studies identified Sp1 binding sites at −58/−55 bp as the critical cis-elements for sodium butyrate induced rNHE3 promoter activity. Sp1 and Sp3 were found to interact with this motif at basal growth conditions, however, DAPA studies with butyrate treated nuclear proteins showed decreased Sp1 and increased Sp3 protein co-precipitation with this motif. Furthermore, butyrate treatment led to increased phosphorylation of Sp1 and acetylation of Sp3, with the later suggested to be involved in mediating the effects of butyrate on the rNHE3 promoter activity [69]. To date, no detailed studies are available on the effects of butyrate on the NHE2 or NHE8 gene expression.
Glucocorticoids
Glucocorticoids increase intestinal salt and water absorption and increase NHE3 activity and mRNA abundance in the intestine and kidney [70, 71]. Methylprednisolone, a commonly used treatment for inflammatory bowel diseases, has been shown to reverse the inhibition of electrolyte and nutrient transport in animal models of IBD or patients with Crohn’s disease and ulcerative colitis [72, 73]). Putative cis-elements for half-sites for GRE, were identified in the 5′-flanking region of hNHE3 promoter [50]. These glucocorticoid response elements (GRE) were also found in the rNHE3 promoter and dexamethasone, a synthetic glucocorticoid, was shown to activate rNHE3 promoter in transiently transfected OK and LLC-PK1 renal cells [51, 52]. Studies by Yun et al. [70], showed that long-term treatment of rabbits with methylprednisolone resulted in enhanced NHE3 mRNA levels in the ileum, whereas NHE1 and NHE2 expression did not change [70]. These data suggested that glucocorticoid effects on NHE3 was mediated by a transcriptional mechanism.
In contrast, recent studies showed that NHE8 expression was decreased in response to glucocorticoids in jejunal and ileal mucosa from rats and in Caco-2 cells [74]. Functional analyses of the hNHE8 promoter in Caco-2 cells localized the GRE to −89/−32 bp region. Site-directed mutagenesis and EMSA demonstrated that Pax-5, a member of the paired box family of transcription factors, interacts with the promoter region at −75/−50 bp and represses the promoter activity [74].
Epidermal Growth Factor
EGF administration to rats has been demonstrated to increase NHE2 mRNA in jejunal mucosa and also enhance NHE2 activity in BBMV prepared from rat jejunum. [75]. Similar results were obtained in vitro in the rat intestinal epithelial cells (RIE). Furthermore, actinomycin D, a transcription inhibitor, blocked not only NHE2 mRNA expression, but also EGF-induced NHE2 activity indicating that a transcriptional mechanism is likely responsible for EGF effect on NHE2 activity. Consistently, EGF also enhanced rNHE2 promoter reporter activity in transiently transfected RIE cells [75]. Similarly, the hNHE2 transcription was also stimulated by EGF [33]. In contrast, EGF reduced the hNHE8 mRNA expression and promoter activity in Caco-2 cells [76]. EGF-mediated repression of NHE8 promoter activity in Caco-2 cells was shown to involve MAP kinases ERK1/2 and Sp3 DNA-binding.
Phorbol 12-myristate 13-acetate
PMA, a diacylglycerol analogue, modulates diverse cellular functions through stimulation of signaling pathways such as protein kinase C (PKC) and mitogen-activated protein kinases (MAPK). Long-term exposure to low concentration of PMA was shown to stimulate the expression of both NHE2 and NHE3 mRNA and enhance their promoter activity in C2BBE1 cells [37, 53]. Functional analyses and DNA-binding assays showed localization of the PMA response elements (PRE) to the −339/ 324 bp region of NHE2 and −89/ 69 bp of NHE3 promoters (Figure 1). The PRE, 5′-GCGGGGGCGGG-3′, were composed of overlapping binding sites for Egr-1 and Sp1 transcription factors, which were also identified in the promoters of other PMA target genes [77, 78]. DNA binding assays showed Sp1 and Sp3 in the basal growth conditions and Egr-1 in PMA-treated nuclear extract interact with PRE of both NHE2 and NHE3 promoters [37, 53]. Binding of PMA-induced Egr-1 to the NHE2 and NHE3 PREs eliminated the Sp1/Sp3 interactions with these motifs indicating that newly synthesized Egr-1 could displace the prebound Sp1/Sp3 and interact with the overlapping Egr-1/Sp1 motif. Egr-1 overexpression studies showed that Egr-1 was sufficient to induce NHE2 and NHE3 promoter activities. siRNA targeted to Egr-1 diminished the stimulatory effects of PMA on NHE3 mRNA expression [37, 53].
Pathophysiological Stimuli
Inflammatory mediators
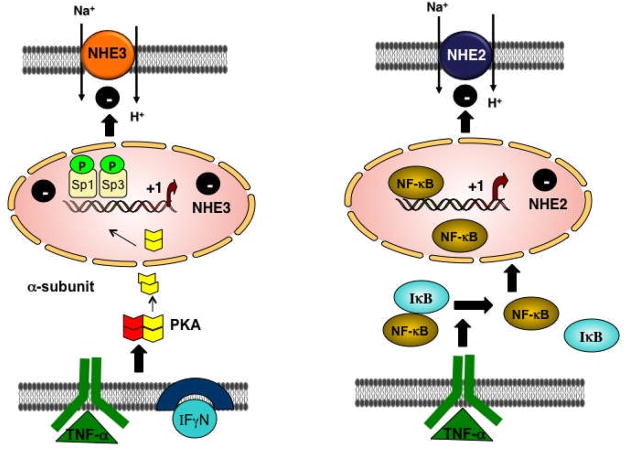
The production of various proinflammatory cytokines such as IFNγ, TNFα, and IL-1β in the intestine is one of the major contributing factors to the pathogenesis of inflammation-associated diarrhea in IBD. The expression and activity of NHE3 has been shown to be suppressed by colonic inflammation, [79, 80] and proinflammatory mediators TNFα and IFNγ [81]. Rocha et al. [81] demonstrated that both in rats and C2BBE1 cells, the expression and activity of NHE2 and NHE3 were downregulated by IFNγ. Recently, it has been shown that simultaneous addition of both TNFα and IFNγ resulted in significant reduction of NHE3 promoter activity that involved the 95/+5 bp region of the NHE3 promoter [54]. TNFα/IFNγ treatment led to diminished binding of Sp1/Sp3 to the Egr-1/Sp1 motif at −89/−69 bp, while the level of Sp1/Sp3 protein expression was not altered. Sp1/Sp3 binding to NHE3 proximal promoter region was also confirmed by ChIP assays [54]. The inhibitory effect of TNFα/IFNγ on the NHE3 promoter was completely blocked by inhibitors of PKA, implicating cytokine-dependent activation of PKA in the repression of NHE3 [54]. Sp1 is known to be phosphorylated by different protein kinases including PKA [79] and dephosphorylated by protein phosphatases [82]. In this study, binding affinity of Sp1 and Sp3 proteins for NHE3 promoter region was found to be significantly decreased after in vitro phosphorylation of nuclear proteins by α-catalytic subunit of PKA [54]. These results suggested that Sp1/Sp3 were essential for NHE3 promoter activity and their phosphorylation by PKA may be involved in repression of NHE3 transcription by cytokines (Figure 2). Along the same lines, butyrate was found to restore the cytokine-induced inhibition of the NHE3 transcriptional activity and mRNA expression [55]. These findings are important as restoration of the NHE3 function in conditions associated with decreased NHE3 activity could have therapeutic relevance if defective Na+ absorptive process could be corrected. A recent study by Yeruva et al. [83], found no significant difference in NHE3 mRNA and protein abundance, but NHE3 function was reduced in UC patients. However, this is in contrast to earlier studies showing decreased NHE3 mRNA and protein levels in UC [84]. These data suggest that transcriptional regulation, in part, may be responsible for impaired Na+ absorption in IBD.
Figure 2. Potential mechanisms regulating the human NHE2 and NHE3 expression by pro-inflammatory cytokines in C2BBE1 cells: NHE2.
TNFα-mediated degradation of IκB-αin the cytoplasm and subsequent activation and translocation of NFκB to the nucleus results in its interaction with the NHE2 promoter. This leads to repressed NHE2 transcription and reduced levels of NHE2 mRNA, protein and function. NHE3. A combination of IFNγ/TNFαrepresses the NHE3 gene expression by PKA activation and translocation of its catalytic subunit to the nucleus where it phosphorylates the Sp1/Sp3. As a result, binding of Sp1/Sp3 to the promoter region is decreased leading to reduced NHE3 transcription, mRNA and protein expression and its function.
The NHE2 mRNA and protein levels as well as its activity is repressed by long-term exposure to IFNγ [81] in the rat intestine and C2BBE1 cells and TNFα [33] in C2BBE1 cells. Studies utilizing actinomycin D plus TNFα further confirmed transcriptional regulation of NHE2 [33]. Consistent with these data, NHE2 promoter activity was also diminished by TNFα treatment. Functional characterization of the 5′-regulatory region of the NHE2 promoter localized the TNFα responsive region and identified an NFκB motif as the TNFα response element (−578 to −568 bp) [33] (Figure 1). TNFα-mediated repression of NHE2 mRNA was accompanied by reduced NHE2 protein abundance as well as its activity (Figure 2).
NHE8 mRNA and protein levels were also reduced in jejunal mucosa from TNBS or LPS-treated rats [62] suggesting that this isoform is also down-regulated by inflammation. In vitro studies in Caco-2 cells demonstrated TNFα-induced downregulation of hNHE8 promoter activity via reduction of DNA-binding activity of Sp3 [62]. Reduced expression of NHEs in inflammation suggests that this may contribute to the diarrhea associated with inflammatory conditions.
Serotonin
Serotonin (5-HT), an important neurotransmitter predominantly present in the gut and brain, plays a critical role in motility, secretion and absorption in the gut [85]. Increased levels of serotonin in the gut have been implicated in pathogenesis of diarrhea associated with IBS and UC [86]. Under acute conditions serotonin decreases NHE2 and NHE3 activities in Caco-2 cells [87]. Long-term treatment of C2BBE1 cells with 5-HT resulted in a significant reduction in NHE3 mRNA and protein expression as well as its promoter activity [56]. This reduction in promoter activity appears to be due to reduced DNA-binding activity of Sp1/Sp3 transcription factors to the −89/−69 bp in the NHE3 proximal promoter region. Negative impact of serotonin on NHE3 could be blocked by PKCα antagonist, Gö6976, implicating PKCα pathway in regulation of the repressive effects of serotonin on NHE3 [56]. To date, no data is available on the long-term effects of serotonin on the NHE2 and NHE8.
CL−/HCO3− EXCHANGERS
Studies over the past decade have shown that DRA and PAT1 are the candidate genes mediating intestinal luminal Cl−/HCO3− exchange [3, 6]. DRA, identified as a gene downregulated in colonic adenomas [88] was shown to be the major Cl−/HCO3 − exchanger involved in electroneutral NaCl absorption in the intestine [12, 19]. Mutations in hDRA have been shown to result in congenital chloride diarrhea (CLD), an autosomal recessive disorder characterized by a watery stool with high chloride concentration and metabolic alkalosis [89–91]. Similarly, the phenotype of DRA knockout mice was similar to CLD in humans [17]. PAT-1 knockout mice showed a decrease in apical Cl− /HCO3 − exchange activity but did not exhibit diarrheal phenotype [92]. It appears that while PAT-1 is involved in mediating Cl− absorption, it is not directly coupled to the water movement as seen with DRA.
Studies have shown that expression of both DRA and PAT-1 varies along the length of the intestine [19, 93] and crypt-villus axis [93–96]. DRA protein is predominantly expressed in the colon compared to much lower levels in the small intestine [3, 19, 93] and is localized to the apical membranes of IECs [19, 90, 96]. However, the expression pattern of PAT-1 is opposite to that of DRA expression in the intestine. PAT-1 is abundantly expressed in the small intestine compared to the colon [95], suggesting that PAT1 is important for the apical Cl−/HCO3− exchange in the small intestine, while DRA is important in the colon [3]. The protein product encoded by DRA gene is a glycoprotein of 764 amino acids [97]. PAT-1 gene encodes an integral membrane protein of 738 amino acids [98]. With respect to the crypt-villus/surface axis distribution, in situ hybridization studies showed expression of DRA in the surface epithelium and upper crypt of the rat and human colon [96]. These observations correlate well with the recent studies showing that DRA Cl−/HCO3− exchange activity was higher in the upper region (surface) of the colonic crypts compared to the lower region (middle & base) [93]. However, PAT-1 was expressed at very low levels in the crypts [95]. Ontogenic studies in mice showed that DRA mRNA expression in colon is low at birth, but increases during postnatal development [99]. However, the small intestine DRA mRNA expression remained lower throughout the postnatal development [99]. Similarly, well-differentiated post-confluent Caco2 cells showed higher expression of DRA with no expression in pre-confluent cells [99]. The above studies suggest that regional, crypt-villus axis or ontogenic expression of DRA and/or PAT1 in the intestine must be under transcriptional regulation. However, very limited studies are available regarding transcriptional regulation (long-term) of DRA and PAT-1 via changes in gene transcription. For example, a few studies have shown an inhibition of DRA expression in intestinal inflammation and in response to C. rodentium infection [93, 96, 100]. PAT-1 expression was also decreased in response to the pro-inflammatory cytokine, IFNγ [101]. In contrast, DRA expression was shown to be increased in response to anti-inflammatory/pro-absorptive agents such as probiotics [102] and butyrate [103]. New insights into the transcriptional mechanisms of DRA and PAT1 regulation are beginning to emerge with the cloning and characterization of the promoter regions of the human DRA and PAT1. The following sections will discuss in detail: 1) the basal characterization and regulation of DRA and PAT-1 promoters; and 2) the transcriptional regulation of DRA & PAT-1 under both physiological and pathophysiological conditions.
DRA
DRA Gene
DRA gene consists of 21 exons and is localized to chromosome 7q22-q31.1 [104]. The DRA cDNA encodes for a protein of 764 amino acids with a predicted molecular weight of 85 kDa [104].
DRA Promoter
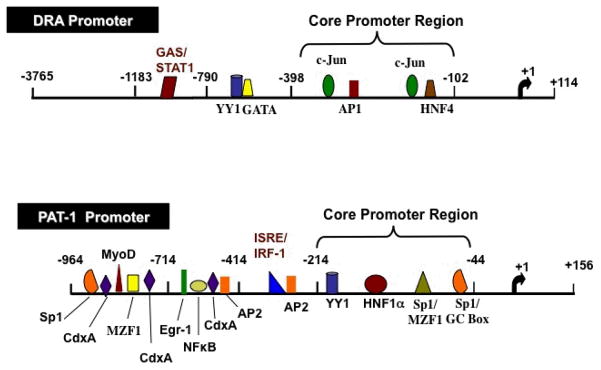
A 3765 bp of the 5′-regulatory region of DRA gene (promoter) was cloned by hybridization screening of a human bacterial artificial chromosome (BAC) genomic library [103]. DRA promoter (p-3765/Luc) was found to be significantly active in LS174T colonic cells [103] and intestinal Caco2 cells [100]. However, in the non-intestinal cells, HepG2 & NIH3T3 fibroblasts, p-3765/Luc DRA promoter construct showed no activity indicating that DRA promoter activity is intestinal cell line dependent. Sequence analysis of the DRA promoter and comparison with transcription factor databases revealed the presence of potential binding sites for transcription factors such as AP1, c-Jun & HNF4, GATA, YY1 and STAT1 (Figure 3).
Figure 3. Schematics of the human DRA and PAT1 promoters.
The transcription initiation site is marked as +1. Predicted transcription factor binding sites are indicated. Transcription factors shown in red text indicate IFNγ response elements and in blue text indicate butyrate response elements.
Core Promoter Region
Luciferase reporter assays of the progressive 5′-deletions of hDRA promoter suggested that cis-elements required for basal transcriptional activity are located in the DNA region between −398 and −102 bp [103]. Sequence analysis of the core promoter region indicated the presence of potential binding sites for transcription factors such as AP1, c-Jun & HNF4 (Figure 3). DNase I footprint analysis and EMSA showed HNF4 may be important in driving the basal activity of DRA promoter in LS174T human colonic cells [103]. HNF4 binds to the consensus sequence 5′-AGGTCANAGGTCA-3′ in order to activate transcription. The role of AP1 & c-Jun in the basal regulation of DRA promoter activity has not been studied and future studies are needed to elucidate their involvement in the basal regulation of DRA promoter in detail. The physiological significance of regulation of DRA promoter activity was further validated in an in vivo mouse model [103]. Expression of human growth hormone (hGH) driven by DRA promoter in transgenic mice was seen specifically in the villus epithelial cells of the small intestine and in surface epithelial cells of the colon [103] but not in other non-intestinal tissues suggesting that expression of DRA is specific to the intestine and differentiation status of the epithelium.
PAT1
PAT-1 Gene
PAT-1 gene is composed of 21 exons, 21 introns and spans 9.7 kb of chromosome 3p21.3 [105]. SLC26A6 gene encodes an integral membrane protein of 738 amino acid with a predicted topology of 11 transmembrane helices and an intracellular –NH2 and -COOH terminus [98, 105]. Human SLC26A6 gene was also found to have three alternatively spliced variants, named SLC26A6 a, c and d. With regard to their tissue distribution, RT-PCR studies indicated that SLC26A6a but not c or d is the spliced variant expressed in the human small intestine and colon [98]. Human PAT1 isoforms a, c and d comprise of 12, 8 and 12 membrane spanning domains, respectively.
PAT-1 Promoter
The 5′-flanking region of PAT-1 gene was cloned by PCR utilizing human genomic DNA and gene specific primers [101]. TIS was localized to Guanine residue 99 nucleotides upstream from the ATG translation start codon. Studies showed that PAT-1 promoter activity (1120 bp; p-964/+156 bp) was high in Caco2 cell line (which on differentiation exhibits small intestinal phenotype) compared to colonic NCM460 and kidney HEK 293 cells consistent with the fact that PAT-1 is predominantly expressed in the small intestine [95]. Sequence analysis of PAT1 promoter showed consensus sites for various transcription factors such as HNF1α, YY1, Sp1, CdxA, AP-2, Mzf1, MyoD & Egr1 (Figure 3).
Core Promoter Region
Progressive deletions from the 5′-flanking region demonstrated that deletions from −714 to −214 bp did not exhibit any change in promoter activity compared to full length PAT-1 promoter construct (−964/+156). Further deletion to −44 bp significantly decreased promoter activity by ~80% indicating that the region between −214 and −44 bp harbors cis elements important for the basal activity of PAT-1 promoter [101]. Sequence analysis of this region revealed the presence of potential binding sites for HNF1α, YY1 & Sp1 (Figure 3). More detailed studies are needed to further define the regulation of PAT1 by these transcription factors. Besides human, there are no reports on the cloning & characterization of DRA or PAT1 promoters in other species.
MODULATION OF DRA AND PAT1 GENE EXPRESSION
Long-term regulation of gene expression generally mimics the prolonged physiological or pathophysiological changes that occur in the intestine such as in response to luminal SCFA butyrate, or intestinal diseases (e.g. IBD). The following section will focus on the transcriptional regulation of DRA & PAT1 gene expression in response to both physiological and pathophysiological stimuli (Table 1).
Physiological Stimuli
Butyrate
Recent studies have shown that butyrate stimulates DRA expression and promoter activity in LS174T colonic cells [103]. Progressive 5′-deletions of DRA promoter showed that the butyrate-responsive region was located between −688 to −398 bp region. DNase I footprint analysis and EMSA identified a region from −468 to −487 bp that harbored two overlapping potential binding sites for YY1 and GATA indicating their involvement in butyrate-induced stimulation of DRA promoter activity [103]. Competition studies showed that YY1 consensus oligonucleotide completely abolished YY1 binding and also decreased GATA binding. On the other hand, GATA consensus oligo did abolish GATA binding but increased YY1 binding indicating that YY1 and GATA may influence the binding of each other. Mutations in the potential YY1 & GATA binding sites attenuated the stimulatory effects of butyrate on promoter activity further suggesting that the binding of YY1 and GATA transcription factors is crucial for butyrate-induced stimulation of DRA promoter [103]. Interestingly, YY1 binding in the DRA promoter was competed by a 20 bp fragment from the promoter region of fatty acid binding protein fabp gene that was previously known to silence the expression of FABP in the crypt epithelium of the small intestine [106]. It seems that YY1 may be critical for silencing the expression of DRA in proliferating undifferentiated intestinal epithelial cells and may play an important role in determining their expression pattern along the crypt-surface axis in the colon. GATA-5 was shown to modulate the promoter activity of rat NHE3 and suggested to play a role in its differential expression along the crypt-villus axis [57]. The GATA isoform involved in butyrate-induced effects on DRA promoter activity is not known [103]. Future studies are needed to elucidate the interplay of GATA & YY1 and to identify the isoform of GATA involved in butyrate-induced effects on DRA promoter activity. Also, regulation of PAT-1 gene expression by butyrate is not known.
Lactobacillus acidophilus
Lactobacilli are one ofthe predominant commensal bacteria in the gut microflora and have previously been used for the prevention and treatment of diarrheal disorders [107]. Recent studies utilizing Caco2 monolayers have shown that Lactobacillus acidophilus (LA) stimulated DRA mRNA and protein expression [102]. However, PAT-1 mRNA & protein expression remained unaltered upon LA treatment. In vivo mouse studies also demonstrated a significant increase in DRA mRNA and protein levels [102] in response to LA. Culture supernatants (CS) of live LA bacteria showed similar effects as live bacteria and stimulated both DRA expression and function in Caco2 monolayers [102]. The effects of LA CS were also seen at the transcriptional level. LA CS significantly increased DRA promoter activity in Caco2 cells [102]. These studies suggested that stimulation of DRA promoter activity and expression by LA secreted soluble effector molecules may contribute to the upregulation of intestinal Cl− absorption and provide novel insights underlying thepotential antidiarrheal effects of LA. However, the molecular mechanisms involved in the transcriptional regulation of DRA by LA secreted soluble factors are not known. Further studies are needed to identify the cis elements and transcription factors involved in the regulation of DRA promoter activity by LA secreted soluble factors and to elucidate the identity of soluble factors involved in the stimulation of DRA promoter activity.
Pathophysiological Stimuli
Inflammatory Mediators
Recent studies have demonstrated that chloride absorption is reduced in patients with ulcerative colitis [93]. The reduction in Cl− absorption in the surface of the colonic crypts was consistent with a decrease in DRA mRNA expression in UC patients compared to normal [93]. A decrease in DRA mRNA expression has also been reported in the cecum and colonic regions of two different animal models of colitis: IL10 knockout mice and HLA-B27/β2M transgenic rat [96]. In vitro studies in response to the pro-inflammatory cytokines utilizing Caco2 cells have also shown a decrease in DRA expression by IL1β [96] and a decrease in both DRA & PAT-1 expression by IFNγ [100, 101]. The transcriptional mechanisms underlying repression of DRA and PAT1 by IFNγ have been investigated in detail as outlined below.
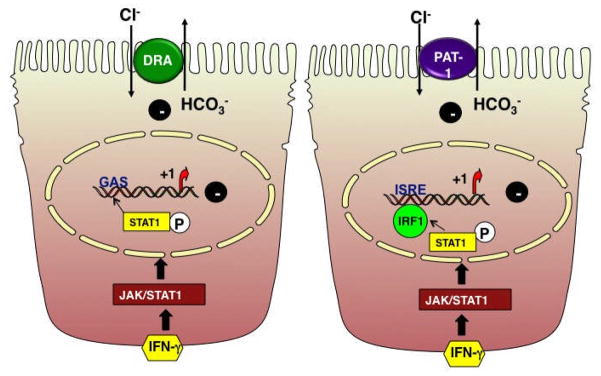
IFNγ elicits its effects via the induction of signal transduction pathway involving JAK 1 & 2 and STAT1. The binding of IFNγ to its surface receptor activates the receptor-associated tyrosine kinases, JAK1 and JAK2. JAKs tyrosine phosphorylate and activate the latent cytosolic STAT1, which then dimerizes, translocates to the nucleus [108] where it binds to IFNγ-response elements [109, 110] IFNγ decreased DRA promoter activity in Caco2 cells via a JAK/STAT1 pathway [100]. Progressive deletions from the 5′-flanking region of DRA promoter showed that the IFNγ-responsive region was located between −1183 to −790 bp region [100]. Sequence analysis of this region identified one potential GAS cis element flanking the region of −933 to −925 bp. GAS element is a 8- to 10-bp inverted repeat DNA element with a consensus sequence of 5′-TT(N4–6) AA-3′[109]. Additionally, EMSA showed increased protein binding to the oligonucleotide spanning the potential GAS element (−933/−925 bp) in response to IFNγ, which was blocked by STAT1 antibody, suggesting that the identified protein is STAT1 [100]. Also, mutational studies and in vivo ChIP assays showed that GAS element is required for the inhibitory effects of IFNγ on DRA promoteractivity. These findings provided evidence for the role of STAT1 in the inhibition of DRA promoter activity by IFNγ [100] (Figure 4). Increased expression and activation of STAT1 has been observed in the colonic mucosa of patients with UC and to a lesser degree in CD. [111]. Infact, the anti-inflammatory effects of glucocorticoids have been suggested to occur via inhibition of STAT1 in IBD patients [111]. Therefore, the development of novel therapeutic modalities against STAT1 may be of importance in the treatment of colonic inflammation where DRA expression and function are downregulated.
Figure 4. Proposed model for inhibition of DRA and PAT-1 promoter activity by IFNγ.
IFNγ binds to the IFNγ-receptor to activate JAK 1 & 2. Activated JAK 1 & 2 phosphorylate the latent cytoplasmic STAT1, which gets activated, dimerizes and is translocated to the nucleus. In the nucleus, phospho STAT1 binds to the GAS element of DRA promoter or activates IRF-1 that binds to ISRE site of PAT-1 promoter, which in turn leads to a decrease in DRA or PAT-1 mRNA, protein expression and function.
In contrast to the role of STAT1 in IFNγ-mediated effects on DRA promoter activity, PAT1 promoter activity was also decreased by IFNγ albeit via distinct cis element/transcription factor [101]. Progressive deletions from the 5′-flanking region of PAT1 promoter showed the presence of potential IFNγ response element (s) region between −414 to −214 bp [101]. Sequence analysis of the IFNγ responsive region identified one potential ISRE (Interferon Stimulated Response Element) binding site flanking the region of −318 to −300 bp. The ISREs, usually defined by pallindromic TTTC sequences separated by two or three nucleotides are located in the promoter region of many IFNγ inducible genes [110]. EMSA also showed increased protein binding to the oligonucleotide spanning the potential ISRE site (−318/−300 bp) in the presence of IFNγ. Additionally, supershift assays with antibody against IRF-1 transcription factor showed that the DNA-protein binding was blocked indicating that IRF-1 binds to the ISRE site [101]. This was further confirmed by ChIP assays showing that the association of IRF-1 with endogenous PAT1 promoter (containing the ISRE site) was significantly enhanced by IFNγ. Mutations in the potential ISRE cis element completely attenuated the inhibitory effects of IFNγ on promoter activity further suggesting the involvement of IRF-1 in the regulation of PAT1 promoter activity by IFNγ [101] (Figure 4). IRF-1 expression is increased in the lamina propria of patients with active Crohn’s disease [112]. Since, CD commonly affects the ileum, it is plausible to speculate that IRF-1 plays a key role in the inflammation of ileum. Immunosuppressive treatment (azathioprine or 6-mercaptopurine for 24h) of patients with CD has been shown to reduce IRF-1 [112]. Although, the role of PAT1 in intestinal inflammation has not been established, targeting of IRF-1 as a therapeutic alternative may be important in the treatment of ileal inflammation where PAT1 expression and function (Cl− absorption) are inhibited.
CONCLUSIONS
The advent of cloning and the use of transgenic mouse models have significantly enhanced our understanding of the molecular mechanisms underlying NaCl absorption and its regulation in health and disease. It is now accepted that NHE2, NHE3 and DRA, PAT1 are the predominant luminal membrane Na+ and Cl− absorbing proteins involved in vectorial NaCl absorption in the adult mammalian intestine. Recent studies have also provided strong evidence that in addition to short-term regulation of these transporters, long-term regulation has strong implications in tissue, region and differentiation specific expression of these genes under physiological conditions. Studies from disease models and patients have further shown that repression of NHE3 and DRA expression are associated with infectious diarrhea or inflammatory bowel diseases. Until recently, detailed mechanistic understanding of the long-term regulation of these transporters was very limited. However, with cloning of the promoters of these transporters, new data on their transcriptional regulation has now started to emerge. These data have identified and characterized the core promoter, the predicted TF binding sites and the TFs involved in basal transcription and regulation of these genes by various physiological and pathophysiological agents. These studies demonstrate that basal transcriptional regulation of both the human NHE2 and NHE3 involves transcription factors Sp1 and Sp3. Although, AP2 was also found to interact with NHE3 promoter, the functional significance of this binding remains to be investigated. The studies further demonstrated that diverse effector molecules e.g. butyrate, phobol esters, IFNγ, TNFα and serotonin converge at a site containing the overlapping Egr-1/Sp1 motif in NHE3 promoter to trigger distinct outcomes. However, the detailed molecular mechanisms as to how these agents exert distinct effects on NHE3 promoter remains to be investigated. In contrast to NHE3, NHE2 repression by inflammatory cytokines was found to involve activation of the NFκB pathway. It is, however, not clear whether NFκB represses NHE2 promoter activity via direct binding or through a putative downstream transcription factor. With respect to transcriptional regulation of DRA and PAT1, the available data is even more sketchy. This has been mainly due to identification of their role in intestinal chloride absorption only very recently and the cloning of their promoters was accomplished only during the last 3 years. It is now known that HNF4 is involved in basal transcription of DRA, however, its stimulation by butyrate involved GATA and YY1 factors. However, the exact GATA isoform involved in the regulation of DRA promoter and the interplay between GATA and YY1 needs more extensive investigation. The basal transcriptional regulation of PAT1 has not been characterized, however, sequence analysis of the promoter region predicted that HNF1α, YY1 and Sp1 might play important roles. Studies have further shown that the cytokine IFNγ down-regulated the expression of both the DRA and PAT1 genes, albeit, via distinct mechanisms. While the activation of STAT1 via JAK-STAT pathway and its direct binding to GAS element mediated the effect of IFNγ on DRA promoter, the activation of IRF-1 by STAT1 and subsequent binding of IRF-1 to ISRE was involved in the suppression of PAT1 promoter by IFNγ. Repression of both PAT1 or DRA and NHE3 promoter activities in response to IFNγ may lead to decreased NaCl absorption in the ileum and colon. The overall decrease in NaCl absorption may play a crucial role in the pathophysiology of diarrhea associated with IBD. Taken, together, these data indicate that down-regulation of NHE2, 3 and DRA and PAT1 expression and the decrease in the promoter activities in response to inflammatory mediators may contribute to pathophysiology of diarrhea associated with inflammatory disorders of the gut. On the other hand, mechanisms involved in the induction of gene expression of NHE3 or DRA by agents e.g. butyrate, soluble factors secreted by probiotics or glucocorticoids need to be investigated in detail as these could be exploited to improve the treatment modalities of the inflammatory bowel diseases. Furthermore, a detailed characterization of the newly identified NHE8 may also add more to our understanding of NaCl absorption and its regulation in health and disease as well as in developing and adult intestine. Finally, the role of epigenetic mechanisms in regulating the expression of intestinal Na+ and Cl− transporters is not fully understood and warrants future investigations.
Acknowledgments
The studies in the authors laboratories in part were supported by the Department of Veterans Affairs and the NIDDK grants, DK 33349 (J. Malakooti), DK 54016 & DK 81858 (PK. Dudeja), P01 DK 067887 (PK. Dudeja, J. Malakooti), DK 74458 (RK. Gill) and CCFA grant Ref # 1942 (S. Saksena). We thank Alip Borthakur, PhD and Waddah A Alrefai, MD for critical editing of the review.
ABBREVIATIONS USED
- 5-HT
5-hydroxytryptamine
- AP
activator protein
- C2BBE1
subclone of Caco2 cells
- Caco2
human colon adenocarcinoma
- Cdx2
caudal-related homeobox 2
- CLD
Congenital Chloride Diarrhea
- CREB
cAMP response element binding
- DRA
Down Regulated in Adenoma
- EGF
epidermal growth factor
- Egr-1
early growth response protein 1
- ERK1/2
extracellular signal regulated kinase 1/2
- GAS
γ-activated site
- GRE
glucocorticoid response element
- h
human
- HEK293
human embryonic kidney
- HepG2
human hepatocellular liver carcinoma
- HNF
hepatocyte nuclear factor
- IBD
Inflammatory Bowel Diseases
- IBS
Irritable Bowel Syndrome
- IEC
intestinal epithelial cell
- IFNγ
interferon γ
- IL1β
interleukin 1β
- IRF-1
interferon regulatory factor 1
- ISRE
interferon stimulated response element
- JAK
janus kinase
- LLC-PCK1
pig kidney epithelial
- LS174T
human colon cancer
- Luc
luciferase
- MAPK
mitogen-activated protein kinase
- MAZ
myc-associated zinc finger
- MyoD
myogenic differentiation 1
- Mzf1
myeloid zinc finger 1
- NCM460
normal human colon mucosal epithelial
- NFB
nuclear factor kappa B
- NHE
sodium hydrogen exchanger
- NIH3T3
mouse embryonic fibroblast
- Oct-1
octameric transcription factor 1
- OK
opossum kidney
- PAT1
Putative Anion Transporter 1
- PKA
protein kinase A
- PKC
protein kinase C
- PMA
phorbol 12 myristate 13-acetate
- PRE
PMA response element
- PS120
Chinese hamster lung fibroblast
- QT-6
avian (quail origin-6)
- r
rat
- SL2
schneider cell line 2
- SLC
solute carrier
- Sp
specificity protein
- STAT
signal transducer and activator transcription factor
- T3R
thyroid hormone receptor
- TF
transcription factor
- TIS
transcription initiation site
- TNFα
tumor necrosis factorα
- USF
upstream stimulatory factor
- VDR
vitamin D3 receptor
- YY1
Ying Yang 1
- ZBP-89
zinc-binding protein-89
References
- 1.Dudeja PK, Ramaswamy K. Physiology of the Gastrointestinal Tract. Elsevier Academic; 2006. Intestinal Anion Aborption. [Google Scholar]
- 2.Gill R, Alrefai W, Ramaswamy K, Dudeja P. Mechanisms and regulation of NaCl absorption in the human intestine. Recent Res Devel Physiol. 2003;1:643–677. [Google Scholar]
- 3.Kato A, Romero MF. Regulation of Electroneutral NaCl Absorption by the Small Intestine. Annu Rev Physiol. 2010 doi: 10.1146/annurev-physiol-012110-142244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donowitz M, Mohan S, Zhu CX, Chen TE, Lin R, Cha B, Zachos NC, Murtazina R, Sarker R, Li X. NHE3 regulatory complexes. J Exp Biol. 2009;212:1638–1646. doi: 10.1242/jeb.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiela PR, Xu H, Ghishan FK. Apical NA+/H+ exchangers in the mammalian gastrointestinal tract. J Physiol Pharmacol. 2006;57(Suppl 7):51–79. [PubMed] [Google Scholar]
- 6.Mount DB, Romero MF. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 2004;447:710–721. doi: 10.1007/s00424-003-1090-3. [DOI] [PubMed] [Google Scholar]
- 7.Bobulescu IA, Moe OW. Luminal Na(+)/H (+) exchange in the proximal tubule. Pflugers Arch. 2009;458:5–21. doi: 10.1007/s00424-008-0595-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander RT, Grinstein S. Tethering, recycling and activation of the epithelial sodium-proton exchanger, NHE3. J Exp Biol. 2009;212:1630–1637. doi: 10.1242/jeb.027375. [DOI] [PubMed] [Google Scholar]
- 9.Donowitz M, Li X. Regulatory binding partners and complexes of NHE3. Physiol Rev. 2007;87:825–872. doi: 10.1152/physrev.00030.2006. [DOI] [PubMed] [Google Scholar]
- 10.Hassan HA, Mentone S, Karniski LP, Rajendran VM, Aronson PS. Regulation of anion exchanger Slc26a6 by protein kinase C. American journal of physiology. 2007;292:C1485–1492. doi: 10.1152/ajpcell.00447.2006. [DOI] [PubMed] [Google Scholar]
- 11.Lissner S, Nold L, Hsieh CJ, Turner JR, Gregor M, Graeve L, Lamprecht G. Activity and PI3-kinase dependent trafficking of the intestinal anion exchanger downregulated in adenoma depend on its PDZ interaction and on lipid rafts. Am J Physiol Gastrointest Liver Physiol. 2010;299:G907–920. doi: 10.1152/ajpgi.00191.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musch MW, Arvans DL, Wu GD, Chang EB. Functional coupling of the downregulated in adenoma Cl-/base exchanger DRA and the apical Na+/H+ exchangers NHE2 and NHE3. Am J Physiol Gastrointest Liver Physiol. 2009;296:G202–210. doi: 10.1152/ajpgi.90350.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saksena S, Tyagi S, Goyal S, Gill RK, Alrefai WA, Ramaswamy K, Dudeja PK. Stimulation of Apical Cl-/HCO3-(OH-) Exchanger, SLC26A3 by Neuropeptide Y (NPY) is Lipid Raft Dependent. Am J Physiol Gastrointest Liver Physiol. 2010 doi: 10.1152/ajpgi.00039.2010. [ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachmann O, Riederer B, Rossmann H, Groos S, Schultheis PJ, Shull GE, Gregor M, Manns MP, Seidler U. The Na+/H+ exchanger isoform 2 is the predominant NHE isoform in murine colonic crypts and its lack causes NHE3 upregulation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G125–133. doi: 10.1152/ajpgi.00332.2003. [DOI] [PubMed] [Google Scholar]
- 15.Broere N, Chen M, Cinar A, Singh AK, Hillesheim J, Riederer B, Lunnemann M, Rottinghaus I, Krabbenhoft A, Engelhardt R, Rausch B, Weinman EJ, Donowitz M, Hubbard A, Kocher O, de Jonge HR, Hogema BM, Seidler U. Defective jejunal and colonic salt absorption and alteredNa(+)/H (+) exchanger 3 (NHE3) activity in NHE regulatory factor 1 (NHERF1) adaptor protein-deficient mice. Pflugers Arch. 2009;457:1079–1091. doi: 10.1007/s00424-008-0579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gawenis LR, Stien X, Shull GE, Schultheis PJ, Woo AL, Walker NM, Clarke LL. Intestinal NaCl transport in NHE2 and NHE3 knockout mice. Am J Physiol Gastrointest Liver Physiol. 2002;282:G776–784. doi: 10.1152/ajpgi.00297.2001. [DOI] [PubMed] [Google Scholar]
- 17.Schweinfest CW, Spyropoulos DD, Henderson KW, Kim JH, Chapman JM, Barone S, Worrell RT, Wang Z, Soleimani M. slc26a3 (dra)-deficient mice display chloride-losing diarrhea, enhanced colonic proliferation, and distinct up-regulation of ion transporters in the colon. J Biol Chem. 2006;281:37962–37971. doi: 10.1074/jbc.M607527200. [DOI] [PubMed] [Google Scholar]
- 18.Simpson JE, Schweinfest CW, Shull GE, Gawenis LR, Walker NM, Boyle KT, Soleimani M, Clarke LL. PAT-1 (Slc26a6) is the predominant apical membrane Cl-/HCO3- exchanger in the upper villous epithelium of the murine duodenum. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1079–1088. doi: 10.1152/ajpgi.00354.2006. [DOI] [PubMed] [Google Scholar]
- 19.Walker NM, Simpson JE, Yen PF, Gill RK, Rigsby EV, Brazill JM, Dudeja PK, Schweinfest CW, Clarke LL. Down-regulated in adenoma Cl/HCO3 exchanger couples with Na/H exchanger 3 for NaCl absorption in murine small intestine. Gastroenterology. 2008;135:1645–1653. e1643. doi: 10.1053/j.gastro.2008.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudeja PK, Rao DD, Syed I, Joshi V, Dahdal RY, Gardner C, Risk MC, Schmidt L, Bavishi D, Kim KE, Harig JM, Goldstein JL, Layden TJ, Ramaswamy K. Intestinal distribution of human Na+/H+ exchanger isoforms NHE-1, NHE-2, and NHE-3 mRNA. Am J Physiol. 1996;271:G483–493. doi: 10.1152/ajpgi.1996.271.3.G483. [DOI] [PubMed] [Google Scholar]
- 21.Xu H, Chen R, Ghishan FK. Subcloning, localization, and expression of the rat intestinal sodium-hydrogen exchanger isoform 8. Am J Physiol Gastrointest Liver Physiol. 2005;289:G36–41. doi: 10.1152/ajpgi.00552.2004. [DOI] [PubMed] [Google Scholar]
- 22.Sardet C, Franchi A, Pouyssegur J. Molecular cloning, primary structure, and expression of the human growth factor-activatable Na+/H+ antiporter. Cell. 1989;56:271–280. doi: 10.1016/0092-8674(89)90901-x. [DOI] [PubMed] [Google Scholar]
- 23.Wormmeester L, Sanchez de Medina F, Kokke F, Tse CM, Khurana S, Bowser J, Cohen ME, Donowitz M. Quantitative contribution of NHE2 and NHE3 to rabbit ileal brush-border Na+/H+ exchange. Am J Physiol. 1998;274:C1261–1272. doi: 10.1152/ajpcell.1998.274.5.C1261. [DOI] [PubMed] [Google Scholar]
- 24.Schultheis PJ, Clarke LL, Meneton P, Miller ML, Soleimani M, Gawenis LR, Riddle TM, Duffy JJ, Doetschman T, Wang T, Giebisch G, Aronson PS, Lorenz JN, Shull GE. Renal and intestinal absorptive defects in mice lacking the NHE3 Na+/H+ exchanger. Nat Genet. 1998;19:282–285. doi: 10.1038/969. [DOI] [PubMed] [Google Scholar]
- 25.Schultheis PJ, Clarke LL, Meneton P, Harline M, Boivin GP, Stemmermann G, Duffy JJ, Doetschman T, Miller ML, Shull GE. Targeted disruption of the murine Na+/H+ exchanger isoform 2 gene causes reduced viability of gastric parietal cells and loss of net acid secretion. J Clin Invest. 1998;101:1243–1253. doi: 10.1172/JCI1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goyal SVHG, Aronson PS. Renal expression of novel Na+/H+ exchanger isoform NHE8. Am J Physiol Renal Physiol. 2003;284:F 467–473. doi: 10.1152/ajprenal.00352.2002. [DOI] [PubMed] [Google Scholar]
- 27.Goyal S, Mentone S, Aronson PS. Immunolocalization of NHE8 in rat kidney. Am J Physiol Renal Physiol. 2005;288:F530–538. doi: 10.1152/ajprenal.00229.2004. [DOI] [PubMed] [Google Scholar]
- 28.Xu H, Chen H, Dong J, Lynch R, Ghishan FK. Gastrointestinal distribution and kinetic characterization of the sodium-hydrogen exchanger isoform 8 (NHE8) Cell Physiol Biochem. 2008;21:109–116. doi: 10.1159/000113752. [DOI] [PubMed] [Google Scholar]
- 29.Greig E, Sandle GI. Diarrhea in ulcerative colitis. The role of altered colonic sodium transport. Ann N Y Acad Sci. 2000;915:327–332. doi: 10.1111/j.1749-6632.2000.tb05260.x. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Augustin O, Romero-Calvo I, Suarez MD, Zarzuelo A, de Medina FS. Molecular bases of impaired water and ion movements in inflammatory bowel diseases. Inflamm Bowel Dis. 2009;15:114–127. doi: 10.1002/ibd.20579. [DOI] [PubMed] [Google Scholar]
- 31.Malakooti J, Dahdal RY, Dudeja PK, Layden TJ, Ramaswamy K. The human Na(+)/H(+) exchanger NHE2 gene: genomic organization and promoter characterization. Am J Physiol Gastrointest Liver Physiol. 2001;280:G763–773. doi: 10.1152/ajpgi.2001.280.4.G763. [DOI] [PubMed] [Google Scholar]
- 32.Malakooti J, Dahdal RY, Schmidt L, Layden TJ, Dudeja PK, Ramaswamy K. Molecular cloning, tissue distribution, and functional expression of the human Na(+)/H(+) exchanger NHE2. Am J Physiol. 1999;277:G383–390. doi: 10.1152/ajpgi.1999.277.2.G383. [DOI] [PubMed] [Google Scholar]
- 33.Amin MR, Orenuga T, Tyagi S, Dudeja PK, Ramaswamy K, Malakooti J. Tumor necrosis factor-alpha represses the expression of NHE2 through NF-kappaB activation in intestinal epithelial cell model, C2BBe1. Inflamm Bowel Dis. 2010 doi: 10.1002/ibd.21419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller YL, Collins JF, Bai L, Xu H, Ghishan FK. Molecular cloning and characterization of the rat NHE-2 gene promoter. Biochim Biophys Acta. 1998;1442:314–319. doi: 10.1016/s0167-4781(98)00191-2. [DOI] [PubMed] [Google Scholar]
- 35.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 36.Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 37.Malakooti J, Sandoval R, Memark VC, Dudeja PK, Ramaswamy K. Zinc finger transcription factor Egr-1 is involved in stimulation of NHE2 gene expression by phorbol 12-myristate 13-acetate. Am J Physiol Gastrointest Liver Physiol. 2005;289:G653–663. doi: 10.1152/ajpgi.00010.2005. [DOI] [PubMed] [Google Scholar]
- 38.Pearse I, Zhu YX, Murray EJ, Dudeja PK, Ramaswamy K, Malakooti J. Sp1 and Sp3 control constitutive expression of the human NHE2 promoter by interactions with the proximal promoter and the transcription initiation site. Biochem J. 2007;407:101–111. doi: 10.1042/BJ20070364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding gene. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 40.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy AL, Du H, Gregor PD, Novina CD, Martinez E, Roeder RG. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. Embo J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JS, Galvin KM, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Briggs MR, Kadonaga JT, Bell SP, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 44.Kadonaga JT, Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai L, Collins JF, Xu H, Ghishan FK. Transcriptional regulation of rat Na(+)/H(+) exchanger isoform-2 (NHE-2) gene by Sp1 transcription factor. American journal of physiology. 2001;280:C1168–1175. doi: 10.1152/ajpcell.2001.280.5.C1168. [DOI] [PubMed] [Google Scholar]
- 46.Hua P, Xu H, Uno JK, Lipko MA, Dong J, Kiela PR, Ghishan FK. Sp1 and Sp3 mediate NHE2 gene transcription in the intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2007;293:G146–153. doi: 10.1152/ajpgi.00443.2006. [DOI] [PubMed] [Google Scholar]
- 47.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 48.Brant SR, Bernstein M, Wasmuth JJ, Taylor EW, McPherson JD, Li X, Walker S, Pouyssegur J, Donowitz M, Tse CM, et al. Physical and genetic mapping of a human apical epithelial Na+/H+ exchanger (NHE3) isoform to chromosome 5p15.3. Genomics. 1993;15:668–672. doi: 10.1006/geno.1993.1122. [DOI] [PubMed] [Google Scholar]
- 49.Brant SR, Yun CH, Donowitz M, Tse CM. Cloning, tissue distribution, and functional analysis of the human Na+/H+ exchanger isoform, NHE3. Am J Physiol. 1995;269:C198–206. doi: 10.1152/ajpcell.1995.269.1.C198. [DOI] [PubMed] [Google Scholar]
- 50.Malakooti J, Memark VC, Dudeja PK, Ramaswamy K. Molecular cloning and functional analysis of the human Na(+)/H(+) exchanger NHE3 promoter. Am J Physiol Gastrointest Liver Physiol. 2002;282:G491–500. doi: 10.1152/ajpgi.00273.2001. [DOI] [PubMed] [Google Scholar]
- 51.Cano A. Characterization of the rat NHE3 promoter. Am J Physiol. 1996;271:F629–636. doi: 10.1152/ajprenal.1996.271.3.F629. [DOI] [PubMed] [Google Scholar]
- 52.Kandasamy RA, Orlowski J. Genomic organization and glucocorticoid transcriptional activation of the rat Na+/H+ exchanger Nhe3 gene. J Biol Chem. 1996;271:10551–10559. doi: 10.1074/jbc.271.18.10551. [DOI] [PubMed] [Google Scholar]
- 53.Malakooti J, Sandoval R, Amin MR, Clark J, Dudeja PK, Ramaswamy K. Transcriptional stimulation of the human NHE3 promoter activity by PMA: PKC independence and involvement of the transcription factor EGR-1. Biochem J. 2006;396:327–336. doi: 10.1042/BJ20051391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amin MR, Malakooti J, Sandoval R, Dudeja PK, Ramaswamy K. IFN-{gamma} and TNF-{alpha} Regulate the Human NHE3 Gene Expression By Modulating the Sp Family Transcription Factors in Human Intestinal Epithelial Cell Line, C2BBe1. American journal of physiology. 2006;291:C887–896. doi: 10.1152/ajpcell.00630.2005. [DOI] [PubMed] [Google Scholar]
- 55.Amin MR, Dudeja PK, Ramaswamy K, Malakooti J. Involvement of Sp1 and Sp3 in differential regulation of human NHE3 promoter activity by sodium butyrate and IFN-gamma/TNF-alpha. Am J Physiol Gastrointest Liver Physiol. 2007;293:G374–382. doi: 10.1152/ajpgi.00128.2007. [DOI] [PubMed] [Google Scholar]
- 56.Amin MR, Ghannad L, Othman A, Gill RK, Dudeja PK, Ramaswamy K, Malakooti J. Transcriptional regulation of the human Na+/H+ exchanger NHE3 by serotonin in intestinal epithelial cells. Biochem Biophys Res Commun. 2009;382:620–625. doi: 10.1016/j.bbrc.2009.03.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiela PR, LeSueur J, Collins JF, Ghishan FK. Transcriptional regulation of the rat NHE3 gene. Functional interactions between GATA-5 and Sp family transcription factors. J Biol Chem. 2003;278:5659–5668. doi: 10.1074/jbc.M209473200. [DOI] [PubMed] [Google Scholar]
- 58.Su HW, Yeh HH, Wang SW, Shen MR, Chen TL, Kiela PR, Ghishan FK, Tang MJ. Cell confluence-induced activation of signal transducer and activator of transcription-3 (Stat3) triggers epithelial dome formation via augmentation of sodium hydrogen exchanger-3 (NHE3) expression. J Biol Chem. 2007;282:9883–9894. doi: 10.1074/jbc.M606754200. [DOI] [PubMed] [Google Scholar]
- 59.Collins JF, Xu H, Kiela PR, Zeng J, Ghishan FK. Functional and molecular characterization of NHE3 expression during ontogeny in rat jejunal epithelium. Am J Physiol. 1997;273:C1937–1946. doi: 10.1152/ajpcell.1997.273.6.C1937. [DOI] [PubMed] [Google Scholar]
- 60.Collins JF, Kiela PR, Xu H, Zeng J, Ghishan FK. Increased NHE2 expression in rat intestinal epithelium during ontogeny is transcriptionally mediated. Am J Physiol. 1998;275:C1143–1150. doi: 10.1152/ajpcell.1998.275.4.C1143. [DOI] [PubMed] [Google Scholar]
- 61.Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. American journal of physiology. 2005;288:C223–239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 62.Xu H, Chen H, Dong J, Li J, Chen R, Uno JK, Ghishan FK. Tumor necrosis factor-{alpha} downregulates intestinal NHE8 expression by reducing basal promoter activity. American journal of physiology. 2009;296:C489–497. doi: 10.1152/ajpcell.00482.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheppach W, Weiler F. The butyrate story: old wine in new bottles? Curr Opin Clin Nutr Metab Care. 2004;7:563–567. doi: 10.1097/00075197-200409000-00009. [DOI] [PubMed] [Google Scholar]
- 64.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 65.Binder HJ, Mehta P. Short-chain fatty acids stimulate active sodium and chloride absorption in vitro in the rat distal colon. Gastroenterology. 1989;96:989–996. doi: 10.1016/0016-5085(89)91614-4. [DOI] [PubMed] [Google Scholar]
- 66.Krishnan S, Rajendran VM, Binder HJ. Apical NHE isoforms differentially regulate butyrate-stimulated Na absorption in rat distal colon. American journal of physiology. 2003;285:C1246–1254. doi: 10.1152/ajpcell.00598.2002. [DOI] [PubMed] [Google Scholar]
- 67.Musch MW, Bookstein C, Xie Y, Sellin JH, Chang EB. SCFA increase intestinal Na absorption by induction of NHE3 in rat colon and human intestinal C2/bbe cells. Am J Physiol Gastrointest Liver Physiol. 2001;280:G687–693. doi: 10.1152/ajpgi.2001.280.4.G687. [DOI] [PubMed] [Google Scholar]
- 68.Kiela PR, Hines ER, Collins JF, Ghishan FK. Regulation of the rat NHE3 gene promoter by sodium butyrate. Am J Physiol Gastrointest Liver Physiol. 2001;281:G947–956. doi: 10.1152/ajpgi.2001.281.4.G947. [DOI] [PubMed] [Google Scholar]
- 69.Kiela PR, Kuscuoglu N, Midura AJ, Midura-Kiela MT, Larmonier CB, Lipko M, Ghishan FK. Molecular mechanism of rat NHE3 gene promoter regulation by sodium butyrate. American journal of physiology. 2007;293:C64–74. doi: 10.1152/ajpcell.00277.2006. [DOI] [PubMed] [Google Scholar]
- 70.Yun CH, Gurubhagavatula S, Levine SA, Montgomery JL, Brant SR, Cohen ME, Cragoe EJ, Jr, Pouyssegur J, Tse CM, Donowitz M. Glucocorticoid stimulation of ileal Na+ absorptive cell brush border Na+/H+ exchange and association with an increase in message for NHE-3, an epithelial Na+/H+ exchanger isoform. J Biol Chem. 1993;268:206–211. [PubMed] [Google Scholar]
- 71.Baum M, Moe OW, Gentry DL, Alpern RJ. Effect of glucocorticoids on renal cortical NHE-3 and NHE-1 mRNA. Am J Physiol. 1994;267:F437–442. doi: 10.1152/ajprenal.1994.267.3.F437. [DOI] [PubMed] [Google Scholar]
- 72.Sundaram U, Wisel S, Coon S. Neutral Na-amino acid cotransport is differentially regulated by glucocorticoids in the normal and chronically inflamed rabbit small intestine. Am J Physiol Gastrointest Liver Physiol. 2007;292:G467–474. doi: 10.1152/ajpgi.00503.2005. [DOI] [PubMed] [Google Scholar]
- 73.Sandle GI, Binder HJ. Corticosteroids and intestinal ion transport. Gastroenterology. 1987;93:188–196. doi: 10.1016/0016-5085(87)90333-7. [DOI] [PubMed] [Google Scholar]
- 74.Xu H, Zhang B, Li J, Chen H, Wang C, Ghishan FK. Transcriptional inhibition of intestinal NHE8 expression by glucocorticoids involves Pax5. Am J Physiol Gastrointest Liver Physiol. 2010;299:G921–927. doi: 10.1152/ajpgi.00227.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu H, Collins JF, Bai L, Kiela PR, Lynch RM, Ghishan FK. Epidermal growth factor regulation of rat NHE2 gene expression. American journal of physiology. 2001;281:C504–513. doi: 10.1152/ajpcell.2001.281.2.C504. [DOI] [PubMed] [Google Scholar]
- 76.Xu H, Zhang B, Li J, Chen H, Tooley J, Ghishan FK. Epidermal growth factor inhibits intestinal NHE8 expression via reducing its basal transcription. American journal of physiology. 2010;299:C51–57. doi: 10.1152/ajpcell.00081.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khachigian LM, Collins T. Early growth response factor 1: a pleiotropic mediator of inducible gene expression. J Mol Med. 1998;76:613–616. doi: 10.1007/s001090050258. [DOI] [PubMed] [Google Scholar]
- 78.Vidal F, Aragones J, Alfranca A, de Landazuri MO. Up-regulation of vascular endothelial growth factor receptor Flt-1 after endothelial denudation: role of transcription factor Egr-1. Blood. 2000;95:3387–3395. [PubMed] [Google Scholar]
- 79.Rohlff C, Ahmad S, Borellini F, Lei J, Glazer RI. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J Biol Chem. 1997;272:21137–21141. doi: 10.1074/jbc.272.34.21137. [DOI] [PubMed] [Google Scholar]
- 80.Barmeyer C, Harren M, Schmitz H, Heinzel-Pleines U, Mankertz J, Seidler U, Horak I, Wiedenmann B, Fromm M, Schulzke JD. Mechanisms of diarrhea in the interleukin-2-deficient mouse model of colonic inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;286:G244–252. doi: 10.1152/ajpgi.00141.2003. [DOI] [PubMed] [Google Scholar]
- 81.Rocha FMM, Lishanskiy L, Bookstein C, Sugi K, Xie Y, Chang EB. IFN-gamma downregulates expression of Na(+)/H(+) exchangers NHE2 and NHE3 in rat intestine and human Caco-2/bbe cells. Am J Physiol Cell Physiol. 2001;280:1224–1232. doi: 10.1152/ajpcell.2001.280.5.C1224. [DOI] [PubMed] [Google Scholar]
- 82.Chu S, Cockrell CA, Ferro TJ. Expression of alpha-ENaC2 is dependent on an upstream Sp1 binding motif and is modulated by protein phosphatase 1 in lung epithelial cells. Biochem Biophys Res Commun. 2003;303:1159–1168. doi: 10.1016/s0006-291x(03)00497-2. [DOI] [PubMed] [Google Scholar]
- 83.Yeruva S, Farkas K, Hubricht J, Rode K, Riederer B, Bachmann O, Cinar A, Rakonczay Z, Molnar T, Nagy F, Wedemeyer J, Manns M, Raddatz D, Musch MW, Chang EB, Hegyi P, Seidler U. Preserved Na(+)/H(+) exchanger isoform 3 expression and localization, but decreased NHE3 function indicate regulatory sodium transport defect in ulcerative colitis. Inflamm Bowel Dis. 2010;16:1149–1161. doi: 10.1002/ibd.21183. [DOI] [PubMed] [Google Scholar]
- 84.Sullivan S, Alex P, Dassopoulos T, Zachos NC, Iacobuzio-Donahue C, Donowitz M, Brant SR, Cuffari C, Harris ML, Datta LW, Conklin L, Chen Y, Li X. Downregulation of sodium transporters and NHERF proteins in IBD patients and mouse colitis models: potential contributors to IBD-associated diarrhea. Inflamm Bowel Dis. 2009;15:261–274. doi: 10.1002/ibd.20743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tobe T, Fujiwara M, Tanaka C. Distribution of serotonin (5-hydroxytryptamine) in the human gastroinetestinal tract. Am J Gastroenterol. 1966;46:34–37. [PubMed] [Google Scholar]
- 86.Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, Crowell MD, Sharkey KA, Gershon MD, Mawe GM, Moses PL. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology. 2004;126:1657–1664. doi: 10.1053/j.gastro.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 87.Gill RK, Saksena S, Tyagi S, Alrefai WA, Malakooti J, Sarwar Z, Turner JR, Ramaswamy K, Dudeja PK. Serotonin inhibits Na+/H+ exchange activity via 5-HT4 receptors and activation of PKC alpha in human intestinal epithelial cells. Gastroenterology. 2005;128:962–974. doi: 10.1053/j.gastro.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 88.Schweinfest CW, Henderson KW, Suster S, Kondoh N, Papas TS. Identification of a colon mucosa gene that is down-regulated in colon adenomas and adenocarcinomas. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:4166–4170. doi: 10.1073/pnas.90.9.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de la Chapelle A. Disease gene mapping in isolated human populations: the example of Finland. J Med Genet. 1993;30:857–865. doi: 10.1136/jmg.30.10.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoglund P, Haila S, Socha J, Tomaszewski L, Saarialho-Kere U, Karjalainen-Lindsberg ML, Airola K, Holmberg C, de la Chapelle A, Kere J. Mutations of the Down-regulated in adenoma (DRA) gene cause congenital chloride diarrhoea. Nat Genet. 1996;14:316–319. doi: 10.1038/ng1196-316. [DOI] [PubMed] [Google Scholar]
- 91.Makela S, Kere J, Holmberg C, Hoglund P. SLC26A3 mutations in congenital chloride diarrhea. Hum Mutat. 2002;20:425–438. doi: 10.1002/humu.10139. [DOI] [PubMed] [Google Scholar]
- 92.Wang Z, Wang T, Petrovic S, Tuo B, Riederer B, Barone S, Lorenz JN, Seidler U, Aronson PS, Soleimani M. Renal and intestinal transport defects in Slc26a6-null mice. American journal of physiology. 2005;288:C957–965. doi: 10.1152/ajpcell.00505.2004. [DOI] [PubMed] [Google Scholar]
- 93.Farkas K, Yeruva S, Rakonczay Z, Jr, Ludolph L, Molnar T, Nagy F, Szepes Z, Schnur A, Wittmann T, Hubricht J, Riederer B, Venglovecz V, Lazar G, Kiraly M, Zsembery A, Varga G, Seidler U, Hegyi P. New therapeutic targets in ulcerative colitis: The importance of ion transporters in the human colon. Inflamm Bowel Dis. 2010 doi: 10.1002/ibd.21432. [ahead of print] [DOI] [PubMed] [Google Scholar]
- 94.Walker NM, Simpson JE, Brazill JM, Gill RK, Dudeja PK, Schweinfest CW, Clarke LL. Role of down-regulated in adenoma anion exchanger in HCO3- secretion across murine duodenum. Gastroenterology. 2009;136:893–901. doi: 10.1053/j.gastro.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Z, Petrovic S, Mann E, Soleimani M. Identification of an apical Cl(-)/HCO3(-) exchanger in the small intestine. Am J Physiol Gastrointest Liver Physiol. 2002;282:G573–579. doi: 10.1152/ajpgi.00338.2001. [DOI] [PubMed] [Google Scholar]
- 96.Yang H, Jiang W, Furth EE, Wen X, Katz JP, Sellon RK, Silberg DG, Antalis TM, Schweinfest CW, Wu GD. Intestinal inflammation reduces expression of DRA, a transporter responsible for congenital chloride diarrhea. Am J Physiol. 1998;275:G1445–1453. doi: 10.1152/ajpgi.1998.275.6.G1445. [DOI] [PubMed] [Google Scholar]
- 97.Byeon MK, Westerman MA, Maroulakou IG, Henderson KW, Suster S, Zhang XK, Papas TS, Vesely J, Willingham MC, Green JE, Schweinfest CW. The down-regulated in adenoma (DRA) gene encodes an intestine-specific membrane glycoprotein. Oncogene. 1996;12:387–396. [PubMed] [Google Scholar]
- 98.Lohi H, Lamprecht G, Markovich D, Heil A, Kujala M, Seidler U, Kere J. Isoforms of SLC26A6 mediate anion transport and have functional PDZ interaction domains. American journal of physiology. 2003;284:C769–779. doi: 10.1152/ajpcell.00270.2002. [DOI] [PubMed] [Google Scholar]
- 99.Silberg DG, Wang W, Moseley RH, Traber PG. The Down regulated in Adenoma (dra) gene encodes an intestine-specific membrane sulfate transport protein. J Biol Chem. 1995;270:11897–11902. doi: 10.1074/jbc.270.20.11897. [DOI] [PubMed] [Google Scholar]
- 100.Saksena S, Singla A, Goyal S, Katyal S, Bansal N, Gill RK, Alrefai WA, Ramaswamy K, Dudeja PK. Mechanisms of transcriptional modulation of the human anion exchanger SLC26A3 gene expression by IFN-{gamma} Am J Physiol Gastrointest Liver Physiol. 2010;298:G159–166. doi: 10.1152/ajpgi.00374.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Saksena S, Dwivedi A, Singla A, Gill RK, Tyagi S, Borthakur A, Alrefai WA, Ramaswamy K, Dudeja PK. Characterization of the 5′-flanking region and regulation of expression of human anion exchanger SLC26A6. J Cell Biochem. 2008;105:454–466. doi: 10.1002/jcb.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Raheja G, Singh V, Ma K, Boumendjel R, Borthakur A, Gill RK, Saksena S, Alrefai WA, Ramaswamy K, Dudeja PK. Lactobacillus acidophilus stimulates the expression of SLC26A3 via a transcriptional mechanism. Am J Physiol Gastrointest Liver Physiol. 2010;298:G395–401. doi: 10.1152/ajpgi.00465.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alrefai WA, Wen X, Jiang W, Katz JP, Steinbrecher KA, Cohen MB, Williams IR, Dudeja PK, Wu GD. Molecular cloning and promoter analysis of downregulated in adenoma (DRA) Am J Physiol Gastrointest Liver Physiol. 2007;293:G923–934. doi: 10.1152/ajpgi.00029.2007. [DOI] [PubMed] [Google Scholar]
- 104.Kere J, Sistonen P, Holmberg C, de la Chapelle A. The gene for congenital chloride diarrhea maps close to but is distinct from the gene for cystic fibrosis transmembrane conductance regulator. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:10686–10689. doi: 10.1073/pnas.90.22.10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Waldegger S, Moschen I, Ramirez A, Smith RJ, Ayadi H, Lang F, Kubisch C. Cloning and characterization of SLC26A6, a novel member of the solute carrier 26 gene family. Genomics. 2001;72:43–50. doi: 10.1006/geno.2000.6445. [DOI] [PubMed] [Google Scholar]
- 106.Simon TC, Roberts LJ, Gordon JI. A 20-nucleotide element in the intestinal fatty acid binding protein gene modulates its cell lineage-specific, differentiation-dependent, and cephalocaudal patterns of expression in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:8685–8689. doi: 10.1073/pnas.92.19.8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Collado MC, Isolauri E, Salminen S, Sanz Y. The impact of probiotic on gut health. Curr Drug Metab. 2009;10:68–78. doi: 10.2174/138920009787048437. [DOI] [PubMed] [Google Scholar]
- 108.Liu KD, Gaffen SL, Goldsmith MA. JAK/STAT signaling by cytokine receptors. Curr Opin Immunol. 1998;10:271–278. doi: 10.1016/s0952-7915(98)80165-9. [DOI] [PubMed] [Google Scholar]
- 109.Decker T, Lew DJ, Mirkovitch J, Darnell JE., Jr Cytoplasmic activation of GAF, an IFN-gamma-regulated DNA-binding factor. Embo J. 1991;10:927–932. doi: 10.1002/j.1460-2075.1991.tb08026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Levy DE. Analysis of interferon-regulated proteins binding the interferon-alpha-stimulated response element. Methods. 1998;15:167–174. doi: 10.1006/meth.1998.0621. [DOI] [PubMed] [Google Scholar]
- 111.Schreiber S, Rosenstiel P, Hampe J, Nikolaus S, Groessner B, Schottelius A, Kuhbacher T, Hamling J, Folsch UR, Seegert D. Activation of signal transducer and activator of transcription (STAT) 1 in human chronic inflammatory bowel disease. Gut. 2002;51:379–385. doi: 10.1136/gut.51.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Clavell M, Correa-Gracian H, Liu Z, Craver R, Brown R, Schmidt-Sommerfeld E, Udall J, Jr, Delgado A, Mannick E. Detection of interferon regulatory factor-1 in lamina propria mononuclear cells in Crohn’s disease. J Pediatr Gastroenterol Nutr. 2000;30:43–47. doi: 10.1097/00005176-200001000-00016. [DOI] [PubMed] [Google Scholar]