Abstract
Eubacteria inactivate their ribosomes as 100S dimers or 70S monomers upon entry into stationary phase. In Escherichia coli, 100S dimer formation is mediated by ribosome modulation factor (RMF) and hibernation promoting factor (HPF), or alternatively, the YfiA protein inactivates ribosomes as 70S monomers. Here, we present high-resolution crystal structures of the Thermus thermophilus 70S ribosome in complex with each of these stationary-phase factors. The binding site of RMF overlaps with that of the messenger RNA (mRNA) Shine-Dalgarno sequence, which prevents the interaction between the mRNA and the 16S ribosomal RNA. The nearly identical binding sites of HPF and YfiA overlap with those of the mRNA, transfer RNA, and initiation factors, which prevents translation initiation. The binding of RMF and HPF, but not YfiA, to the ribosome induces a conformational change of the 30S head domain that promotes 100S dimer formation.
Bacteria slow down protein synthesis during nutrient starvation by converting ribosomes into translationally inactive 100S dimers (1) or 70S monomers (2), enter a stationary phase, and cease to grow. In this phase, bacterial cells are resistant to external stresses, which allows them to resist antimicrobial agents (3) and to engage in increased mutagenesis (4). Expression of stationary-phase proteins, such as ribosome modulation factor (RMF) (1) and hibernation promoting factor (HPF) (5), results in formation of 100S dimer, which leads to “ribosome hibernation” that aids cell survival.
In Escherichia coli (Eco) RMF and HPF proteins are encoded by the rmf and hpf genes, respectively. In contrast to the rmf gene, deletion of the hpf gene neither affects cell viability nor affects dimer formation in vivo during the stationary phase (5). The binding of RMF both in vivo and in vitro causes dimerization of 70S ribosomes into 90S particles (6), which are then stabilized as 100S dimers upon HPF binding (6). The 90S and 100S particles are both dimers but have different sedimentation coefficients. Negative staining (7) and cryo–electron microscopy (cryo-EM) studies (8, 9) showed that the two 70S ribosomes in a 100S dimer are linked together via their small subunits in vivo. Unlike RMF, HPF alone cannot induce ribosome dimerization (6).
An additional stationary-phase protein, YfiA, encoded by the yfiA gene, promotes the formation of translationally inactive monomeric 70S ribosomes (10), prevents the recycling of ribosomes for translation initiation. Although YfiA and HPF share a 40% sequence similarity (6), they have different effects on 100S dimer formation: HPF converts 90S into 100S particles, whereas YfiA prevents RMF-dependent 90S formation (6). The low-resolution electron density maps of YfiA (11.5 Å) and PSRP1 (8.5 Å), a chloroplast-specific YfiA homolog, bound to the ribosome, suggest that they can interfere with protein synthesis because their binding sites overlap with those of transfer RNAs (tRNAs) on the 30S subunit (11, 12). Although the structural models of these stationary-phase proteins exist, high-resolution structures of each of these proteins in complex with the ribosome are required to understand their mechanisms of action.
Here, we present three high-resolution crystal structures of the Thermus thermophilus (Tth) 70S ribosome in complex with RMF, HPF, or YfiA that were refined by using data extending to 3.0 Å (I/σI = 1), 3.1 Å (I/σI = 1), and 2.75 Å (I/σI = 1) resolution, respectively. The resolutions at which I/σI = 2 are 3.2 Å, 3.4 Å, and 2.9 Å, respectively. The structure of each complex was solved by molecular replacement using, as the starting model, the atomic coordinates of both ribosomal subunits from the published structure of the Tth 70S ribosome with ligands removed (13). For all complexes, the initial unbiased difference electron density maps calculated with the Fobs – Fcalc amplitudes showed positive density corresponding to the known structures of each of the hibernation factors (Fig. 1, A to C, and fig. S1, A, C, and E). The statistics for data processing and refinement of all complexes are shown in table S1 (14).
Fig. 1.
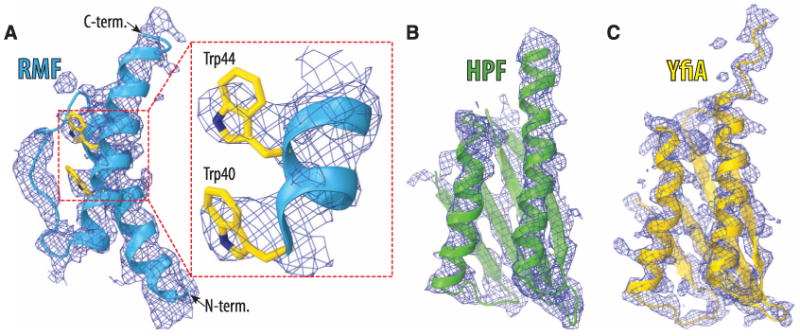
Unbiased (Fobs – Fcalc) difference Fourier maps of hibernation factors in complex with the T. thermophilus 70S ribosomes. The unbiased difference electron densities are contoured at 2.2, 2.5, and 2.7 σ for RMF (A), HPF (B) and YfiA (C), respectively. The refined models of RMF (blue), HPF (green), and YfiA (yellow) are displayed in their respective electron densities for clarity. The inset in (A) is a close-up view of the RMF electron density in the region between Trp40 and Trp44.
The 3.0 Å resolution crystal structure of RMF bound to the Tth 70S ribosome reveals that RMF binds next to the 3′ end of the 16S ribosomal RNA (rRNA) at the anti–Shine-Dalgarno (anti-SD) region between the head and the platform domains in the mRNA exit channel (Fig. 2, A and B, and movie S1). It interacts with three nucleotides of the 16S rRNA that are upstream of the anti-SD sequence (Fig. 2C). This position of RMF is incompatible with the SD of the mRNA making interactions with the anti-SD (Fig. 2D). During the first stages of initiation of protein synthesis, the mRNA forms a double helix with the anti-SD sequence of the 3′ end of the 16S rRNA (15, 16). RMF thus prevents initiation of protein synthesis by sterically interfering with the formation of the double helix between the anti-SD and the SD sequence of an incoming mRNA.
Fig. 2.
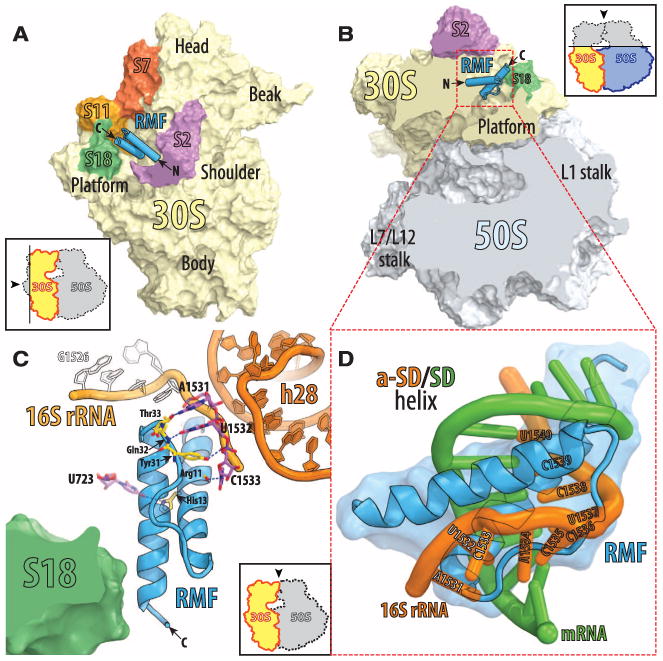
The structure of RMF bound to the 70S ribosome. (A) RMF (blue) bound to the 70S ribosome viewed from the cytosolic side of the 30S subunit (light yellow). As indicated by the inset, the 50S subunit and part of ribosomal protein S2 (magenta) are omitted for clarity. The ribosomal proteins surrounding the RMF binding site are colored red for S7, orange for S11, and green for S18. (B) The same as (A) but viewed from the top after removing the head of the 30S subunit (light yellow) and the protuberances of the 50S subunit (light blue) as indicated by the inset. (C) Detailed interactions of RMF with components of the 30S subunit. Different hues of orange indicate parts of the 16S rRNA: light orange for the 3′ end and dark orange for helix 28. C-terminal helix of the RMF comes close enough to ribosomal protein S18 to allow direct contacts, although specific interactions were not identified because of disorder of the S18 side chains. (D) A close-up view of the sterical clash between an mRNA (green) involved in the Shine-Dalgarno interactions (25) (PDB entry: 2HGR) and ribosome-bound RMF (blue). The anti-SD part of 16s rRNA is in orange. Residues 1531 to 1540 of the 16S rRNA are indicated.
The position of RMF in our complex contradicts the conclusions from previous cross-linking (7) and chemical-probing experiments (17), which were interpreted to suggest that the RMF binding site is located in the vicinity of the peptidyl transferase center (PTC) on the 50S sub-unit (fig. S2, A and B). Our structural data are not consistent with the formation of cross-links between RMF and ribosomal proteins S13, L2, and L13 that were reported. However, no cross-links were observed between RMF and other ribosomal proteins that are located closer to the PTC and are accessible to a cross-linking agent.
The results from chemical probing suggested that binding of RMF caused conformational changes to several bases in the 50S subunit that are located around the PTC (17). However, in that study, only regions of 23S rRNA closer to the PTC were selected for analysis; the 16S rRNA was not analyzed. Therefore, those experiments do not exclude the possibility that RMF does bind to the 30S subunit. Moreover, we observed that the 30S subunits dimerize in vitro upon binding RMF, which is in agreement with the presumed location of the RMF binding on the 30S and not the 50S subunit (fig. S3B). Additionally, the only unassigned electron density in both recent cryo-EM reconstructions of 100S dimers (8, 9) coincides with the binding site of RMF on the 30S subunit, as observed in our structure.
The binding of RMF to the ribosome induces a conformational change (Fig. 3) that facilitates the formation of 100S ribosome dimers in which the 30S subunits of the two 70S ribosomes interact to form a two-fold symmetrical particle (8, 9) with two prominent contact regions: One is centered around ribosomal protein S2, and the other is between the head domains (fig. S4A). The dimerization is not mediated by the direct contact between the RMF molecules, as they are ∼100 Å apart in our 100S dimer model and are not located at the dimer interface (fig. S4B). Therefore, we hypothesize that RMF promotes the formation of a 100S dimer by inducing the movement of the head domains of the 30S subunits toward each other (movie S1). In our structure, the head domain of the 30S subunit is repositioned away from the central protuberance of the 50S subunit to an orientation that increases the contact surface between the head domains, and this increased contact promotes formation of the 100S dimer (Fig. 3). The rotation axis of the head domain is perpendicular to the one observed in recent studies of the ratcheted ribosome (18) (movie S2). The binding of HPF to the ribosome induces the head domain of the 30S subunit to adopt a conformation similar to the one observed with bound RMF, consistent with its role in stabilizing 100S dimer formation. Neither the apo-state nor the YfiA-bound state of the ribosome stabilizes a dimerization-competent orientation of the small subumit.
Fig. 3.
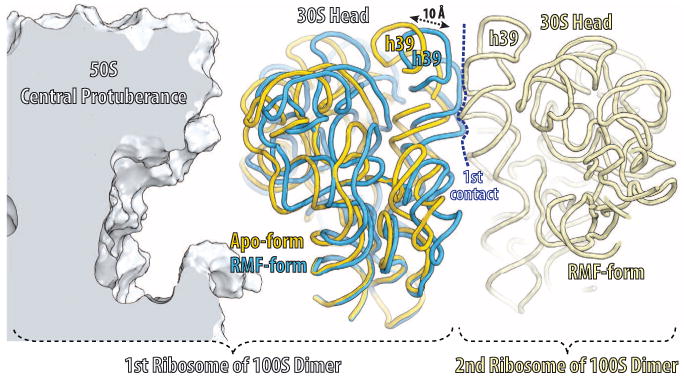
RMF-induced reorientation of the 30S-subunit head domain. The orientation of the two RMF-bound 70S monomers in the 100S dimer was derived from the published electron density for the 100S particle (9) (Electron Microscopy Data Bank entry: 1750). Parts of the first 70S monomer are displayed: 50S subunit (light blue), apo or YfiA-bound form of the 30S subunit (yellow); RMF-bound form of the 30S subunit (dark blue). Parts of the second 70S monomer are displayed in light yellow.
The electron density is observed for RMF bound to both copies of the ribosome in the asymmetric unit. Although the conformation of the head domain in one copy is compatible with 100S dimer formation, its orientation in the other copy is similar to the classical state (13), presumably because of important crystal contacts made by the head domain in this copy. The equilibrium between the apoand dimer-forming conformations of the head domain can be altered by a number of factors, e.g., monomeric 70S apo-ribosomes can be converted to 100S dimers simply by increasing the Mg2+ concentration in vitro (19). Note that one of the two contacts between the 30S subunits in the dimer (fig. S4, A and B) is similar to a crystal contact observed in crystals of the Tth 30S subunit (20).
The observed location of HPF (Fig. 4, A and B, and fig. S5) and YfiA (Fig. 4, C and D, and fig. S5) in our 3.1 Å and 2.75 Å resolution maps, respectively, is in general agreement with the published models derived from an 11.5 Å resolution map of the analogous complex between YfiA and the Eco ribosome (11), as well as with 8.5 Å resolution cryo-EM density map of a similar complex between chloroplast-specific YfiA homolog, PSRP1, bound to the Eco ribosome (12). In our model, HPF and YfiA are bound in the channel that lies between the head and the body of the 30S subunit where tRNAs and mRNA bind during protein synthesis (Fig. 4, E and F). Although the globular domains of HPF and YfiA have overlapping binding sites, HPF stabilizes the 100S ribosome dimer, whereas YfiA inhibits its formation (6). The inhibition is due to the extended C-terminal tail of YfiA, which HPF does not have, that blocks the binding of RMF and, thus, the RMF-induced dimer formation. The visible portion of the YfiA tail follows the mRNA channel, whereas the C-terminal end of the tail, which presumably projects into the RMF binding site (Fig. 4D), could not be modeled. This observation is consistent with previous biochemical studies suggesting mutually exclusive binding of YfiA and RMF to the 70S ribosome (5). Additionally, YfiA also prevents the formation of the 100S dimer by stabilizing the head domain of the small subunit in its apo-conformation.
Fig. 4.
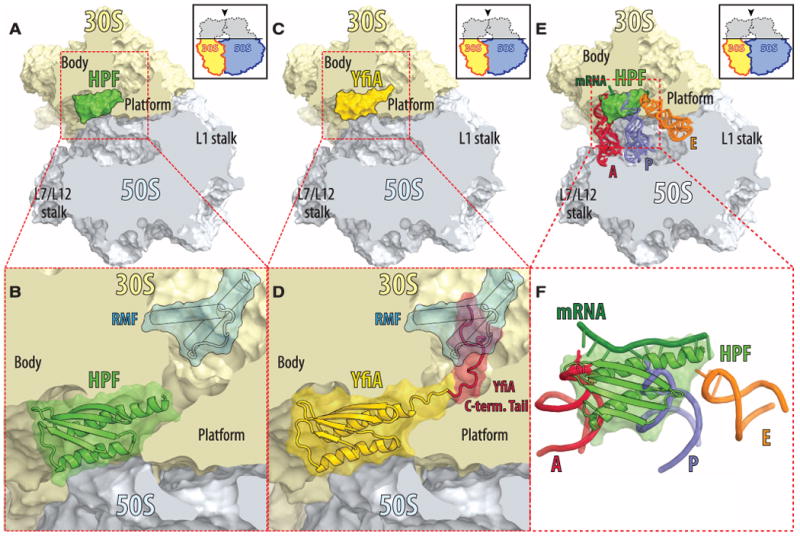
The structures of HPF and YfiA bound to the 70S ribosome. (A) HPF (green) bound in the mRNA channel between the head and the body of the 30S subunit (light yellow), viewed from the top after removing the head of the 30S subunit (light yellow) and the protuberances of the 50S subunit (light blue), as indicated by the inset. (B) Close-up view of (A) with the RMF protein superimposed (blue). (C) YfiA (yellow) bound to the 70S ribosome. The view is the same as in (A). (D) Close-up view of (C) with the RMF protein superimposed (blue) and the solution structure–derived model of the C-terminal tail of YfiA (red) (26). Electron density was observed only for the N-terminal part (residues 90 to 96) of the 24-residue tail; the remaining 17 residues are disordered. (E) Steric interference of HPF with the binding of mRNA and tRNAs. The structure of HPF (green) bound to the ribosome with superimposed mRNA (dark green) and tRNAs in the A site (red), P site (blue), and E site (orange) (13) (PDB entry: 2J00). View is the same as in (A). (F) A close-up view of (E) but the ribosome is omitted for clarity.
Our structures reveal that RMF and HPF can bind simultaneously and function together to interfere with the initiation of protein synthesis, which is consistent with the biochemical data (5). The HPF-YfiA binding site not only overlaps with all of the tRNA binding sites (Fig. 4, E and F) but also with the binding sites of the initiation factors IF1 and IF3 (fig. S6, A and B), which are directly involved in dissociation of the ribosomes into subunits (21), and elongation factor G (fig. S6, C and D), which assists ribosome recycling factor in dissociating posttermination complexes of 70S ribosomes (22). The inability of these factors to perform their function as a result of blocking by HPF or YfiA of their binding sites explains the reduced dissociation of the stationary-phase ribosomes into sub-units (10). Because RMF, HPF, and YfiA bind exclusively to the 30S subunit, they might not only interfere with initiation of protein synthesis starting on the 70S ribosomes, as in the case of leaderless mRNA (23) or during reinitiation along polycistronic mRNAs (24) but also with canonical initiation starting on the 30S subunits.
These studies show that these stationary-phase proteins, when bound to the ribosome, sterically clash with mRNA and tRNAs, and therefore, they cannot act on actively translating ribosomes. This ensures that the stationary-phase factors function only after the completion of the ongoing translation cycles and can only act effectively during stress and/or starvation conditions when the availability of the mRNA and/or tRNAs is limiting.
Acknowledgments
We thank the staff at the National Synchrotron Light Source (beamlines X29 and X25) and at the Advanced Photon Source (beamline 24ID) for help during data collection, the staff at the Center for Structural Biology at Yale University for computational support, and S. Seetharaman and M. Gagnon for valuable discussions. This work was supported by NIH grant GM022778 awarded to T.A.S. The structure factors and coordinates for both copies of the 70S ribosome in the asymmetric unit of all complexes have been deposited in the Research Collaboration for Structural Biology Protein Data Bank with the following accession codes: 3V22, 3V23, 3V24, and 3V25 for the RMF-ribosome complex; 3V26, 3V27, 3V28, and 3V29 for the HPF-ribosome complex; and 3V2C, 3V2D, 3V2F, and 3V2E for the YfiA-ribosome complex. T.A.S. owns stock in and is on the advisory board of Rib-X Pharmaceuticals, Inc., which does structure-based drug design targeted at the ribosome.
References and Notes
- 1.Wada A, Yamazaki Y, Fujita N, Ishihama A. Proc Natl Acad Sci USA. 1990;87:2657. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agafonov DE, Kolb VA, Spirin AS. EMBO Rep. 2001;2:399. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez JL, Baquero F. Antimicrob Agents Chemother. 2000;44:1771. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kivisaar M. Environ Microbiol. 2003;5:814. doi: 10.1046/j.1462-2920.2003.00488.x. [DOI] [PubMed] [Google Scholar]
- 5.Ueta M, et al. Genes Cells. 2005;10:1103. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 6.Ueta M, et al. J Biochem. 2008;143:425. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida H, et al. J Biochem. 2002;132:983. doi: 10.1093/oxfordjournals.jbchem.a003313. [DOI] [PubMed] [Google Scholar]
- 8.Kato T, et al. Structure. 2010;18:719. doi: 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Ortiz JO, et al. J Cell Biol. 2010;190:613. doi: 10.1083/jcb.201005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agafonov DE, Kolb VA, Nazimov IV, Spirin AS. Proc Natl Acad Sci USA. 1999;96:12345. doi: 10.1073/pnas.96.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JH. Nat Struct Mol Biol. 2004;11:1054. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 12.Sharma MR, et al. J Biol Chem. 2010;285:4006. doi: 10.1074/jbc.M109.062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selmer M, et al. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 14.Materials and methods are available as supplementary materials on Science Online.
- 15.Shine J, Dalgarno L. Proc Natl Acad Sci USA. 1974;71:1342. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steitz JA, Jakes K. Proc Natl Acad Sci USA. 1975;72:4734. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida H, Yamamoto H, Uchiumi T, Wada A. Genes Cells. 2004;9:271. doi: 10.1111/j.1356-9597.2004.00723.x. [DOI] [PubMed] [Google Scholar]
- 18.Jin H, Kelley AC, Ramakrishnan V. Proc Natl Acad Sci USA. 2011;108:15798. doi: 10.1073/pnas.1112185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tissières A, Watson JD, Schlessinger D, Hollingworth BR. J Mol Biol. 1959;1:221. [Google Scholar]
- 20.Wimberly BT, et al. Nature. 2000;407:327. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 21.Pavlov MY, Antoun A, Lovmar M, Ehrenberg M. EMBO J. 2008;27:1706. doi: 10.1038/emboj.2008.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janosi L, Hara H, Zhang SJ, Kaji A. Adv Biophys. 1996;32:121. doi: 10.1016/0065-227x(96)84743-5. [DOI] [PubMed] [Google Scholar]
- 23.Moll I, Hirokawa G, Kiel MC, Kaji A, Bläsi U. Nucleic Acids Res. 2004;32:3354. doi: 10.1093/nar/gkh663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karamyshev AL, Karamysheva ZN, Yamami T, Ito K, Nakamura Y. Biochimie. 2004;86:933. doi: 10.1016/j.biochi.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Nature. 2006;444:391. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 26.Rak A, Kalinin A, Shcherbakov D, Bayer P. Biochem Biophys Res Commun. 2002;299:710. doi: 10.1016/s0006-291x(02)02721-3. [DOI] [PubMed] [Google Scholar]


