Abstract
Targeting the tumor suppressor p53 to the mitochondria triggers a rapid apoptotic response as efficiently as transcription-dependent p53.1, 2 p53 forms a complex with the anti-apoptotic Bcl-XL, which leads to Bak and Bax oligomerization resulting in apoptosis via mitochondrial outer membrane permeabilization.3, 4 Although p53 performs its main role in the mitochondrial outer membrane it also interacts with different proteins in the mitochondrial inner membrane and matrix.5, 6 To further investigate mitochondrial activity of p53, EGFP-p53 was fused to different mitochondrial targeting signals (MTSs) directing it to the mitochondrial outer membrane (“XL-MTS” from Bcl-XL; “TOM-MTS” from TOM20), the inner membrane (“CCO-MTS” from cytochrome c oxidase) or matrix (“OTC-MTS” from ornithine transcarbamylase). Fluorescence microscopy and a p53 reporter dual luciferase assay demonstrated that fusing MTSs to p53 increased mitochondrial localization and nuclear exclusion depending on which MTS was used. To examine if the MTSs initiate mitochondrial damage, we fused each individual MTS to EGFP (a non-toxic protein) as negative controls. We performed caspase-9, TUNEL, Annexin-V, and 7-AAD apoptosis assays on T47D breast cancer cells transfected with mitochondrial constructs. Except for EGFP-XL, apoptotic potential was observed in all MTS-EGFP-p53 and MTS-EGFP constructs. In addition, EGFP-p53-XL showed the greatest significant increase in programmed cell death compared to its non-toxic MTS control (EGFP-XL). The apoptotic mechanism for each construct was further investigated using pifithrin-α (an inhibitor of p53 transcriptional activity), pifithrin-μ (a small molecule that reduces binding of p53 to Bcl-2 and Bcl-XL), and over-expressing the anti-apoptotic Bcl-XL. Unlike the MTSs from TOM, CCO, and OTC, which showed different apoptotic mechanisms, we conclude that p53 fused to the MTS from Bcl-XL performs its apoptotic potential exclusively through p53/Bcl-XL specific pathway.
Keywords: p53, mitochondria, Bcl-XL, apoptosis, pifithrin, T47D
Introduction
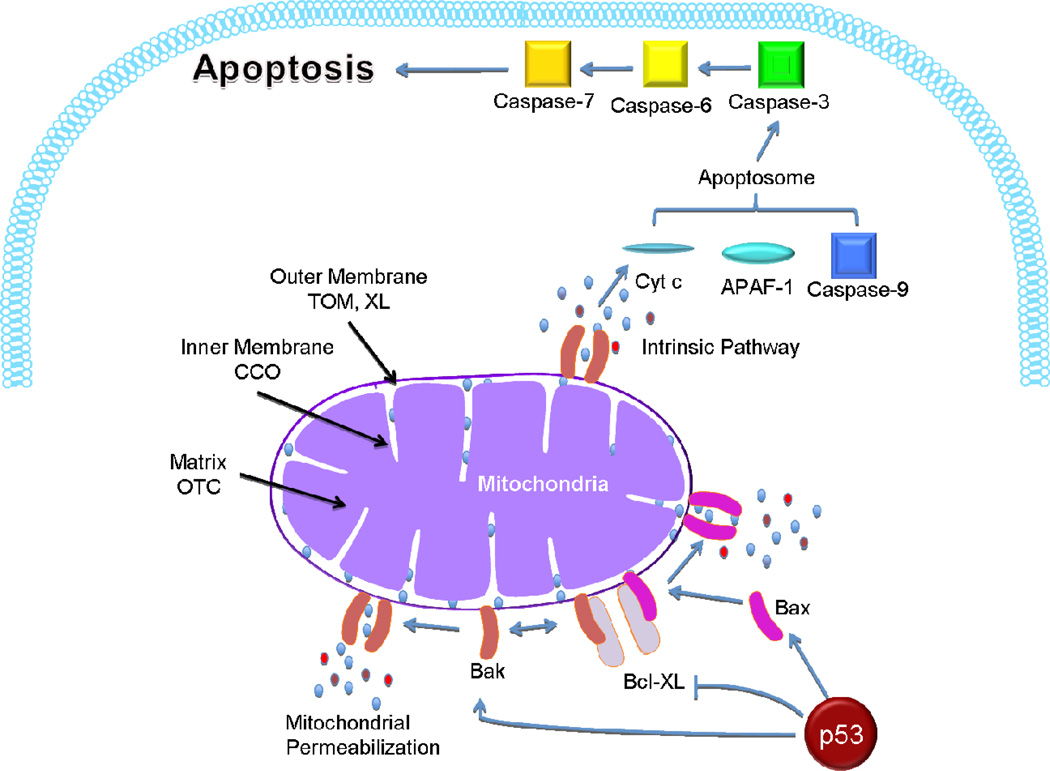
The tumor suppressor p53 stimulates a wide network of signals involved in DNA repair, cell cycle arrest, senescence and apoptosis.7–9 Although most of these effects can be linked to its role as a transcription factor, recent work has clearly demonstrated that p53 can cause apoptosis through its transcription-independent mitochondrial pathway.3, 10 A small but highly reproducible fraction of p53 translocates to the mitochondria at the onset of p53-dependent apoptosis.10 Translocation of p53 to the mitochondrial outer membrane triggers the release of cytochrome c and procaspase-3 activation. The DNA binding domain of p53 (DBD, residues 239–248) forms inhibitory complexes with anti-apoptotic Bcl-XL and Bcl-2 proteins (Figure 1), which are located in the mitochondrial outer membrane.11 This induces oligomerization of Bak and Bax allowing them to form supramolecular pores.4, 12, 13 In addition, p53 activates, directly binds to, and induces oligomerization of Bak and Bax.4, 13, 14 The formation of permeability transition pores causes outer membrane rupture and releases cytochrome c from the intramembranous space into the cytosol triggering apoptosis via the apoptosome formation (Figure 1).15–17 In addition, targeting p53 to the mitochondria triggers apoptosis faster than the transcription-dependent nuclear pathway.1, 2 In most cancer cells, p53 is not able to bind to Bcl-2 proteins due to missense mutations in the DBD of p53, demonstrating the importance of the DBD in mitochondrial apoptosis.11
Figure 1.
The mitochondrial apoptotic pathway of p53. When p53 is targeted to the mitochondria, it interacts with anti-apoptotic Bcl-XL, enables Bax and Bak oligomerization, and activates the intrinsic apoptotic pathway. The apoptosome (cytochrome c, APAF-1 and caspase-9) is triggered, leading to apoptosis via activation of caspases-3, -6, and -7. The left side of the diagram indicates the mitochondrial signals (MTSs) used and the different subsections of the mitochondria targeted, including the outer membrane (“XL-MTS” from Bcl-XL; “TOM-MTS” from TOM20), the inner membrane (“CCO-MTS” from cytochrome c oxidase), and the matrix (“OTC-MTS” from ornithine transcarbamylase).
The therapeutic effect of pharmaceutical agents such as p53 may differ depending on their intracellular delivery.18 Precise compartmentalization and sub-compartmentalization of proteins are essential for their biological activity. The majority of endogenous mitochondrial p53 localizes to the outer surface of the mitochondria,19 which occurs after either DNA damage or hypoxic stress.10 A detailed mechanism on how p53 translocates into the mitochondria is still under investigation since it is a nuclear protein due to its nuclear localization signals.20 A study has shown that Bad (Bcl-2 antagonist of cell death) physically interacts with cytoplasmic p53 directing it to the surface of the mitochondria.21, 22 Moll has suggested that p53 is imported via mtHsp70 targeting the membranous compartments.10 The interaction with mtHsp70 and Hsp60 is increased with the R72P polymorphism in p53 due to elevation in nuclear export of p53.23 MDM2 protein is responsible for the nuclear exclusion and ubiquitylation of p53. The monoubiquitylated p53 is targeted to the mitochondria under stress, which disrupts the p53/MDM2 complex.24
Despite what is known, exogenous mitochondrial targeting of p53 is still under investigation. To improve apoptosis via the p53/Bcl-XL pathway, p53 was delivered to different mitochondrial compartments. In this study, p53 was fused to different mitochondrial targeting signals (MTSs) targeting the mitochondrial outer membrane [MTS from Bcl-XL and MTS from the translocase of the outer membrane (TOM 20)], inner membrane (MTS from cytochrome c oxidase subunit VIII) and matrix (MTS from ornithine transcarbamylase) (Figure 1). These MTSs (Table 1) are generally thought to form α-helices, and are important for their recognition by translocation machineries in the mitochondrial outer (TOM complex) and inner (TIM complex) membranes.25–28 Since MTSs are mainly found on the amino terminus of mitochondrial proteins.25 All MTSs used were cloned on the amino terminus except for XL, which is inherently found on the carboxy terminus.29 XL targets the outer surface of the mitochondria29–32 while TOM is inserted into the outer membrane;29 CCO translocates the outer membrane and then becomes imbedded into the inner membrane33, 34 while the OTC crosses both membranes and translocates to the matrix.3, 35 The purpose of this work is to determine the ideal MTS for p53 mitochondrial targeting to induce the intrinsic apoptotic pathway.
Table 1.
A list of the mitochondrial targeting signals used in this paper. TOM and XL target the mitochondrial outer membrane, CCO translocates to the mitochondrial inner membrane, and the OTC localizes to the mitochondrial matrix.
| Protein | Compartment | MTS Sequence |
|---|---|---|
| XL | Outer Membrane | RGPGIQKGPGEIQQMVPDRHDRGRRGAAGQPVQQKX |
| TOM | Outer Membrane | MVGRNSAIAAGVCGALFIGYCIYFDRKRRSDPN |
| CCO | Inner Membrane | MSVLTPLLLRGLTGSARRLPVPRAKIHSL |
| OTC | Matrix | MLFNLRILLNNAAFRNGHNFMVRNFRCGQPLQ |
Materials and Methods
EGFP-p53 Plasmid (pEGFP-p53)
The DNA encoding p53 was amplified through PCR from pCMV-p53 wt (a generous gift from Dr. S. J. Baker, Addgene, Cambridge, MA) using the primers 5′-GCGCGCGCGCTCCGGAGCCATGGAGGAGCCGCAGT-3′ and 5′-GCGCGCGCGCGGTACCTCAGTCTGAGTCAGGCCCTTCTGTC-3′. This was subcloned into the BspEI and KpnI restriction enzyme sites in pEGFP-C1 (Clontech, Mountain View, CA).
pOTC-EGFP-p53 Plasmid
An oligonucleotide encoding the mitochondrial targeting signal from OTC (including the Kozak region), 5′-CCGGTCGCCACCATGCTGTTTAATCTGAGGATCCTGTTAAACAATGCAGCTTTTAGAAATGGTCACAACTTCATGGTTCGAAATTTTCGGTGTGGACAACCACTACAAAATAAAGTGCAGCGA-3′ was annealed to its complementary strand. This was then cloned at the amino terminus of EGFP-p53 at the AgeI site.
pTOM-EGFP-p53 and pCCO-EGFP-p53 Plasmids
The OTC region from pOTC-EGFP-p53 was replaced with annealed oligonucleotides encoding the mitochondrial targeting signal from the TOM20 complex (TOM) at the AgeI site, 5′-GATCCCCGGTCGCCACCATGGTGGGTCGGAACAGCGCCATCGCCGCCGGTGTATGCGGGGCCCTTTTCATTGGGTACTGCATCTACTTCGACCGCAAAAGACGAAGTGACCCCAACCGA-3′ and its reverse complement, or with oligonucleotides encoding the mitochondrial targeting signal from the cytochrome c oxidase (CCO) at the AgeI site, 5′-CCGGTCGCCACCATGTCCGTCCTGACGCCGCTGCTGCTGCGGGGCTTGACAGGCTCGGCCCGGCGGCTCCCAGTGCCGCGCGCCAAGATCCATTCGTTGA-3′ and its reverse complement.
pEGFP-p53-XL Plasmid
The mitochondrial signal from Bcl-XL was fused to the carboxy terminus of EGFP-p53 using the BamHI restriction site as follows. The XL oligonucleotide 5′-AGAAAGGGCCAGGAGAGATTCAACAGATGGTTCCTGACCGGCATGACCGTGGCCGGCGTGGTGCTGCTGGGCAGCCTGTTCAGCAGAAAGTGA-3′ was annealed to its complimentary strand (with BamHI sticky ends). The p53 stop codon was then mutated (TGA to TTA) in pEGFP-p53-XL using the primers 5′-GAAGGGCCTGACTCAGACTTAGGTACCGCGGGCCCGGGAT-3′ and the reverse complement.
pEGFP Constructs with MTSs
Plasmids encoding OTC-EGFP, TOM-EGFP, CCO-EGFP, and EGFP-XL were constructed using the same oligonucleotides and restriction sites mentioned above but inserted in pEGFP-C1 instead of pEGFP-p53.
MTS-EGFP-p53NLSmut
Mutations (K319T and K320T) in the nuclear localization signal (NLS) of p53 were introduced in all mitochondrial p53 constructs via QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA) using the primers 5′-CTCTCCCCAGCCAACGACGAAACCACTGG -3′ and its reverse complement.
Cell Lines and Transient Transfections
1471.1 murine adenocarcinoma cells (gift of G. Hager, NCI, NIH), MCF-7 human breast adenocarcinoma cells (ATCC, Manassas, VA), and T47D human ductal breast epithelial tumor cells (ATCC) were grown as monolayers in DMEM (1471.1) or RPMI (MCF-7 and T47D) (Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (Invitrogen), 1% penicillin-streptomycin-glutamine (Invitrogen), and 0.1% gentamicin (Invitrogen). In addition, T47D and MCF-7 media was supplemented with 4 mg/L insulin (Sigma, St. Louis, MO). The cells were maintained in a 5% CO2 incubator at 37°C. 7.5 × 104 cells for 1471.1 and 3.0 × 105 cells for MCF-7 and T47D were seeded in 6-well plates (Greiner Bio-One, Monroe, NC) or 2-well live cell chambers (Nalgene Nunc, Rochester, NY). Transfections were carried out 24 hours after seeding using Lipofectamine 2000 (Invitrogen) following the manufacturer’s recommendations. Unless otherwise indicated, 1 pmol DNA was transfected per well for all assays.
Mitochondrial Staining, Microscopy, and Image Analysis
Prior to live-cell imaging and mitochondrial staining, media in live cell chambers was replaced with phenol red-free DMEM (Invitrogen) for 1471.1 cells or phenol red-free RPMI (Invitrogen) for T47D and MCF-7 cells containing 10% charcoal-stripped fetal bovine serum (CS-FBS, Invitrogen). Cells were incubated with 250 nM MitoTracker Red FM (Invitrogen) for 15 min at 37°C and protected from light. Images were acquired as previously,36 using an Olympus IX71F fluorescence microscope (Scientific Instrument Company, Aurora, CO) with high-quality narrow band GFP filter (ex: HQ480/20 nm, em: HQ510/20 nm) and HQ:TRITC filter (ex: HQ545/30, em: HQ620/60) from Chroma Technology (Brattleboro, VT) with a 40X PlanApo oil immersion objective (NA 1.00) on an F-View Monochrome CCD camera. Images were analyzed for mitochondrial stain overlap with EGFP fusion constructs using ImageJ software and the JACoP plugin.37 JACoP was used to generate the colocalization statistic [i.e., Pearson’s correlation coefficient (PCC) post Costes’ automatic threshold algorithm],38, 39 as we have done before.40 PCC evaluates correlation between pairs of individual pixels from EGFP and MitoTracker stained cells. The higher the PCC value the higher the correlation. For increased visual clarity of mitochondrial localization of the EGFP-fused constructs, spatial representations of pixel intensity correlation have been generated using Colocalization Colormap (ImageJ).41 Microscopy was repeated in triplicate (n=3) and 10 cells were analyzed for each construct.
Luciferase Assay
All constructs (3.5 µg of DNA) were co-transfected with 3.5 µg of p53-Luc Cis-Reporter (encoding for firefly luciferase, Agilent) in T47D and MCF-7 cells. To normalize for transfection efficiency, 0.35 µg of pRL-SV40 plasmid (encoding for renilla luciferase, gift from Dr. Philip Moos, University of Utah) was co-transfected. The Dual-Glo Luciferase Assay System (Promega, Madison, WI) was used to determine firefly luciferase activity and renilla luciferase per manufacturer's instructions. Luciferase activity was detected 24 hours after transfection using PlateLumino (Stratec Biomedical Systems, Birkenfeld, Germany). Firefly luciferase values were normalized for renilla luciferase. EGFP-p53 served as a positive control and EGFP as a negative control. The Dual-Glo Luciferase Assay was run three times independently, each in triplicate.
Caspase-9 Assay
T47D cells were probed 19 hours after transfection using CaspaLux®9-M2D2 kit (OncoImmunin, Inc., Gaithersburg, MD) per manufacturer’s recommendations. The cells were then suspended in flow cytometry buffer (OncoImmunin, Inc.) and analyzed via the FACSAria-II (BD-BioSciences, University of Utah Core Facility) utilizing 488 nm (for EGFP) and 563 nm (for cleaved caspase 9 substrate) lasers. FACSDiva software was used as an evaluation tool. Only EGFP transfected cells at 507 nm emission were analyzed. All constructs were gated at the same EGFP intensity levels to ensure equal expression of proteins. The samples were detected in the PE (phycoerythrin) channel with the 580 nm emission peak. Each construct was assayed three times (n=3).
TUNEL Assay
T47D cells were prepared 48 hours after transfection using In Situ Death Detection Kit, TMR red (Roche, Mannheim, Germany) per the company’s protocol. The kit, labels the DNA single strand breaks (TUNEL reaction) in apoptotic cells. The FACSAria-II was used to analyze the cells suspended in PBS (Invitrogen). The same FACS settings mentioned above with the caspase-9 assay were used. Only EGFP positive cells were analyzed for DNA segmentation. Each construct was analyzed three times (n=3).
Annexin-V Assay
At 48 hours post transfection, T47D cells were assayed for Annexin-V binding. The cells were suspended in 100 µl Annexin binding buffer (Invitrogen) and incubated with 5 µl Annexin-APC (Annexin-V conjugated to allophycocyanin, Invitrogen) for 15 minutes. The incubated cells were then diluted in 400 µl Annexin binding buffer and analyzed using the FACSCanto-II (BD-BioSciences, University of Utah Core Facility) with FACSDiva software. EGFP was excited at 488 nm wavelength and detected at 507 nm. APC was excited with 635 nm laser and detected at 660 nm. Analysis was based on EGFP positive cells. All constructs were gated at the same EGFP intensity levels. Each construct was tested three times (n=3).
7-AAD Assay
T47D and MCF-7 cells were stained with 7-aminoactinomycin D (7-AAD, Invitrogen) according to manufacturer’s instructions 48 hours after transfection. The samples were analyzed using the FACSCanto-II (BD-BioSciences). Analyzed cells were gated for EGFP (as mentioned in Annexin-V assay). In addition, EGFP and 7-AAD were excited with the 488 nm laser. EGFP and 7-AAD were detected at 507 nm and 660 nm, respectively. Each construct was assayed three times (n=3).
Rescue Experiment Using Pifithrin-α
Six hours after transfection, T47D cells were incubated with previously optimized concentration of 40 µM pifithrin-α (Cayman Chemical, Ann Arbor, MI) for 42 hours and compared to transfected cells without pifithrin-α. At the 48 hour time point, the 7-AAD assay was performed as above.
Rescue Experiment Using Pifithrin-μ
Six hours after transfection, T47D cells were incubated with previously optimized concentration of 5 nM pifithrin-μ (Tocris Bioscience, Ellisville, MO) for 42 hours and compared to transfected cells without pifithrin-μ. At the 48 hour time point, the 7-AAD assay was performed as detailed above.
Rescue Experiment Using Bcl-XL
T47D Cells were co-transfected with 1 pmol of MTS constructs and 1 pmol of pBcl-XL (Addgene). After 48 hours, cells were pelleted and assayed with 7-AAD as described above.
Statistical Analysis
All experiments were repeated in triplicate (n=3). The data were presented as the mean ± standard error. Statistical differences between each MTS-EGFP-p53 and its MTS-EGFP were resolved via unpaired t-test using GraphPad Prism software. The MTS-EGFP controls were compared to EGFP by one-way ANOVA with Tukey’s post test. The degree of colocalization was analyzed using odds ratio with Pearson’s Chi-square. A p value <0.05 was considered significant.
Results
Mitochondrial localization of MTS-EGFP-p53
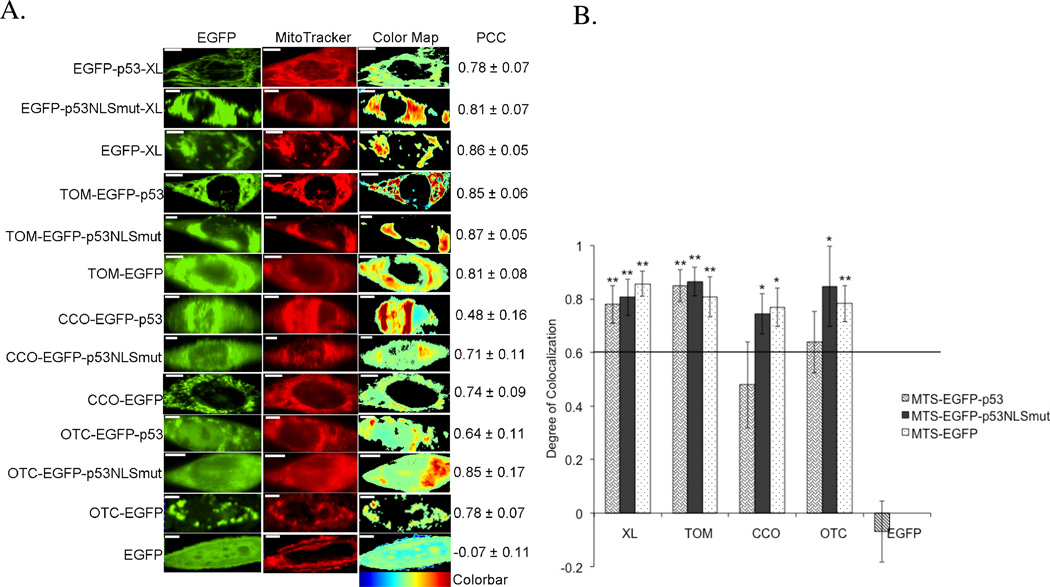
Mitochondrial targeting of all constructs was verified using fluorescence microscopy. Figure 2A shows representative 1471.1 cells, which are larger in size, spread well, and are generally easier to visualize versus T47D or MCF-7 cells. However, irrespective of the cell line, similar microscopy results were observed in T47D and MCF-7 cells (data not shown). To better illustrate the colocalization of the EGFP fused constructs, PCC values were generated and graphed for each construct (Figure 2B). PCC values range from +1 to −1; perfect correlation is represented by +1, anti-correlation by −1, and a PCC value of zero denotes random distribution.37 Following the example of Bolte and Cordelières, a PCC of 0.6 or greater will define colocalization, or co-compartmentalization (Figure 2B).37, 40 Fusing different MTSs to EGFP and p53 showed a high degree of colocalization with the mitochondria. EGFP-C1 served as negative control for colocalization analysis and as expected, there was no colocalization between EGFP alone and the mitochondria. EGFP and p53 tagged to TOM and XL targeted the mitochondria better than CCO-EGFP-p53 and OTC-EGFP-p53. CCO and OTC were the “weakest” MTSs since there was some nuclear targeting of both CCO-EGFP-p53 and OTC-EGFP-p53. Therefore we mutated K319T and K320T in the nuclear localization signal (NLS) of p5320 which resulted in increased mitochondrial targeting for CCO-EGFP-p53 and OTC-EGFP-p53. For easier visualization of colocalization among constructs, a spatial depiction of pixel overlap and intensity correlation are provided in the ‘Color Map’ column (Figure 2A). The Color Map spectrum moves from cold to warm colors as pixel correlation increases.41
Figure 2.
Colocalization of MTS constructs and MitoTracker Red mitochondrial stain in 1471.1 cells. A) Representative images of MTS-EGFP-p53, MTS-EGFP-p53 NLS mutation, MTS-EGFP and EGFP-C1 are shown in the left column with images of MitoTracker Red distribution in the middle column. The ‘EGFP’ and ‘MitoTracker’ columns have been false colored green and red, respectively. Enhanced visualization of colocalized pixels is rendered in the ‘Color Map’ column. Warm colors depict pixels with highly correlated intensity and spatial overlap while cool colors are indicative of anti- or random correlation (colorbar for interpretation is shown below column). Corresponding PCC values are shown in the right column. White scale bars are all 10 µm. B) The degree of colocalization is represented by PCC following Costes’ approach. 38, 39 All constructs with values higher than 0.6 are considered highly colocalized with mitochondrial stain MitoTracker Red. Statistical analysis was performed by using odds ratio with Pearson’s Chi-square. The adjusted odds ratio for PCC value of 0.6 were compared with each sample. *p<0.05, and **p<0.01 comparing odds ratio of lowest value for samples with odds ratio of 1 for PCC of 0.6.
Testing the transcriptional activity of MTS-EGFP-p53
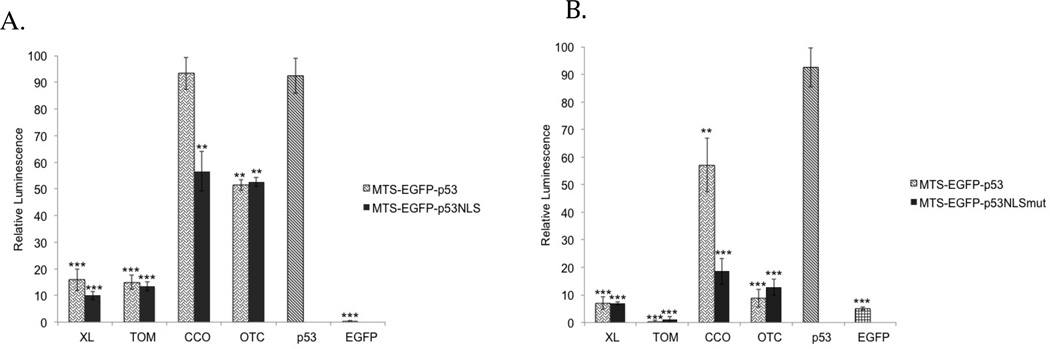
To demonstrate the lack of transcriptional activity of these p53 constructs, a p53 reporter dual luciferase assay was performed in T47D (Figure 3A) and MCF-7 cells (Figure 3B). T47D cells contain a mutation in p53 (in the DBD which renders it inactive) that is also localized in the cytoplasm42, 43 while MCF-7 cells express wild-type p53 mislocalized to the cytoplasm.44 TOM and XL fused to EGFP-p53 showed no nuclear activity in either cell lines. CCO-EGFP-p53 expressed similar transcriptional activity to EGFP-p53 (positive control) in T47D (Figure 3A) and significant activity in MCF-7 (Figure 3B) compared to the EGFP negative control. Introducing NLS mutations (K3319T and K320T) in CCO-EGFP-p53 resulted in major reduction of nuclear activity in both cell lines. OTC-EGFP-p53 showed low transcriptional activity in MCF-7 (Figure 3B) and a significant activation in T47D cells (Figure 3A). Surprisingly, introduction of NLS mutations into OTC-EGFP-p53 did not result in any changes in nuclear activity in either cell line (Figures 3A and B).
Figure 3.
Luciferase assay: All MTS-EGFP-p53 and MTS-EGFP-p53 NLS mutation constructs were tested for their ability to activate a p53 reporter in A) T47D cells and B) MCF-7 cells. EGFP-p53 serves as a positive control and EGFP as a negative control. All constructs were corrected to EGFP-p53 control, which is set at 100%. Statistical analysis was performed by using one-way ANOVA with Tukey’s post test. **p<0.005 and ***p<0.0005 compared to EGFP-p53.
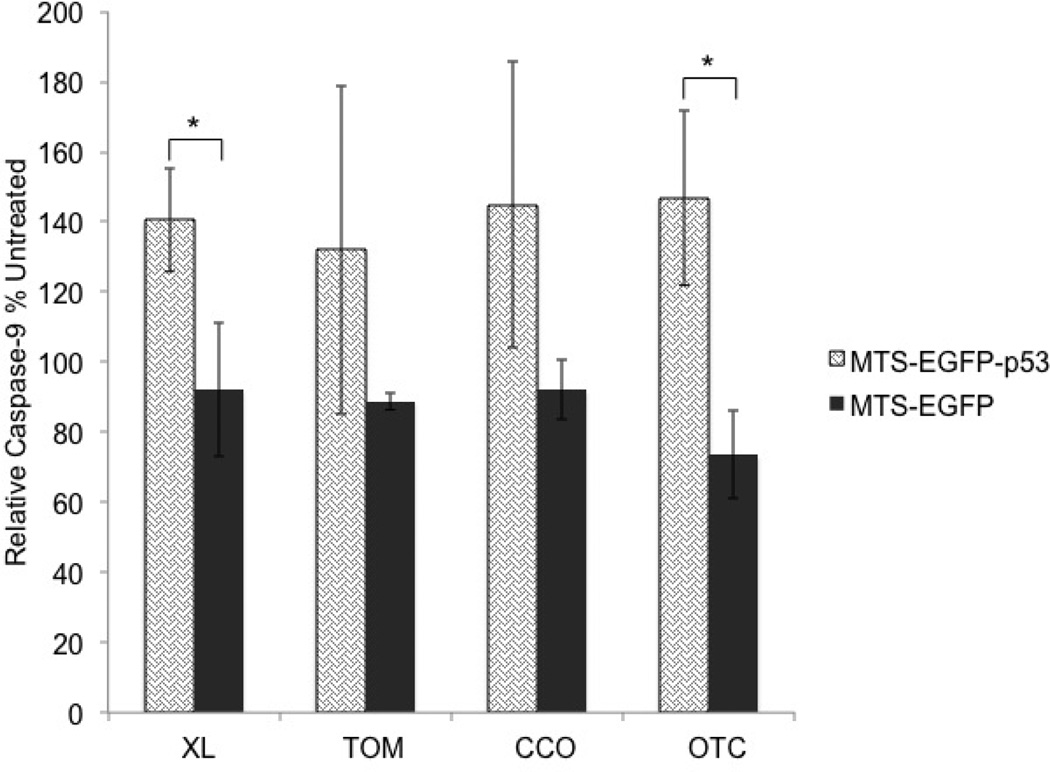
The effect of MTS-EGFP-p53 on early apoptosis (caspase-9)
In this paper we focused on T47D cells because they are more resistant to apoptosis compared to MCF-7.45 The apoptotic potential of p53 fused to different MTSs targeting the mitochondrial matrix, outer, and inner membranes was evaluated via caspase-9, TUNEL, Annexin-V and 7-AAD assays. Caspases are a group of proteolytic enzymes that are directly involved in apoptosis by cleaving proteins such as lamin and PARP. Its inactive form procaspase-9 is activated through cytochrome c release and APAF-1 which occurs after mitochondrial outer membrane disruption (known as the intrinsic apoptotic pathway).46 Caspase-9 itself cleaves the peptide sequence LEHD, which was used in the caspase-9 assay to measure the intrinsic apoptotic pathway.47 Only EGFP-p53-XL (p<0.05) and OTC-EGFP-p53 (p<0.05) were significantly different from their corresponding MTS-EGFP controls as shown in figure 4.
Figure 4.
The activation of caspase-9 was analyzed 19 hours following transfection of T47D cells. All constructs were corrected to untreated control, which is set at 100%. Statistical analysis was performed by using unpaired t-test. * p<0.05 for MTS-EGFP-p53 compared to its MTS-EGFP.
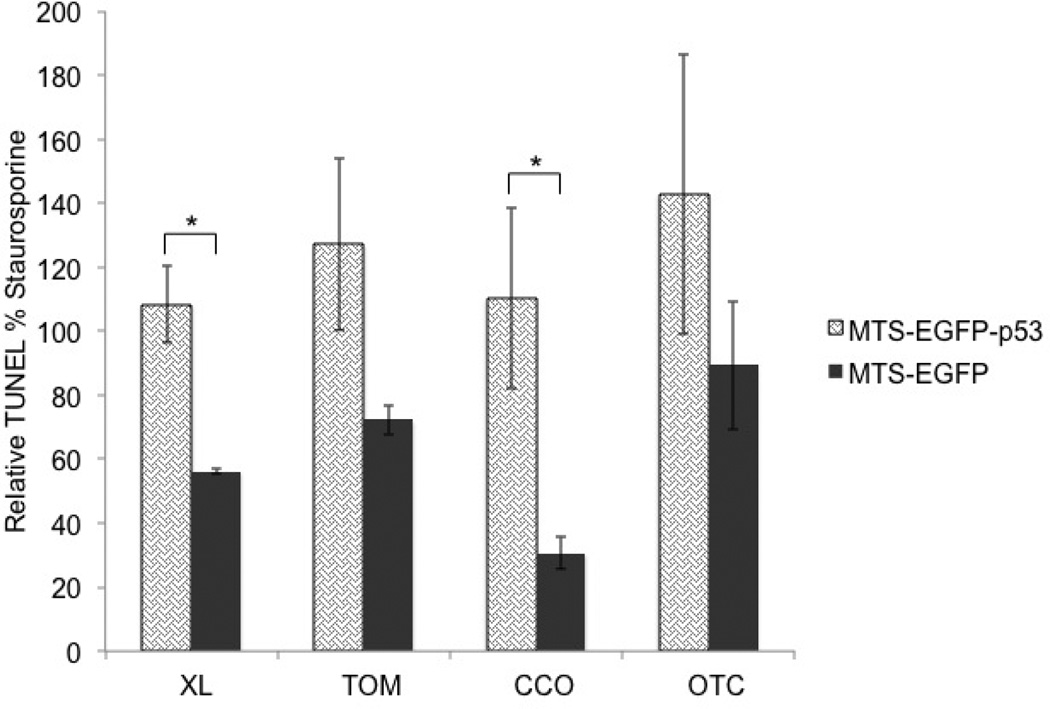
The effect of MTS-EGFP-p53 on DNA fragmentation
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) was measured to detect DNA fragmentation by labeling the terminal end of nucleic acids. DNA fragmentation is a hallmark of apoptosis and is generated by caspase cleavage.48 Figure 5 illustrates that EGFP-p53-XL and CCO-EGFP-p53 are the only constructs that were statistically significant from their control MTS-EGFP. Constructs with mutations in the NLS of p53 did not differ from constructs without mutation (see the supporting information: S1). Staurosporine, which activates caspase-3/7 and leads to DNA fragmentation,45 served as a positive control.
Figure 5.
T47D cells were tested 48 hours following transfection. DNA fragmentation was analyzed with the TUNEL assay. All constructs were corrected to staurosporine positive control, which is set at 100%. Statistical analysis was performed by unpaired t-test. * p<0.05 for MTS-EGFP-p53 compared to its MTS-EGFP.
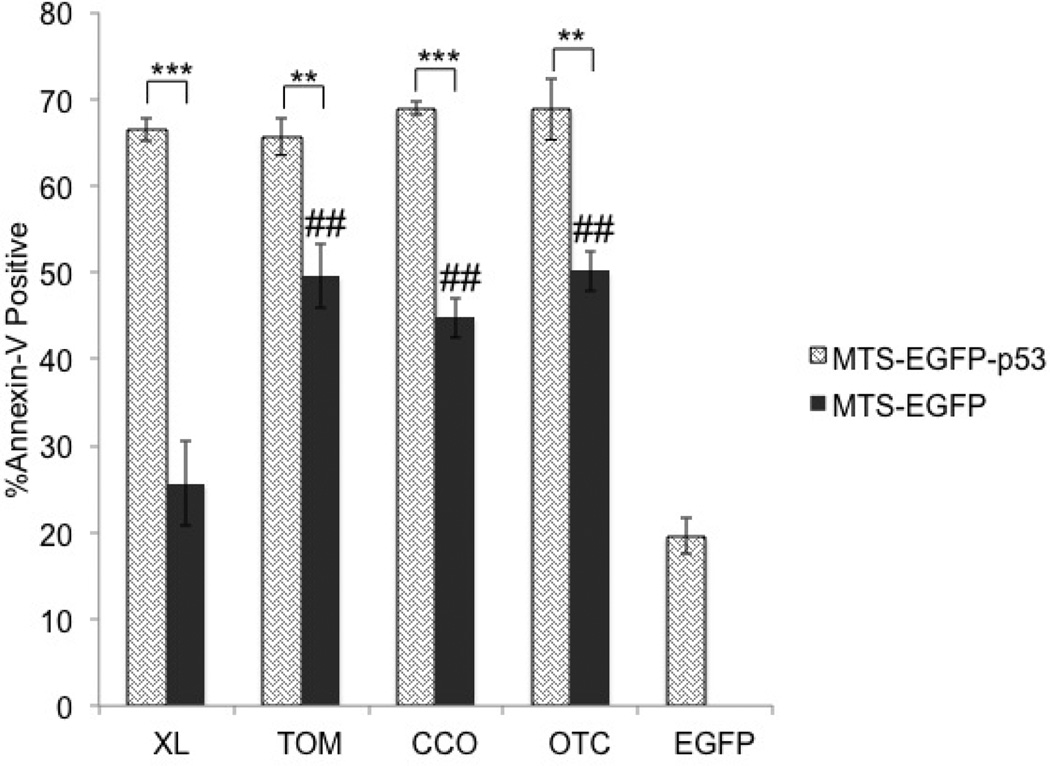
The effect of MTS-EGFP-p53 on plasma membrane
The effects of the mitochondrial constructs were further explored by analyzing externalization of phosphatidylserine and cell membrane rupture. Annexin-V was used to detect the externalization of phosphatidylserine on the cell surface of apoptotic cells via flow cytometry.49, 50 In the majority of healthy cells, the plasma membrane expresses phosphatidylserine on the cytosolic surface while in apoptotic cells, the phosphatidylserine is transported to the outer surface, which allows labeled Annexin-V to bind.51 All MTS-EGFP-p53 constructs showed a significant effect on inducing apoptosis compared to their corresponding MTS-EGFP controls in T47D cells (Figure 6). In addition, EGFP-XL was the only construct that showed minimal activity similar to the non-toxic EGFP negative control (Figure 6).
Figure 6.
Annexin-V assay was conducted in T47D cells 48 hours after transfection. Statistical analysis was performed by one-way ANOVA with Tukey’s post test. **p<0.005 and ***p<0.0005 comparing MTS-EGFP-p53 to their MTS-EGFP controls. The negative controls (MTS-EGFP) were compared to EGFP-C1 using one-way ANOVA with Tukey’s post test ##p<0.005.
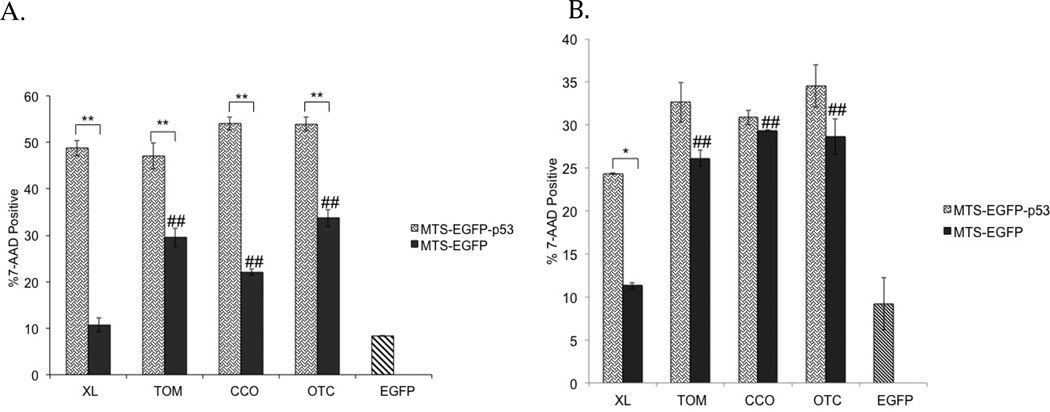
To further validate the effect of mitochondrial p53 on late apoptosis, the 7-AAD assay was performed via flow cytometry (Figure 7). The 7-AAD fluorescent marker cannot stain the DNA in healthy cells due to inability to penetrate an intact cell membrane.52 However, it is able to stain the DNA in apoptotic and necrotic cells because of their disrupted membrane.53 Similar to Annexin-V results, all mitochondrial p53 constructs showed significant 7-AAD intercalation with DNA compared to their MTS-EGFP controls (Figure 7A). In addition, EGFP-XL was the only construct that showed similar activity to EGFP alone (Figure 7A). In a cell line that is less resistant to apoptosis such as MCF-7,45 EGFP-p53-XL is the only construct that was significantly different than its MTS-EGFP control (Figure 7B). In MCF-7 cells, there was no statistical difference between p53 fused to TOM, CCO or OTC and their respective MTS-EGFP controls (Figure 7B). Mutating the NLS of p53 in MTS-EGFP-p53 had no effect on 7-AAD permeation in both cell lines except with CCO-EGFP-p53 in T47D (see the supporting information: S2). In addition, EGFP-XL showed no activity similar to EGFP alone in MCF-7 (Figure 7B). Similar to Annexin V, EGFP-XL was also the only construct with minimum activity as EGFP alone in T47D cells (Figure 7A).
Figure 7.
The 7-AAD assay was tested in (A) T47D and (B) MCF-7 cells 48 hours after transfection. Statistical analysis was performed by one-way ANOVA with Tukey’s post test. *p<0.05, **p<0.005 comparing MTS-EGFP-p53 to their MTS-EGFP controls. The controls (MTS-EGFP) were compared to EGFP-C1 using one-way ANOVA with Tukey’s post test ##p<0.005.
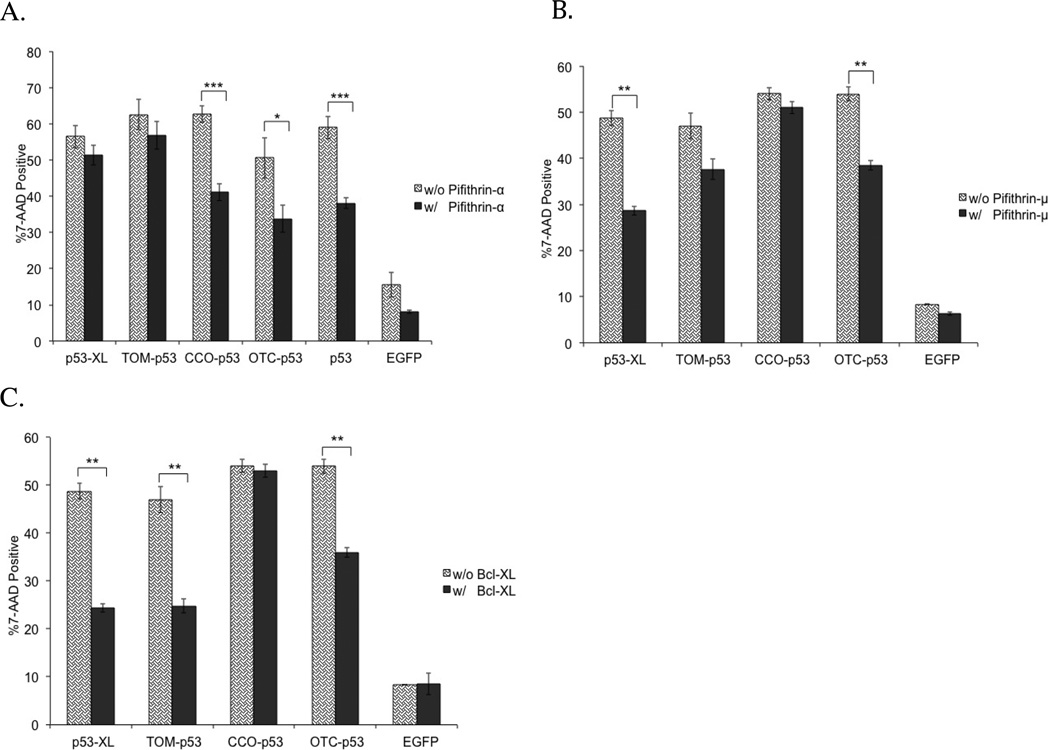
Investigating the apoptotic mechanism
Apoptosis resulting from p53 transcriptional activity of our mitochondrial p53 constructs was examined via a pifithrin-α rescue experiment. Pifithrin-α is a small molecule, which inhibits p53-mediated transcriptional activity.54, 55 The effect of pifithrin-α was measured in a 7-AAD assay (Figure 8A). As expected from the transcriptional activity data, the apoptotic effect of EGFP-p53 fused to either CCO or OTC was reduced significantly after pifithrin-α treatment. In addition, there was no impact on the apoptotic potential of EGFP-p53 fused to either XL or TOM (Figure 8A).
Figure 8.
Rescue experiments using (A) pifithrin-α, (B) pifithrin-μ, or (C) Bcl-XL. 7-AAD assay was performed 48 hours after transfection in T47D cells. All constructs were fused to EGFP. Statistical analysis was performed by unpaired t-test. * p<0.05, **p<0.005, ***<0.0005 comparing treated (with pifithrin-α, pifithrin-μ, or Bcl-XL) to untreated (no drug or Bcl-XL added).
In order to investigate p53’s apoptotic mechanism in the mitochondria, pifithrin-μ was used in a rescue experiment in the 7-AAD assay. Pifithrin-μ is a compound that reduces the binding affinity of p53 to the anti-apoptotic proteins, Bcl-2 and Bcl-XL.56, 57 Pifithrin-μ had a significant impact on the apoptotic potential of p53 fused to both XL and OTC (Figure 8B). However, apoptosis caused by p53 fused to either TOM or CCO was not rescued by pifithrin-μ. In addition, all the MTS-EGFP controls were not altered in this rescue experiment (see the supporting information: S3A).
The apoptotic mechanism was further explored by over-expression of the anti-apoptotic protein, Bcl-XL. The cells were then analyzed in the 7-AAD assay via flow cytometry. Bcl-XL had a significant effect on reducing 7-AAD positive cells treated with EGFP-p53 fused to TOM, XL, and OTC (Figure 8C). Cells treated with EGFP-p53-XL showed the most significant reduction in 7-AAD when co-transfected with Bcl-XL. Meanwhile, Bcl-XL had no effect on cells transfected with CCO-EGFP-p53. In addition, all the MTS-EGFP controls were not altered in this rescue experiment (see the supporting information: S3B).
Discussion
The tumor suppressor p53 was targeted with different MTSs in order to investigate its optimal mitochondrially triggered apoptotic pathway. We hypothesized that it might be possible that sending any protein (even EGFP) to mitochondria could be “toxic” to cells by disruption of mitochondrial function. Fusing p53 to MTSs targeting the mitochondrial outer membrane, inner membrane, and matrix did indeed result in apoptosis. However, sending even EGFP to the mitochondrial matrix (OTC), the inner membrane (CCO), or the TOM complex (TOM) perturbed mitochondrial stability as evidenced by the apoptotic assays. Only p53-XL was capable of inducing mitochondrial apoptosis exclusively via mitochondrial p53/Bcl-XL pathway.
Directing EGFP to the matrix and inner membrane via OTC and CCO, respectively, resulted in late stage apoptosis. This shows that fusing EGFP to OTC or CCO has a toxic effect on the mitochondria. We speculate that sending EGFP to the mitochondrial matrix and inner membrane could cause an imbalance in the sensitive mitochondrial system due to import through mitochondrial membranes. On the other hand, targeting EGFP to the mitochondrial outer membrane using TOM and XL shows minimal toxicity with XL but significant toxicity with TOM. Since EGFP-XL is directed towards Bcl-XL 58 on the surface of the outer membrane,29–31 it is not expected to become imbedded into the mitochondrial membrane. Conversely, the TOM-EGFP may interfere with TOM20 involved in mitochondrial import machinery.18, 59 The TOM complex is responsible for importing proteins across the mitochondrial outer membrane. TOM20 is one of the receptor subunits in the TOM complex.60, 61 Perhaps fusing any protein to the MTS from TOM20 might affect the sensitive import mechanism.
As p53 is a nuclear protein, the MTS fused to it will compete with the protein’s nuclear localization signals (NLSs). p53 contains three NLSs; the most active of them is located at residues 305–322.20 The nuclear import of large proteins is dependent on the availability of a NLS.62–64 To prevent the nuclear targeting of our constructs, we introduced mutations (K319T and K320T) in the strongest NLS of p53.20 Colocalization data and p53 transcriptional activity assay showed an increase in mitochondrial targeting and a decrease in p53 nuclear activity after the introduction of the NLS mutations in CCO-p53. According to our colocalization data, CCO-EGFP showed the lowest mitochondrial targeting compared to the other MTS-EGFP (Figure 2). The weak mitochondrial CCO signal explains the high transcriptional activity when fused to p53 without NLS mutations (Figure 3). The strong NLS in p53 competes with the relatively weak MTS from CCO and shifts the distribution to the nucleus. After mutating the strong NLS, the CCO MTS was also in competition with the other weak NLSs in p53,20 which may explain why the CCO-p53 NLS mutation still showed transcriptional activity (Figure 3). However, the mutations did not have any effect on the mitochondrial targeting or nuclear activity of the TOM, XL, and OTC constructs. EGFP-p53 fused to TOM and XL showed minimal nuclear p53 activity presumably due to their strong mitochondrial signals. Introducing NLS mutations to p53 fused to TOM or XL did not show any reduction on the already low transcriptional activity (Figure 3). On the other hand, OTC-p53 showed significant p53 nuclear activity but was not reduced upon NLS mutation (Figure 3).
In addition, the nuclear activity of MTS-p53 differed between MCF-7 and T47D. These differences might be due to variability in proteins involved with p53 transcriptional activity, mitochondrial shuttling, or number of mitochondria in each cell line. In MCF-7, all MTS-p53 constructs (with or without NLS mutations) showed minimum transcriptional activity except for the CCO-p53, which had half the activity of wild type p53 (Figure 3B). However, in T47D cells all MTS-p53 constructs showed generally higher nuclear activity than in MCF-7, especially CCO-p53, which showed the same nuclear activity as wild type p53. CCO-p53 NLS mutation and OTC-p53 (with and without NLS mutation) showed fifty percent transcriptional activity in T47D (Figure 3A).
Even though the NLS mutations increased mitochondrial targeting of the CCO-p53 construct, it did not have any effect on increasing the apoptotic potential. This was also the case for NLS mutations in all other constructs (see the supporting information: S1 and S2). CCO-p53 was significant compared to its CCO-EGFP control in TUNEL, Annexin-V, and 7-AAD assays. Since CCO-EGFP showed cytotoxicity, the increase in apoptosis when attached to p53 was likely due to nuclear p53 activity. This is reflected in our luciferase assay (Figure 3) and the rescue experiments with pifithrin-α, pifithrin-μ and Bcl-XL (Figure 8). The apoptotic activity of CCO-p53 was reduced in the pifithrin-α (an inhibitor of p53 transcriptional activity) rescue experiment. However, it was not rescued by either over-expression with the anti-apoptotic Bcl-XL or incubation with pifithrin-μ (an inhibitor of p53 binding to Bcl-2 and Bcl-XL).56, 57 This demonstrates that CCO-p53 does not initiate p53/Bcl-XL specific apoptosis.
OTC-p53 also showed transcriptional activity. In addition, OTC-p53 exhibited significant caspase-9 induction, and late stage apoptosis compared to its cytotoxic OTC-EGFP control. To examine if the increase of activity was due to nuclear or mitochondrial p53, the rescue experiments (with pifithrin-μ and Bcl-XL) were conducted and showed reduction in programmed cell death (Figure 8). This indicates that apoptosis was likely initiated through p53 binding to Bcl-XL and Bcl-2. In addition, the transcriptional activity data demonstrates that OTC-p53 has activity in both the nucleus (rescued by pifithrin-α) and the mitochondria (rescued by pifithrin-μ and Bcl-XL). Even though OTC directs p53 to the mitochondrial matrix, p53 is still able to interact with Bcl-XL and Bcl-2 proteins on the outer membrane. This could be due to cleavage of the MTS by endopeptidase, which enables p53 to target the outer membrane.3, 10
Instead of targeting the protein to the matrix then translocating it to the outer membrane, as was the case for OTC, we fused EGFP-p53 to TOM to directly target the outer membrane. Direct targeting of the outer membrane with TOM-p53 was able to initiate apoptosis (Annexin-V and 7-AAD) robustly compared to its TOM-EGFP control. Interestingly this increase in apoptosis was only rescued when Bcl-XL was co-transfected but not when pifithrin-α or pifithrin-μ were added (Figure 8). The pifithrin-α rescue experiment indicates that TOM-p53 has no transcriptional activity. We speculate that TOM-EGFP-p53 is binding to pro-apoptotic Bak and enhancing its oligomerization, which disrupts the mitochondrial outer membrane.13, 14 Bcl-XL forms a heterodimer with Bak and prevents Bak homodimerization.14, 65, 66 Therefore, when Bcl-XL is over-expressed, it competes with TOM-p53 in binding with Bak and hence reduces apoptosis. Since pifithrin-μ reduces the binding of p53 to anti-apoptotic Bcl-XL and Bcl-2 and has no effect on binding to Bak, it did not show reduction in programmed cell death for TOM-p53.
In an effort to directly target the p53/Bcl-XL pathway, we fused XL to EGFP-p53. Directing p53 to the mitochondria via XL showed significant caspase-9, TUNEL, 7-AAD, and Annexin-V activity compared to its EGFP-XL control. This apoptotic response was not due to transcriptional activity of p53 as shown in the luciferase assay data (Figure 3) and the pifithrin-α rescue experiment (Figure 8A). However, the apoptotic response was due to p53/Bcl-XL pathway. To confirm this interaction, rescue experiments using pifithrin-μ and Bcl-XL were conducted and showed reduction in apoptosis (Figure 8B and C). In addition, the EGFP-XL control showed no toxicity compared to the other MTS-EGFP controls especially in MCF-7 cells (Figure 6 and 7). This data demonstrates that sending p53 to a specific protein (Bcl-XL) in the mitochondrial outer membrane causes p53 specific apoptosis. Table 2 is a summary of the results and speculation from this work.
Table 2.
A summary of collected data and speculated mechanism. The table compares the four MTSs in mitochondrial localization, strength (*based on colocalization), mito-toxicity of MTS-EGFP, apoptotic response of MTS-p53 compared to MTS-EGFP, and speculated apoptotic mechanism.
| MTS | XL | TOM | CCO | OTC |
|---|---|---|---|---|
| Mitochondrial compartment | Outer surface of outer membrane | Outer membrane | Inner membrane | Matrix |
| Relative MTS Strength* | Strong | Strong | Weak | Medium/Strong |
| Intrinsic mitotoxicity of MTS-EGFP | Non-toxic | Toxic | Toxic | Toxic |
| p53 apoptotic response | Caspase-9, TUNEL, Annexin-V, and 7-AAD | Annexin-V, and 7-AAD | TUNEL, Annexin-V, and 7-AAD | Caspase-9, Annexin-V, and 7-AAD |
| Speculated apoptotic mechanism | May interact with Bcl-XL | May interact with Bak | Transcriptional p53 | Transcriptional p53 and may interact with Bcl-XL |
In summary, efficiency in targeting the mitochondria depends on the strength of the MTS. In the case of targeting proteins containing relatively strong NLSs such as p53 (residues 305–322),20 mitochondrial targeting can best be achieved by using strong MTSs to counter the NLS. In this study, relatively weaker MTSs are not efficient enough to compete with the strong NLS in p53. In addition, protein targeting to the mitochondria disrupts the sensitive balance in the mitochondria, which initiates intrinsic apoptosis. Except for EGFP-XL, all mitochondrial constructs had apoptotic effects. We conclude that p53-XL was the most specific to p53/Bcl-XL mitochondrial pathway. Our data shows that not all mitochondrial targeting signals are optimal for mitochondrial induction of apoptosis with p53. In conclusion, specific binding of p53 to mitochondrial Bcl-XL (and hence apoptotic activity) is best achieved by directly targeting p53 to Bcl-XL via the XL MTS. This work therefore provides a mechanistic explanation and provides additional speculation towards the understanding of mitochondrial p53 apoptosis. Our future goal is to employ the p53-XL construct as a therapeutic in vivo using viral delivery. Ultimately, p53-XL gene therapy is expected to be beneficial for other types of progressive cancers that currently have no effective therapy.
Supplementary Material
Acknowledgements
This work was funded by NIH R01-CA151847. We acknowledge Katharina Opper, Geoff Miller, Andy Dixon, Ben Bruno, Rian Davis, David Woessner, and Shams Reaz for scientific discussions.
Footnotes
Supporting Information Available
This information is available free of charge via the internet at http://pubs.acs.org/.
References
- 1.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol. Cell. Biol. 2004;24(15):6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erster S, Moll UM. Stress-induced p53 runs a transcription-independent death program. Biochem. Biophys. Res. Commun. 2005;331(3):843–850. doi: 10.1016/j.bbrc.2005.03.187. [DOI] [PubMed] [Google Scholar]
- 3.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11(3):577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 4.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303(5660):1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, St Clair DK. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 2005;65(9):3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 6.Canugovi C, Maynard S, Bayne AC, Sykora P, Tian J, de Souza-Pinto NC, Croteau DL, Bohr VA. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair. 2010;9(10):1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 8.El-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill DE, Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54(5):1169–1174. [PubMed] [Google Scholar]
- 9.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24(17):2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 10.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 2000;275(21):16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 11.Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, Pan H, Kessler H, Pancoska P, Moll UM. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J. Biol. Chem. 2006;281(13):8600–8606. doi: 10.1074/jbc.M507611200. [DOI] [PubMed] [Google Scholar]
- 12.Chipuk JE, Maurer U, Green DR, Schuler M. Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell. 2003;4(5):371–381. doi: 10.1016/s1535-6108(03)00272-1. [DOI] [PubMed] [Google Scholar]
- 13.Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat. Cell. Biol. 2004;6(5):443–40. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- 14.Pietsch EC, Leu JI, Frank A, Dumont P, George DL, Murphy ME. The tetramerization domain of p53 is required for efficient BAK oligomerization. Cancer Biol. Ther. 2007;6(10):1576–1583. doi: 10.4161/cbt.6.10.4719. [DOI] [PubMed] [Google Scholar]
- 15.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281(5381):1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 16.Reed JC. Cytochrome c: can't live with it--can't live without it. Cell. 1997;91(5):559–562. doi: 10.1016/s0092-8674(00)80442-0. [DOI] [PubMed] [Google Scholar]
- 17.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1(1):19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 18.Mossalam M, Dixon AS, Lim CS. Controlling subcellular delivery to optimize therapeutic effect. Ther. Deliv. 2010;1(1):169–193. doi: 10.4155/tde.10.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mihara M, Moll UM. Detection of mitochondrial localization of p53. Methods Mol. Biol. 2003;234:203–209. doi: 10.1385/1-59259-408-5:203. [DOI] [PubMed] [Google Scholar]
- 20.Shaulsky G, Goldfinger N, Ben-Ze'ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol. Cell Biol. 1990;10(12):6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang P, Du W, Heese K, Wu M. The Bad guy cooperates with good cop p53: Bad is transcriptionally up-regulated by p53 and forms a Bad/p53 complex at the mitochondria to induce apoptosis. Mol. and Cel. Biol. 2006;26(23):9071–9082. doi: 10.1128/MCB.01025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488(3):110–115. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- 23.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003;33(3):357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 24.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26(4):923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5(6):1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koehler CM. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057. [DOI] [PubMed] [Google Scholar]
- 27.Roise D, Schatz G. Mitochondrial presequences. J. Biol. Chem. 1988;263(10):4509–4511. [PubMed] [Google Scholar]
- 28.Wiedemann N, Frazier AE, Pfanner N. The protein import machinery of mitochondria. J. Biol. Chem. 2004;279(15):14473–14476. doi: 10.1074/jbc.R400003200. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann T, Schlipf S, Sanz J, Neubert K, Stein R, Borner C. Characterization of the signal that directs Bcl-x(L), but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 2003;160(1):53–64. doi: 10.1083/jcb.200210084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollack M, Leeuwenburgh C. Apoptosis and aging: role of the mitochondria. J. Gerontology. 2001;56(11):B475–B482. doi: 10.1093/gerona/56.11.b475. [DOI] [PubMed] [Google Scholar]
- 31.Lindsay J, Esposti MD, Gilmore AP. Bcl-2 proteins and mitochondria--specificity in membrane targeting for death. Biochim. Biophys. Acta. 2011;1813(4):532–539. doi: 10.1016/j.bbamcr.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 32.Rehling P, Brandner K, Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5(7):519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- 33.Power SD, Lochrie MA, Patterson TE, Poyton RO. The nuclear-coded subunits of yeast cytochrome c oxidase. II. The amino acid sequence of subunit VIII and a model for its disposition in the inner mitochondrial membrane. J. Biol. Chem. 1984;259(10):6571–6574. [PubMed] [Google Scholar]
- 34.Fabrizi GM, Sadlock J, Hirano M, Mita S, Koga Y, Rizzuto R, Zeviani M, Schon EA. Differential expression of genes specifying two isoforms of subunit VIa of human cytochrome c oxidase. Gene. 1992;119(2):307–312. doi: 10.1016/0378-1119(92)90288-z. [DOI] [PubMed] [Google Scholar]
- 35.Isaya G, Fenton WA, Hendrick JP, Furtak K, Kalousek F, Rosenberg LE. Mitochondrial import and processing of mutant human ornithine transcarbamylase precursors in cultured cells. Mol. Cell Biol. 1988;8(12):5150–5158. doi: 10.1128/mcb.8.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon AS, Kakar M, Schneider KM, Constance JE, Paullin BC, Lim CS. Controlling subcellular localization to alter function: Sending oncogenic Bcr-Abl to the nucleus causes apoptosis. J. Control. Release. 2009;140(3):245–249. doi: 10.1016/j.jconrel.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J. Microscopy. 2006;224 (Pt 3):213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 38.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophysical J. 2004;86(6):3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adler J, Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander's overlap coefficient. Cytometry A. 77(8):733–742. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- 40.Dixon AS, Miller GD, Bruno BJ, Constance JE, Woessner DW, Fidler TP, Robertson JC, Cheatham TE, Lim CS. Improved coiled-coil design enhances interaction with bcr-abl and induces apoptosis. Mol. Pharm. 2012;9(1):187–195. doi: 10.1021/mp200461s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaskolski F, Mulle C, Manzoni OJ. An automated method to quantify and visualize colocalized fluorescent signals. J. Neuroscience Methods. 2005;146(1):42–49. doi: 10.1016/j.jneumeth.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Schafer JM, Lee ES, O'Regan RM, Yao K, Jordan VC. Rapid development of tamoxifen-stimulated mutant p53 breast tumors (T47D) in athymic mice. Clin. Cancer Res. 2000;6(11):4373–4380. [PubMed] [Google Scholar]
- 43.Alkhalaf M, El-Mowafy AM. Overexpression of wild-type p53 gene renders MCF-7 breast cancer cells more sensitive to the antiproliferative effect of progesterone. J. Endocrinol. 2003;179(1):55–62. doi: 10.1677/joe.0.1790055. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi K, Sumimoto H, Suzuki K, Ono T. Protein synthesis-dependent cytoplasmic translocation of p53 protein after serum stimulation of growth-arrested MCF-7 cells. Mol. Carcinog. 1993;8(1):58–66. doi: 10.1002/mc.2940080112. [DOI] [PubMed] [Google Scholar]
- 45.Mooney LM, Al-Sakkaf KA, Brown BL, Dobson PR. Apoptotic mechanisms in T47D and MCF-7 human breast cancer cells. Br. J. Cancer. 2002;87(8):909–917. doi: 10.1038/sj.bjc.6600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chowdhury I, Tharakan B, Bhat GK. Caspases - an update. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008;151(1):10–27. doi: 10.1016/j.cbpb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Yin Q, Park HH, Chung JY, Lin SC, Lo YC, da Graca LS, Jiang X, Wu H. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol. Cell. 2006;22(2):259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loo DT, Rillema JR. Measurement of cell death. Meth. Cell Biol. 1998;57:251–264. doi: 10.1016/s0091-679x(08)61583-6. [DOI] [PubMed] [Google Scholar]
- 49.Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84(5):1415–1420. [PubMed] [Google Scholar]
- 50.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immun. Meth. 1995;184(1):39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 51.Metkar SS, Wang B, Catalan E, Anderluh G, Gilbert RJ, Pardo J, Froelich CJ. Perforin rapidly induces plasma membrane phospholipid flip-flop. PLoS One. 2011;6(9):e24286. doi: 10.1371/journal.pone.0024286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmid I, Krall WJ, Uittenbogaart CH, Braun J, Giorgi JV. Dead cell discrimination with 7-amino-actinomycin D in combination with dual color immunofluorescence in single laser flow cytometry. Cytometry. 1992;13(2):204–208. doi: 10.1002/cyto.990130216. [DOI] [PubMed] [Google Scholar]
- 53.Serrano MJ, Sanchez-Rovira P, Algarra I, Jaen A, Lozano A, Gaforio JJ. Evaluation of a gemcitabine-doxorubicin-paclitaxel combination schedule through flow cytometry assessment of apoptosis extent induced in human breast cancer cell lines. Jap. J. Cancer Res. 2002;93(5):559–566. doi: 10.1111/j.1349-7006.2002.tb01291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285(5434):1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 55.Liu X, Chua CC, Gao J, Chen Z, Landy CL, Hamdy R, Chua BH. Pifithrin-alpha protects against doxorubicin-induced apoptosis and acute cardiotoxicity in mice. Am. J. Physiol. 2004;286(3):H933–H939. doi: 10.1152/ajpheart.00759.2003. [DOI] [PubMed] [Google Scholar]
- 56.Hagn F, Klein C, Demmer O, Marchenko N, Vaseva A, Moll UM, Kessler H. BclxL changes conformation upon binding to wild-type but not mutant p53 DNA binding domain. J. Biol. Chem. 2010;285(5):3439–3450. doi: 10.1074/jbc.M109.065391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, Komarova EA, Gudkov AV. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat. Chem. Biol. 2006;2(9):474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 58.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim. Biophys. Acta. 2009;1787(5):414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 2010;11(9):655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 60.Endo T, Kohda D. Functions of outer membrane receptors in mitochondrial protein import. Biochim. Biophys. Acta. 2002;1592(1):3–14. doi: 10.1016/s0167-4889(02)00259-8. [DOI] [PubMed] [Google Scholar]
- 61.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313(5785):314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 62.Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 63.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 64.Davis JR, Kakar M, Lim CS. Controlling protein compartmentalization to overcome disease. Pharm. Res. 2007;24(1):17–27. doi: 10.1007/s11095-006-9133-z. [DOI] [PubMed] [Google Scholar]
- 65.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275(5302):983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 66.Galluzzi L, Morselli E, Kepp O, Tajeddine N, Kroemer G. Targeting p53 to mitochondria for cancer therapy. Cell Cycle. 2008;7(13):1949–1955. doi: 10.4161/cc.7.13.6222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.