Abstract
In bacteria, the hybrid transfer-messenger RNA (tmRNA) rescues ribosomes stalled on defective messenger RNAs (mRNAs). However, certain gram-negative bacteria have evolved proteins that are capable of rescuing stalled ribosomes in a tmRNA-independent manner. Here, we report a 3.2 angstrom–resolution crystal structure of the rescue factor YaeJ bound to the Thermus thermophilus 70S ribosome in complex with the initiator tRNAifMet and a short mRNA. The structure reveals that the C-terminal tail of YaeJ functions as a sensor to discriminate between stalled and actively translating ribosomes by binding in the mRNA entry channel downstream of the A site between the head and shoulder of the 30S subunit. This allows the N-terminal globular domain to sample different conformations, so that its conserved GGQ motif is optimally positioned to catalyze the hydrolysis of peptidyl-tRNA. This structure gives insights into the mechanism of YaeJ function and provides a basis for understanding how it rescues stalled ribosomes.
The rescue of stalled ribosomes ensures efficient protein synthesis and cell viability. In bacteria, the most studied pathway involves a ribonucleoprotein complex comprising the hybrid transfer-mRNA (tmRNA) and the small protein B (SmpB) (1, 2). The tmRNA system is not essential for cell viability in several model bacteria, including Escherichia coli (3), which suggests that these bacteria have alternate ribosome rescue pathways that function independently of the tmRNA system.
Recently, the products of the yhdL and yaeJ genes, known as the alternative ribosome rescue factor A (ArfA) and YaeJ, respectively, which promote the hydrolysis of peptidyl–transfer RNA (peptidyl-tRNA) in stalled ribosomes, have been identified in E. coli (4–7). When YaeJ is over-expressed, it competes with tmRNA to rescue ribosomes that are stalled at the 3′ end of nonstop mRNA and suppresses the lethal phenotype of cells lacking both tmRNA and ArfA (6). YaeJ is classified as a putative release factor because it has sequence similarity with domain III of class I release factors (RFs) 1 and 2. The GGQ motif found in RF1 and RF2 is conserved in YaeJ (fig. S1) and catalyzes the aminoacyl ester hydrolysis of peptidyl-tRNA on the ribosome stalled by either nonstop mRNAs or mRNAs containing rare codon clusters (4, 6, 8). Mutations in the GGQ motif reduce the efficiency of peptidyl-tRNA hydrolysis but do not affect the ability of YaeJ to bind the ribosome (4, 6). However, the 140– amino acid YaeJ is substantially shorter than RF1/2 (∼360 amino acids) because it lacks both the domains that are involved in stop-codon recognition (9). Taken together, these observations indicate that YaeJ functions as a ribosome-dependent, codon-independent peptidyl-tRNA hydrolase that rescues stalled ribosomes independently of the tmRNA and ArfA pathways (4, 6).
YaeJ homologs have been identified in many gram-negative bacteria and are completely conserved in eukaryotic genomes (4). Recent biochemical data, together with sequence and structural alignments of YaeJ [Protein Data Bank (pdb) 2JY9] and its human homolog ICT1 (10) [pdb 1J26 (11)] (figs. S1 and S2), show that the latter also has the GGQ motif, shares the same fold, and harbors a positively charged C-terminal tail. Therefore, ICT1 is also proposed to function as a ribosome-dependent codon-independent peptidyl-tRNA hydrolase (10). Although the GGQ motif is known to function in catalysis, the precise role of the positively charged, unstructured C-terminal tail has remained obscure. Recent biochemical studies suggest that the YaeJ tail is required for its binding to the ribosome (4, 6). In addition, the C-terminal tail of SmpB has a similar charge distribution to that of the YaeJ tail (6), but its specific role in ribosome rescue also remains poorly understood (12, 13).
Here, we report the crystal structure of full-length E. coli YaeJ bound to the Thermus thermophilus 70S ribosome in complex with the initiator tRNAifMet and a short mRNA fragment diffracting to a resolution of 3.2 Å (14). We determined the structure by molecular replacement using a model of the empty 70S ribosome at 2.8 Å resolution (15) as a search model. The initial difference Fourier map calculated using the Fobs – Fcalc amplitudes revealed difference electron density for the initiator tRNA, mRNA, and YaeJ. However, certain beta strands, the GGQ loop, and the C-terminal tail of YaeJ could only be unambiguously modeled from the difference Fourier maps calculated using the Fobs – Fobs(*) amplitudes (fig. S3) in which the Fobs(*) structure factors came from an isomorphous crystal containing the 70S ribosome alone. The final model was refined to an Rwork/Rfree of 18.8/24.5% (14) and is consistent with both the Fobs – Fobs(*) and Fobs – Fcalc difference Fourier maps. The binding of YaeJ to the ribosome does not induce major conformational rearrangements in the ribosomal subunits. The structure of this stalled ribosome model system reveals that the N-terminal globular domain of YaeJ is bound in the A site of the 50S subunit next to the P-site tRNA (Fig. 1B), and its C-terminal tail occupies the mRNA path downstream of the 30S A site (Fig. 1, A and B). The binding of the tail in the mRNA entry channel positions the GGQ motif of YaeJ in the peptidyl-transferase center (PTC) adjacent to the CCA end of the peptidyl-tRNA, consistent with the role of the GGQ motif in peptide release (16, 17).
Fig. 1.
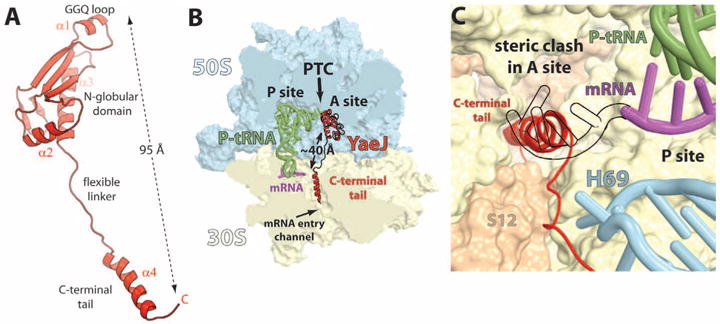
The structure of YaeJ bound to the ribosome. (A) Cartoon representation of YaeJ shown in its ribosome-bound conformation. (B) Overview of YaeJ, P-site tRNA, and mRNA bound to the 70S ribosome. The 50S and 30S subunits are shown in light blue and yellow, respectively. Portions of the ribosome are omitted for clarity. The N-terminal domain of YaeJ is bound in the A site of the 50S subunit, adjacent to the CCA end of the peptidyl-tRNA, and the C-terminal tail occupies the mRNA entry channel in the 30S. (C) Positioning of the α-helical YaeJ tail in the mRNA entry channel. The mRNA in the present complex (magenta) ends in the P site. The mRNA making codon-anticodon interaction in the A site (shown as a black outline) [2J00, (15)] was superimposed on our model to reveal the steric clash with the YaeJ tail.
The C-terminal tail, which is disordered in the nuclear magnetic resonance structure of YaeJ alone (pdb 2JY9), extends from the globular domain and reaches the 30S subunit A site more than 40 Å away (Fig. 1B), which allows it to bind both ribosomal subunits in a manner reminiscent of many ribosome-binding factors. It forms an α helix with the positively charged surface of the tail accommodated inside the mRNA entry channel downstream of the A site. In this position, the basic side chains of the YaeJ tail are free to interact with the phosphate backbone of the ribosomal RNA (rRNA) that forms the walls of the channel. The tail of YaeJ spans the region between the decoding center and the central pseudoknot in the 30S subunit. In this position, it mimics the mRNA in the mRNA entry channel downstream of the A site (Fig. 1B), which is not compatible with the presence of an A-site codon primed for codon-anticodon interaction (Fig. 1C), and this explains why YaeJ does not act on ribosomes during normal translation (4). Because recent biochemical data suggest that YaeJ can catalyze peptidyl-tRNA hydrolysis on ribosomes stalled by a rare codon cluster in vitro (4), the possibility that a codon can occupy the A site cannot be excluded. However, we speculate that the flexibility of the mRNA allows it to avoid a steric clash with YaeJ and still accommodate a few codons downstream from the P site of our model.
Because YaeJ does not have domains that are involved in the recognition of stop codons as in the class I release factors, it uses a different mechanism to bind the ribosome. From previous biochemical data (1, 12), it had been proposed that some rescue factors recognize the empty mRNA entry channel in order to detect stalled ribosomes. Furthermore, nonstop mRNAs and mRNAs with rare codon clusters are cleaved by endonucleases (1, 18), which results in an empty mRNA entry channel, and consequently, ribo-some rescue is almost always preceded by mRNA cleavage (1, 19). Consistent with these observations, the structure presented here suggests that the YaeJ tail functions as a sensor that probes the occupancy of the mRNA entry channel in order to distinguish between stalled and translating ribosomes. The positioning of the YaeJ tail is also consistent with the proposed position of the similar SmpB tail based on recent biochemical, cryo–electron microscopy (cryo-EM), and homology modeling data on the tmRNA-SmpB rescue system (1, 12, 20, 21). Thus, the structure of YaeJ bound to the ribo-some provides direct structural evidence for the use of its tail as a sensor to recognize a stalled ribosome in addition to providing a rationale for the lack of domains required for stop-codon recognition in YaeJ.
The binding of YaeJ to the ribosome induces perturbations in the decoding center and helix 69 (H69) (Fig. 2A). As is seen in standard decoding and in termination that is mediated by stop co-dons, the binding of the YaeJ tail also induces G530 to switch from syn to anti conformation by the stacking of Arg118 (R118) on G530 (Fig. 2B), mimicking the stacking normally made by the third nucleotide of the stop codon (22, 23). The positions of A1492 and A1493 in the present complex appear different from those induced by tRNA binding, presumably to avoid a steric clash with the YaeJ tail (Fig. 2C). The side chain of R117 interacts with C518 and Ser50 (S50) of ribosomal protein S12 (Fig. 2B) and replaces the interaction that the latter normally makes with position N6 of A1492 during regular decoding (Fig. 2C). In their new orientation, A1492 is stacked within h44 and A1493 stacks with A1913 from H69 (Fig. 2A) to avoid a steric clash with the YaeJ tail (Fig. 2C). The observed stacking between A1493 (h44) and A1913 (H69) was previously reported in the structures of RF1 and RF2 bound to the ribosome (22, 23).
Fig. 2.
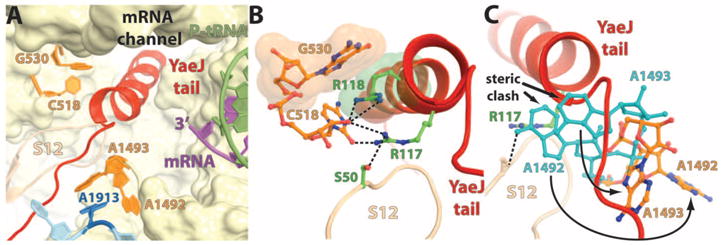
Interactions of the YaeJ tail with the universally conserved nucleotides in the decoding center. (A) Overview of the YaeJ tail in the decoding center and its interaction with the key bases that are labeled. (B) Close-up view of the stacking between G530 (16S rRNA) and R118 of YaeJ shown as space-filling representation in orange and green, respectively. Putative hydrogen bonds of R117 with C518 (16S rRNA) and S50 from ribosomal protein S12 are shown as black dashes. (C) Conformational differences between A1492 and A1493 from our model (shown in orange) relative to their counterparts from a structure with an A site–bound tRNA [2J00 (15)] (shown in light blue) reveal that the latter sterically clash with the YaeJ tail. The positioning of these key bases in the present complex is indicated by the curved arrows.
We propose a model based on this structure for how the binding of the YaeJ tail in the mRNA entry channel influences the accommodation of the GGQ loop in the 50S subunit A site. The binding recognition element (C-terminal tail) and the catalytic GGQ loop of YaeJ are spatially separated by at least 95 Å and span both ri-bosomal subunits (Fig. 1, A and B). However, biochemical data suggest that binding of the YaeJ tail to the ribosome is required for peptide release (4, 6). Hence, the binding of YaeJ to a stalled ribosome is likely coupled to the rearrangements of its catalytic GGQ loop to ensure efficient rescue. The globular domain and the C-terminal tail of YaeJ are physically connected by a short linker that spans residues 101 to 108. Even though the tail is relatively ordered, some of the residues in the linker region are poorly ordered, as indicated by increased temperature factors and their discontinuous electron density in the 2Fobs – Fcalc map. On the basis of these observations, it is tempting to suggest that the binding of the YaeJ tail in the mRNA entry channel allows the movement of its globular domain to position the GGQ loop adjacent to the CCA end of the peptidyl-tRNA. Pro110 (P110), which is anchored by its stacking with the conserved A1493 (Fig. 3A), can act as a hinge to fix the conformation of the tail on one side while allowing free movement of the globular domain through the flexible linker. Notably, the presence of a hook like feature at the C terminus of YaeJ (residues 129 to 133), where the tail turns toward the 16S rRNA central pseudoknot (Fig. 3B), appears to stabilize the position of he tail on the other end. This proposed stabilization is consistent with biochemical data showing that deletion of the last 10 residues in YaeJ abolishes its binding to the ribosome (4). Taken together, the structure suggests communication between two distinct regions of the protein where the rigidity conferred by the anchored tail and the flexibility of the linker allow the N-terminal domain to interact with the 50S sub-unit A site, which results in the proper positioning of the GGQ loop in the PTC primed for catalysis.
Fig. 3.
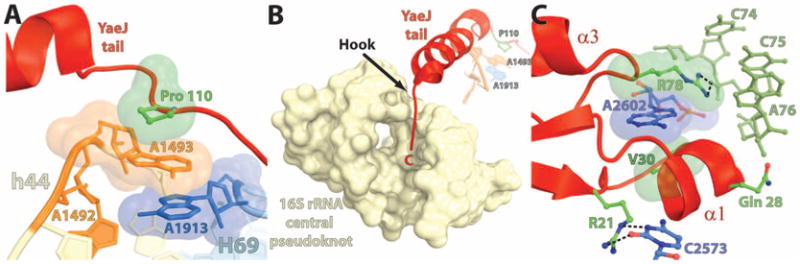
Stabilization of the YaeJ tail in the mRNA entry channel (A and B) and the GGQ loop in the PTC (C). (A) Stacking between Pro110 (P110) from YaeJ (green), A1493 from h44 (orange), and A1913 from H69 (blue). (B) Interaction of the C-terminal end of YaeJ (residues 129 to 133) with the 16S rRNA central pseudoknot consisting of helix 1 (nucleotides 9 to 13 and 21 to 25) and helix 2 (nucleotides 17 to 19 and 916 to 918). A1493 from h44 in the decoding center (orange), A1913 from H69 (blue), and P110 from YaeJ (green) are shown in the background. The interactions shown in (A) and (B) together anchor the YaeJ tail in the mRNA entry channel. (C) The potential hydrogen bonds are shown as black dashes. The H-bonds between C2573 and R21, R78, and C75 of the P-tRNA and the stacking between A2602, R78, and V30 (shown as space-filling representation) help to position the GGQ loop next to the CCA end of the P-tRNA.
The essential GGQ motif containing residues 26 to 28 is positioned in the PTC of the present complex, which is consistent with its involvement in the peptidyl-tRNA hydrolase activity of YaeJ (4, 6). As observed in previous structures of release factors, the unstructured GGQ loop in YaeJ becomes ordered when it binds the ribo-some and adopts a short α-helical structure (Fig. 3C) (22, 23). A network of interactions between the side chains of YaeJ and the rRNA bases in the A site of the 50S subunit seen in this structure plays a pivotal role in positioning the GGQ loop in the PTC to promote efficient catalysis and is also consistent with previous biochemical data (Fig. 3C) (22–24). The positions of all the residues that are involved in catalysis are identical to their counterparts in the structure of the ribosome in complex with RF2, which suggests a similar mechanism of peptide release (22). The side chain of the catalytic Gln28 was modeled with its carbonyl oxygen within hydrogen-bonding distance of the 3′-OH of A76 of the P-site tRNA (Fig. 4A). As proposed earlier (22), a small change in the dihedral angles of the glutamine side chain can accommodate the water molecule that acts as a nucleophile poised for an inline attack on the peptidyl-tRNA ester bond (Fig. 4B). The position of the nucleophilic water can be inferred by a superposition on the structure of a peptidyl-tRNA analog bound to the 50S subunit (Fig. 4B) (25).
Fig. 4.
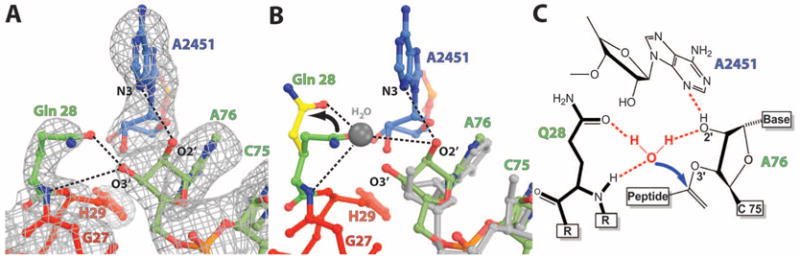
Positioning of the GGQ loop and catalysis. (A) σA-weighted 2Fob s – Fcalc electron density map contoured at 1.2σ showing the interactions of the GGQ loop at the site of catalysis. Putative hydrogen bonds between Gln28, A2451, and the riboseofA76 are shownasblack dashes. (B) Proposed catalytic mechanism reveals that a small change in dihedral angles of Gln28 as indicated by the arrow (green versus yellow) allows it to coordinate the nucleophilic water [shown as a gray ball modeled based on 1VQ7 (25)] that attacks the aminoacyl ester bond. (C) Schematic representation of the proposed catalysis reaction shown in (B).
The structure presented here supports the following model for YaeJ-dependent rescue of stalled ribosomes. The C-terminal tail of YaeJ functions as a sensor to detect a stalled ribosome based on the occupancy of the mRNA channel. Subsequently, the binding of the YaeJ tail in the mRNA entry channel allows its N-terminal globular domain to sample different orientations through its flexible linker region. A network of interactions between the globular domain of YaeJ and the 50S A site can then stabilize the GGQ loop in the PTC in a conformation that promotes catalysis. After pep-tide release, the ribosomes may become a substrate for the ribosome recycling factor (RRF), which, together with the elongation factor G (EF-G), dissociates ribosomes into subunits (26). Beyond addressing the critical role of the protein tail in the recognition of a stalled ribosome, this study raises questions about the unassigned roles of extended tails present in many ribosome-binding factors. Together with the recently determined structures of the eukaryotic ribosome (27, 28), our structure reveals the details of how protein segments can mimic RNA to perform important functions. Additional structural studies of other rescue factors are required to establish common architectural features governing the rescue of stalled ribosomes.
Acknowledgments
We are thankful to the members of the Steitz lab for useful suggestions and discussions. We thank R. Grodzicki for providing the tRNAifMet and the staffs of the Advanced Photon Source beamline 24-ID and the National Synchrotron Light Source beamline X29 for help during data collection and the Center for Structural Biology facility at Yale University for computational support. Special thanks to Y. Polikanov, J. Wang, and W. Meng for advice with crystallographic software. This work was supported by NIH grant GM022778 (to T.A.S.). Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4DH9, 4DHA, 4DHB, and 4DHC. T.A.S. owns stock in and is on the advisory board of Rib-X Pharmaceuticals, which does structure-based design of drugs that target the ribosome.
References and Notes
- 1.Felden B, Gillet R. RNA Biol. 2011;8:440. doi: 10.4161/rna.8.3.15387. [DOI] [PubMed] [Google Scholar]
- 2.Keiler KC, Waller PR, Sauer RT. Science. 1996;271:990. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 3.Withey J, Friedman D. J Bacteriol. 1999;181:2148. doi: 10.1128/jb.181.7.2148-2157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handa Y, Inaho N, Nameki N. Nucleic Acids Res. 2011;39:1739. doi: 10.1093/nar/gkq1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garza-Sánchez F, Schaub RE, Janssen BD, Hayes CS. Mol Microbiol. 2011;80:1204. doi: 10.1111/j.1365-2958.2011.07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chadani Y, Ono K, Kutsukake K, Abo T. Mol Microbiol. 2011;80:772. doi: 10.1111/j.1365-2958.2011.07607.x. [DOI] [PubMed] [Google Scholar]
- 7.Chadani Y, et al. Mol Microbiol. 2010;78:796. doi: 10.1111/j.1365-2958.2010.07375.x. [DOI] [PubMed] [Google Scholar]
- 8.Youngman EM, McDonald ME, Green R. Annu Rev Microbiol. 2008;62:353. doi: 10.1146/annurev.micro.61.080706.093323. [DOI] [PubMed] [Google Scholar]
- 9.Ito K, Uno M, Nakamura Y. Nature. 2000;403:680. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 10.Richter R, et al. EMBO J. 2010;29:1116. doi: 10.1038/emboj.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handa Y, et al. J Mol Biol. 2010;404:260. doi: 10.1016/j.jmb.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 12.Kurita D, Muto A, Himeno H. RNA. 2010;16:980. doi: 10.1261/rna.1916610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurita D, Sasaki R, Muto A, Himeno H. Nucleic Acids Res. 2007;35:7248. doi: 10.1093/nar/gkm677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Materials and methods are available as supporting material on Science Online.
- 15.Selmer M, et al. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 16.Mora L, et al. Mol Microbiol. 2003;47:267. doi: 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- 17.Frolova LY, et al. RNA. 1999;5:1014. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto Y, Sunohara T, Jojima K, Inada T, Aiba H. RNA. 2003;9:408. doi: 10.1261/rna.2174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanova N, Pavlov MY, Felden B, Ehrenberg M. J Mol Biol. 2004;338:33. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 20.Weis F, et al. RNA. 2010;16:299. doi: 10.1261/rna.1757410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutmann S, et al. Nature. 2003;424:699. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 22.Weixlbaumer A, et al. Science. 2008;322:953. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurberg M, et al. Nature. 2008;454:852. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 24.Burakovsky DE, et al. RNA. 2010;16:1848. doi: 10.1261/rna.2185710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmeing TM, Huang KS, Strobel SA, Steitz TA. Nature. 2005;438:520. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 26.Savelsbergh A, Rodnina MV, Wintermeyer W. RNA. 2009;15:772. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Science. 2011;331:730. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 28.Ben-Shem A, et al. Science. 2011;334:1524. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]


