Abstract
The genetic code is continuously expanding with new nucleobases designed to suit specific research needs. These synthetic nucleotides are used to study DNA polymerase dynamics and specificity and may even inhibit DNA polymerase activity. The availability of an increasing chemical diversity of nucleotides allows questions of utilization by different DNA polymerases to be addressed. Much of the work in this area deals with the A family DNA polymerases, for example, Escherichia coli DNA polymerase I, which are DNA polymerases involved in replication and whose fidelity is relatively high, but more recent work includes other families of polymerases, including the Y family, whose members are known to be error prone. This paper focuses on the ability of DNA polymerases to utilize nonnatural nucleotides in DNA templates or as the incoming nucleoside triphosphates. Beyond the utility of nonnatural nucleotides as probes of DNA polymerase specificity, such entities can also provide insight into the functions of DNA polymerases when encountering DNA that is damaged by natural agents. Thus, synthetic nucleotides provide insight into how polymerases deal with nonnatural nucleotides as well as into the mutagenic potential of nonnatural nucleotides.
1. Introduction
Since the structure of DNA was determined [1, 2], biochemists have sought more detailed ways to study DNA and the proteins that interact with it [3, 4]. Solid phase nucleic acid synthesis of DNA molecules facilitates the site-specific incorporation of a wide range of chemically modified bases and sugar-phosphate backbones, allowing the roles of specific atoms in DNA function and recognition to be probed. Synthetic nonnatural nucleobases are useful for a variety of studies of DNA polymerase function, such as studies of DNA polymerase specificity, mutagenesis, and dynamics, as well as fluorescence resonance energy transfer (FRET) analysis of DNA polymerase interactions with DNA. The study of mutagenesis facilitated by DNA polymerases has attracted increasing interest because replication defects can lead to certain human diseases like the cancer-prone syndrome xeroderma pigmentosum variant (XPV) [5, 6] and other diseases [7–9], as well as potentially contribute to antibiotic resistance [10, 11]. Moreover, specialized damage-bypass DNA polymerases are implicated in conferring cellular tolerance to cancer chemotherapy agents that act via DNA damage, thereby decreasing their effectiveness [12–16]. This paper will focus on the ability of DNA polymerases to recognize and accept nonnatural bases either on the template strand or as the incoming triphosphate nucleotide. Much of the work discussed here will deal with A family polymerases (e.g., Klenow fragment (KF) of pol I and Taq DNA polymerase), but more recent work with Y family polymerases [17] and their ability to utilize certain nucleotide analogs will also be discussed.
DNA polymerases generally adopt a right-hand fold, in which the thumb and fingers bind DNA and nucleotide (Figure 1) [18, 19]. DNA polymerases add nucleotides to the growing DNA strand via nucleophilic attack of the free 3′ hydroxyl group of the DNA primer on the alpha phosphate of the incoming deoxynucleotide with release of pyrophosphate. DNA polymerase active site residues, which are usually glutamate or aspartate and are located in the palm domain, coordinate divalent magnesium ions that serve to activate the 3′-OH nucleophile (Figure 1) [20–25]. The catalytic cycle is generally accompanied by conformational changes in the fingers domain. In replicating DNA, DNA polymerases have to be able to form all four base pair combinations specifically and efficiently in order to maintain the integrity of the genome (Figure 2); however, when replication errors occur, the mismatched bases can be removed by the exonucleolytic proofreading function of DNA polymerases [26]. Replicative DNA polymerases possess an exonuclease domain that may be part of the same or a separate polypeptide that utilizes a metal-dependent mechanism to excise mismatched bases [26, 27]. The proofreading process involves translocation of the primer terminus from the polymerase active site to the exonuclease active site; after the phosphodiester bond is hydrolyzed to remove the mismatched base, the primer strand reanneals to the template so that polymerization can continue [27, 28]. Replication errors that escape proofreading can be repaired by the mismatch repair system [29].
Figure 1.
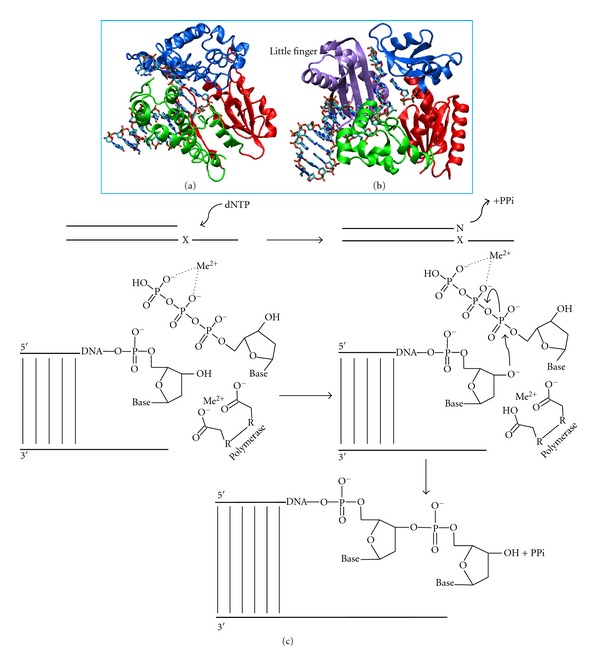
Comparison of the overall folds of (a) a replicative DNA polymerase, Bacillus stearothermophilus DNA pol I [30], and (b) a Y family DNA polymerase, Sulfolobus solfataricus Dpo4 [31]. The respective thumb domains are shown in green, palm domains are in red, and fingers domains are in blue. The little finger domain unique to the Y family DNA polymerases is in purple [31]. The “vestigial” exonuclease domain of Bs pol I has been omitted for clarity [32]. (c) Polymerase catalyzed DNA replication (phosphoryl transfer) reaction. Polymerization of DNA occurs at a free 3′ hydroxyl group of the deoxyribose. Polymerases use a divalent magnesium ion (Me2+) to coordinate the negative charges of both the phosphate groups and the aspartic acid or glutamic acid in the active site of the polymerase [26].
Figure 2.
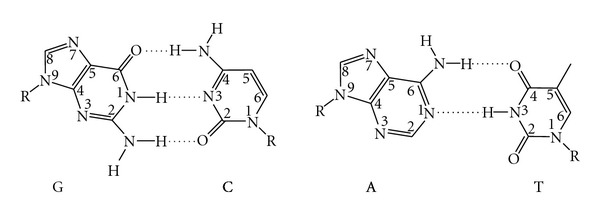
The canonical Watson-Crick base pairs. Standard numbering is indicated. Unless otherwise noted, R indicates the position of the deoxyribose in all figures.
Based on sequence conservation, DNA polymerases are divided into A, B, C, D, X, and Y families. The A and B family DNA polymerases can be involved in replication or repair, whereas members of the C family are involved in DNA replication [33]. X family DNA polymerases are involved in repair, and Y family DNA polymerases are specialized for copying damaged DNA [33] in a process known as translesion synthesis (TLS). In general, replicative DNA polymerases cannot copy damaged DNA; rather, a specialized TLS polymerase must be recruited to extend primers a sufficient distance past distortions in DNA templates to allow replicative DNA polymerases to recover synthesis [34–37]. DNA replication past damage or unusual DNA structures requires the ability to both insert a nucleotide opposite a modification in the template as well as to extend the newly generated primer beyond that position. Some polymerases may be able to insert a nucleotide opposite nonstandard bases but be unable to extend the resulting primer terminus, as discussed below.
The four canonical bases vary in their chemical and geometric properties, but the C1′–C1′ distance of the standard Watson-Crick base pairs and the backbone C–O–P–O–C bonds remain constant regardless of the particular base pair [38]. Expansion of the nucleobase alphabet must take some of these structural considerations into account; usually nonnatural bases need to have similar geometries as the natural bases, usually but not necessarily retain some level of hydrogen bonding capabilities, and usually have π electron systems in order to retain the stability provided by base stacking. Hydrophobicity and base stacking interactions are also important for DNA structure [38].
2. Abasic Sites and Small Molecule Substitutions
Stable, synthetic abasic sites were first introduced into DNA in 1987 [39]. As it is estimated that 10,000 abasic sites form in each human cell per day [29], it was important to develop a stable, synthetic abasic site in order to facilitate the study of DNA polymerase interactions. Furthermore, it is informative to determine the activity of DNA polymerases in the absence of an instructional base. Takeshita et al. introduced 3-hydroxy-2-hydroxymethyl-tetrahydrofuran into DNA, which is a model for the predominant cyclic version of 2′-deoxyribose. Therefore, this analog serves as the sugar lacking the base, or an AP (apurinic/apyrimidinic) site [39]. It was shown that KF of Escherichia coli pol I, as well as calf-thymus DNA polymerase α, add dATP opposite synthetic abasic sites most frequently [39], leading to the proposal that DNA polymerases generally follow the “A-rule,” inserting A in the absence of specific coding information [40]. The cocrystal structure of KlenTaq DNA polymerase with the furan synthetic abasic site in the templating position suggests a mechanism for this, as a protein Tyr side chain fills the space left vacant by the missing base and acts as a pyrimidine base mimic [41]. Some C family DNA polymerases that are error prone and/or involved in mutagenesis can also bypass synthetic abasic sites, in the case of Streptococcus pyogenes by incorporating dA, dG, or to a lesser extent dC, and in the case of Bacillus subtilis by weakly incorporating dG and generating one-nucleotide deletions via a misalignment mechanism [42–44]. Saccharomyces cerevisiae B-family member DNA pol zeta only weakly bypasses abasic sites [45]. Strikingly, African Swine Fever Virus (ASFV) DNA pol X is a highly error-prone DNA polymerase but is unable to copy DNA containing an abasic site [46]. Pol X cannot insert a nucleotide opposite an abasic site, nor can it extend a primer terminus containing an abasic site [46].
Because Y family DNA polymerases are known to copy noncanonical DNA structures, their proficiency at copying synthetic abasic sites has been examined in some detail. The model Y family DNA polymerase Sulfolobus solfataricus Dpo4 copies synthetic abasic sites mainly by incorporating dA but also by generating small deletions [47]. E. coli DinB (DNA pol IV) efficiently copies DNA containing a synthetic abasic site, but primarily by generating (−2) deletions [48]. E. coli DNA pol V (UmuD′2C) bypasses synthetic abasic sites by inserting primarily dA (~70%) or dG (~30%) opposite the modification [49, 50]. Even though both E. coli Y family DNA polymerases can copy DNA containing abasic sites, Pol V is used to bypass abasic sites in vivo, probably because base substitutions are generally less harmful than frameshift mutations [48]. Human DNA pol iota, which, like ASFV pol X, is highly inaccurate when replicating undamaged DNA [51], can efficiently incorporate dG opposite an abasic site but is unable to extend from primer termini containing abasic sites [52]. Human DNA pol eta copies abasic sites by incorporating predominantly dA but also dG [53, 54], whereas human pol kappa incorporates predominantly dA but also generates one nucleotide deletions [55, 56]. Y family member Rev1 from yeast incorporates dC opposite abasic sites, which have been suggested to be the cognate lesion of Rev1 [57–59]. Yeast pol alpha, replicative DNA polymerase pol epsilon, and Y family pol eta are all capable of bypassing abasic sites, whereas replicative DNA polymerase pol delta is less efficient [60, 61]. Intriguingly, yeast pol eta and KF add a pyrene nucleotide opposite the template abasic site more efficiently than adding A, likely in part because pyrene is approximately the same size as a base pair and can engage in base stacking interactions [62, 63]. Bacteriophage T4 DNA polymerase incorporates nucleotide triphosphate versions of 5-nitroindolyl, 5-cyclohexyl-indole, and 5-cyclohexenyl-indole opposite abasic sites more efficiently than it incorporates dAMP [64, 65]. Due to the complicated responses of even the relatively forgiving Y family DNA polymerases to the synthetic model abasic site, it has been demonstrated that multiple DNA polymerases may be used to bypass DNA damage efficiently while minimizing mutations [66, 67].
Y family DNA polymerases are able to copy DNA containing noncanonical structures ranging from abasic sites to bulky DNA adducts [68–73]. Therefore, it was of interest to determine the minimal features of DNA required for replication. Short (three or 12) chains of methylene (CH2) residues in the middle of canonical DNA templates were used to probe tolerance for minimal DNA backbones. E. coli pols I, II, and III were unable to replicate either DNA structure. On the other hand, both pols IV and V could replicate the three- or 12-methylene linker-containing DNA in vitro, although, in an analogous situation to abasic sites, only pol V is observed to replicate these unusual structures in vivo [74]. Human DNA polymerases showed more subtle differences, in that pols eta, kappa, and iota could replicate a three-methylene linker by inserting nucleotides opposite the noninstructional segment, but only pols eta and kappa could fully bypass the modified gap [75]. Pols eta and iota could insert nucleotides opposite the 12-methylene linker, whereas pol kappa had little to no activity, and none of these three polymerases could completely bypass the 12-methylene linker [75]. Clearly, at least some Y family DNA polymerases are capable of replicating non-DNA segments.
In order to probe the size tolerance for bases in the active site, a series of dG analogs with increasingly large substituents at the N 2 position in the minor groove were constructed and used as the template base with a range of DNA polymerases. The N 2 modifications included methyl, ethyl, isobutyl, benzyl, CH2-napthyl, CH2-anthracenyl, and, in some cases, CH2-benzo[a]pyrenyl derivatives [76–80]. Bacteriophage T7 DNA polymerase (exonuclease−) and HIV-1 reverse transcriptase are both able to bypass the N 2-methyl derivative efficiently, although significantly less efficiently than unmodified DNA, but are not able to bypass any of the larger adducts [80]. Moreover, even the methyl substituent caused a high frequency of misincorporation [80]. On the other hand, each of the human Y family DNA polymerases is more tolerant of the size-expanded bases [76–79]. Rev1 is the most tolerant of N 2-dG-substitutions, followed by pol iota and pol kappa, whereas pol eta is the least tolerant, showing a decrease in activity of approximately two orders of magnitude between the CH2-napthyl and CH2-anthracenyl substituents [79]. A similar analysis of O 6-substituted bases showed that only Rev1 and pol iota could tolerate size-expanded substituents up to the benzyl substitution, but pol eta and pol kappa showed decreased activity even with an O 6-methyl substitution [79, 81]. The use of a series of well-defined synthetic base modifications provides insights into the steric constraints of DNA polymerase active sites and allows detailed comparisons to be made between replicative and damage-bypass polymerases.
3. Methyl-Substituted Phenyl Analogs
Efforts have been made to examine how DNA polymerases recognize methyl-substituted phenyl-based analogs that do not appear to be large enough to perturb DNA structure (Figure 3) [82]. There was significant self-base pairing of these substituted phenyl nucleobase analogs, which was not observed with the benzene analog [82]. Generally, in incorporation opposite these analogs, the Klenow fragment discriminates most against dCTP and dGTP, which tend to be the most hydrophilic nucleotides, while dTTP incorporation varies with the extent of methyl substitution, and dATP is added to these bases most efficiently [82]. The 2-substituted methyl-bearing phenyl groups generally favored dATP addition, but with the 3-substituted benzene rings, KF discriminated against dATP [82]. Interestingly, KF inserts dATP opposite MM1, DM2, DM5, and TMB (Figure 3) with a rate comparable to that of template dT [82]. This is hypothesized to be related not just to shape mimicry of dT in the template but to the placement of the specific substituents on the phenyl ring, which when appropriately oriented, can foster hydrophobic packing with the incoming dATP [82]. The most efficiently extended of these small substituted benzene derivatives are the ones that contain a methyl group at the 4-position [82]. Subsequent work using methoxy substituents, which unlike the methyl-substituted phenyl rings can form hydrogen bonds, suggests that positioning a hydrogen bond acceptor in the minor groove enhances both selectivity and efficiency of DNA synthesis by KF [83].
Figure 3.
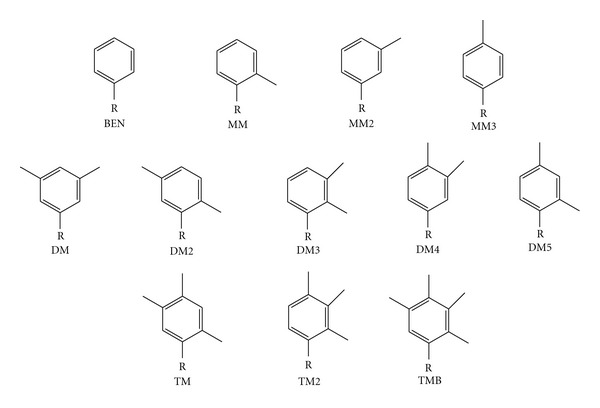
Small molecule analogs based on the benzene parent. BEN is benzene; the MM series is monomethylated at the 2, 3, and 4 positions, respectively; the DM series is dimethylated; the TM series is trimethylated; TMB is the tetramethylated benzene analog [82].
4. Hydrophobic Base Analogs
The use of hydrophobic and van der Waals interactions have been the driving force behind the development of a variety of unnatural nucleobases as possible base pairing partners and to assess polymerase utilization (Figure 4) [84]. The first of these is a self-pairing base known as 7-propynyl isocarbostyril nucleoside (PICS), which stabilizes the DNA helix when paired with itself but is destabilizing when paired with dA, dC, dG, or dT [84]. The PICS base does not demonstrate structural similarity to the natural bases, but the incorporation of dPICSTP opposite PICS in the template strand by KF is more efficient than the natural bases, ranging from 20-fold more efficient than dTTP insertion opposite dPICS to ~140-fold more efficient than dGTP insertion opposite PICS [84]. KF does not extend beyond the PICS : PICS base pair, however, which is postulated to be due to a perturbation in the position of the 3′-OH of the growing primer strand caused by the nonnatural base pair [84].
Figure 4.
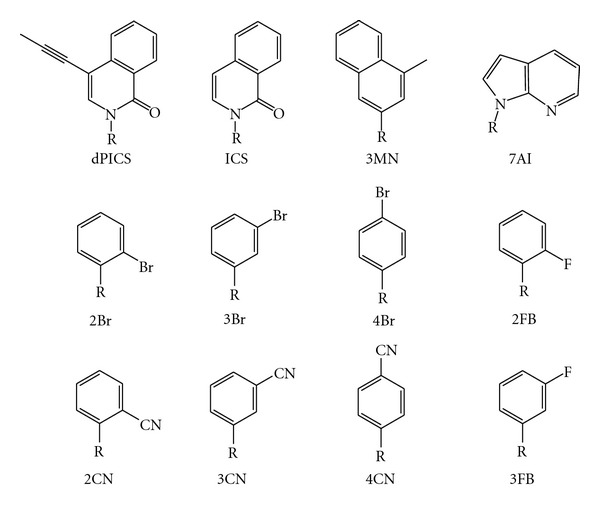
Hydrophobic nucleobase analogs: 7-propynyl isocarbostyril nucleoside (dPICS) [84]; isocarbostyril nucleoside (ICS); 3-methylnaphthalene (3MN); azaindole (7AI) [85]; bromo phenyl derivatives at positions 2, 3, and 4 (2Br, 3Br, and 4Br, resp.); cyano derivatives at positions 2, 3, and 4 (2CN, 3CN, and 4CN, resp.), fluoro derivatives at positions 2 and 3 (2FB, 3FB, resp.) [86].
Other hydrophobic nonnatural nucleobases are based on either the naphthalene system (Figure 4), nitrogenous base-like skeleton (Figure 4), or the skeleton of benzene (Figure 4) substituted with methyl, halide, or cyano groups [85]. While dATP is the nucleoside triphosphate most generally inserted opposite these analogs, the bromo and cyano adducts show interesting differences in KF discrimination, in that dG, dT, and dC are incorporated across from the 2-bromo derivative within threefold of the catalytic efficiency of dATP incorporation [85]. The cyano derivative at the same position leads to incorporation of dGTP ~sevenfold less efficiently than dATP, incorporation of dTTP even less efficiently, with no dCTP incorporation detected [85]. Relative to the benzene parent, only incorporation of dCTP from the 4-bromo derivative and dGTP paired opposite the 2-cyano derivative were more efficient [85]. KF primer extension after unnatural base pairing is more intriguing; specifically, 3-position substituted benzenes showed no detectable extension, with the exception of the 3-fluoro derivative which may be too small to inhibit extension due to steric hindrance (Figure 4) [85]. The base pair 4Br : 2CN was the most efficiently extended, most likely because the CN can act as a hydrogen bond acceptor and can be a driving force in primer extension [85].
5. Purine/Pyrimidine Mimics
Pyrimidine nucleotide analogs can affect polymerase activity in different ways. On one hand, pyrimidine nucleotide analogs lacking the 2-keto group can inhibit DNA polymerase activity [87]. Specifically, 2-amino-5-(2′-deoxy-β-D-ribofuranosyl)pyridine-5′-triphosphate (d*CTP), a cytosine analog, and 5-(2′-deoxy-β-D-ribofuranosyl)-3-methyl-2-pyridone-5′-triphosphate (d*TTP), a thymine analog, completely block Taq DNA polymerase from inserting them along a growing DNA strand (Figure 5) [87]. In these two analogs, in addition to the keto deletion, the C–N glycosidic bond functionality is removed and replaced with a slightly longer C–C bond, which may alter steric and electronic complementarity between the nucleotides and the polymerase [87]. These modified triphosphates, however, are tolerated by T7 RNA polymerase [88]; thus, it was concluded that the lack of the carbonyl functionality of these analogs is more responsible for the inhibition of Taq DNA polymerase than that of the longer C–C bond [87].
Figure 5.
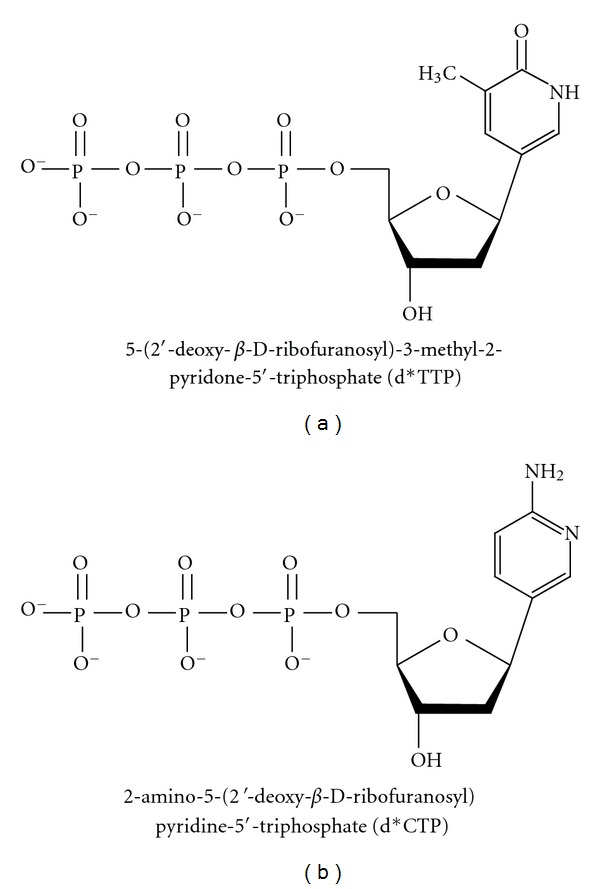
Taq polymerase activity is severely inhibited by (a) 5-(2′-Deoxy-β-D-ribofuranosyl)-3-methyl-2-pyridone-5′-triphosphate d*TTP and (b) 2-Amino-5-(2′-deoxy-β-D-ribofuranosyl)pyridine-5′-triphosphate (d*CTP), dC, and dT analogs, respectively, lacking the 2-keto groups [87].
An effort to probe recognition of purines by Bacillus stearothermophilus DNA pol I utilized a number of aza-purine derivatives and found that substitutions of carbon at N-1 or N-3 caused the most severe defects in efficiency, whereas alterations at N-1 or N 6 resulted in loss of fidelity [89]. A similar type of analysis found that removal of the exocyclic 2-amino group of G had little effect on the efficiency of either T7 DNA polymerase or Dpo4 [90]. However, replacement of the 2-amino group by progressively larger and less electronegative substituents, F, O, and Br, led to decreasing activity by both T7 DNA pol and Dpo4 [90]. This observation led to the suggestion that the trend was due to both the size and charge of the C-2 substituent [90].
Azole heterocyclic carboxamides can act as nucleobase mimics and, in fact, structurally can take on the appearance of either purines or pyrimidines (Figure 6) [91]. Because these analogs are small, they have some molecular mobility and can shift in order to adjust the hydrogen bonding patterns and electronic interactions to allow pairing with different bases [91]. Each of these azole heterocyclic carboxamides show some preference for pairing with specific incoming dNTPs, based on the position of the hydrogen bond donors and acceptors (Figure 6) [91]. For example, (1H)-1,2,3-Triazole-4-carboxamide directs the insertion of dGTP, but others do not [91]. The modified bases 1,2,4-triazole-3-carboxamide and 1,2,3-triazole-4-carboxamide, as well as 1,2-pyrazole-3-carboxamide orient in a way to promote hydrogen bonding to dC [91]. Taq DNA polymerase can utilize these analogs in PCR reactions but has different incorporation efficiencies for the different analog-dNTP pairs [91]. The presence of an azole analog in a DNA template reduces the catalytic efficiency for matched versus mismatched base pairs from 1000-fold discrepancy for natural base pairs to ~50-fold difference for base pairs involving azole analogs [91]. Therefore, these analogs are treated less stringently, but also incorporated less efficiently than natural bases by Taq DNA pol I, and demonstrate the complexity of the process of nucleotide addition, which involves electrostatic interactions, hydrogen bonding, and shape recognition [91].
Figure 6.

Classes of purine/pyrimidine mimics. (a) Skeleton structure of azole heterocyclic carboxamides. Depending on the position of the heteroatoms and their ability to donate or accept hydrogen bonds, these analogs can form base pairs with either a purine or a pyrimidine [91]. (b) General structure of the furo/thieno pyridinones; different heteroatoms in each position give the following compounds: furo[2,3-c]pyridin-7(6H)-one: 7OFP, thieno[2,3-c]pyridin-7(6H)-one: 7OTP, furo[2,3-c]pyridin-7-thiol: 7TFP, furo[3,2-c]pyridin-4(5H)-one: 4OFP, thieno[3,2-c]pyridin-4(5H)-one: 4OTP, furo[3,2-c]pyridin-4-thiol: 4TFP [86, 92].
Other scaffolds for unnatural self-pairing heteroatom-containing purine mimics have been developed, known as furo or thieno pyridinones (furo[2,3-c]pyridin-7(6H)-one: 7OFP, thieno[2,3-c]pyridin-7(6H)-one: 7OTP, furo[2,3-c]pyridin-7-thiol: 7TFP, furo[3,2-c]pyridin-4(5H)-one: 4OFP, thieno[3,2-c]pyridin-4(5H)-one: 4OTP, furo[3,2-c]pyridin-4-thiol: 4TFP) (Figure 6) [86]. The goal of using these analogs is to increase the ability of the DNA polymerase to continue to extend after the analog is bypassed, which is an important step in DNA polymerization, especially for DNA damage tolerance [34–37]. The most stable base pairing of these analogs is self-pairing followed by dA, dG, dC in that order, with the sulfur moiety providing more stabilization than that of oxygen [86]. KF does not discriminate strongly when synthesizing the furo versus the thieno pyridinones as self-pairs but does exhibit differences when extending beyond the unnatural bases when they are self-paired [86]. Most of these analogs disrupted the addition of dCTP to dG at the next nucleotide position after the pyridinone self-pair, with the exception of 4TFP [86]. No natural nucleotide triphosphate is found to be inserted by KF opposite 7TFP making it the most selective. The pyridinone 4OTP is the second most selective for its self-pairing, with only dTTP a modest 1.7-fold more efficiently incorporated, and selectivity drops in the following order: 7OTP, 4TFP, 4OFP, with 7OFP being the least selective [86]. Each of these analogs, with the exception of 4OFP nucleotide triphosphate, is efficiently incorporated by KF opposite dG, with the other templating bases having lower incorporation efficiencies but that are within 20-fold of the natural DNA pairs being synthesized [86]. The extension beyond these analogs by KF polymerase increases by at least fivefold over the PICS-type analogs [86]. The purine mimic 5-nitro-indolyl-2′-deoxyribose-5′-triphosphate is known to block E. coli DNA replication, not by inhibiting the polymerase directly but by inhibiting the ability of the clamp loader to assemble the entire replisome by blocking ATP binding and hydrolysis [93]. However, in the Taq system, a directed evolution experiment led to the identification of a DNA polymerase variant containing multiple mutations that facilitates bypass of the 5-nitro-indole analog, while polymerization by wild-type Taq was strongly blocked [94]. The mutations were concentrated in and near the active site but were also found throughout the DNA polymerase, indicative of the multiple mechanisms by which this Taq DNA polymerase variant is able to copy unusual DNA structures [94].
There is evidence that some unnatural dA mimics paired with abasic sites are proofread. Purines are generally added opposite abasic sites; unnatural nucleotides based on the indole scaffold substituted at the five position (Figure 7) were used to probe insertion by T4 DNA polymerase [95]. Despite the difference in size and shape, both 5-phenyl-indolyl-2′deoxyriboside triphosphate (5-PhITP) and 5-nitro-indolyl-2′-deoxyriboside triphosphate (5-NITP) are rapidly incorporated opposite an abasic site, whereas the 5-fluoro (dFITP) and 5-amino (dAITP) analogs have a very low efficiency of incorporation; the increase in π electrons of the former is apparently a key contributor to catalytic efficiency [95]. Two of these analogs, dNITP and dPhITP, are used as chain terminators (Figure 7) [96] but are excised more efficiently when inserted opposite a natural nucleoside as opposed to an abasic site [96]. Evidence also exists for structural changes to allow these chain terminators to be readily incorporated. Furthermore, KF proofreads bases paired with the template purine analog 4-methylbenzimidazole as efficiently as it proofreads natural mismatches; however, it is less efficient at removing 4-methylbenzimidazole from a primer terminus, suggesting that natural bases may be specifically recognized by the exonuclease active site [97].
Figure 7.
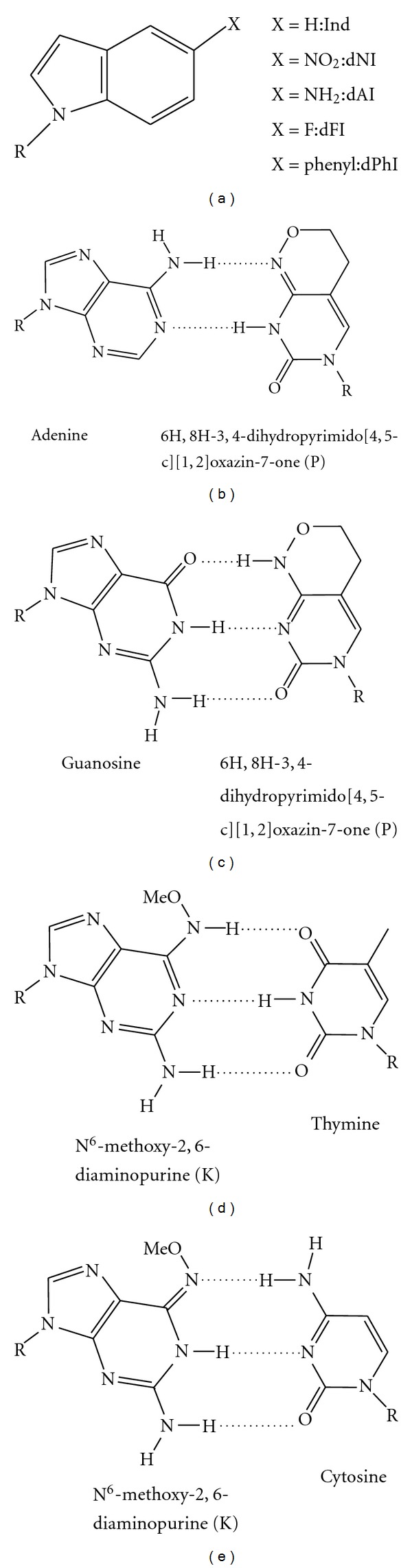
Purine and pyrimidine mimics. (a) Purine analogs based on the indole scaffold: 5-substituted indolyl-2′deoxyriboside triphosphates where X = H, indole (Ind); X = NO2, 5-nitro-1-indole (dNI); X = NH2, 5-amino-1-indole (dAI); X = F, 5-fluoro-1-indole (dFI); X = phenyl, 5-phenyl-1-indole (dPhI) [95, 96]. ((b)–(e)) Pyrimidine mimic 6H,8H-3,4-dihydropyrimido[4,5-c][1,2]oxazin-7-one (P), and purine mimic N 6-methoxy-2,6-diaminopurine (K) base pairing partners. (b) Adenine : P. (c) Guanosine : P. (d) Thymine : K. (e) Cytosine : K. The ability of these analogs to form different tautomers can result in mutations [98].
Modified bases 6H,8H-3,4-dihydropropyrimido[4,5-c][1,2]oxazin-7-one (P) and N 6-methoxy-2,6-diaminopurine (K) are generic pyrimidine and purine mimics, respectively (Figure 7) [99]. Taq DNA polymerase copies each of these as expected: P is treated generically as a pyrimidine in the template strand, pairing with either dG or dA, and K is treated by Taq as a general templating purine, pairing with either dC or dT [99]. Taq shows a preference to use P as dT in PCR reactions, giving a dT : dC ratio of 3 : 2, while preferring to use K as dA, with an dA : dG ratio of 7 : 1 [99]. These analogs are effective as universal bases due to the prevalence of tautomeric forms, observed in nuclear magnetic resonance (NMR) experiments, that allow base pairing to multiple partners [98–100].
6. isoC and isoG
isoC and isoG were recognized as forming base pairs in DNA and RNA in the late eighties and early nineties (Figure 8) [101, 102] and then were accepted as a third base pair of DNA in 2003 [103]. The isoC : isoG base pair is different from its natural counterparts in the transposition of the amine and carbonyl groups on both dG and dC; however, standard Watson-Crick hydrogen-bonding is still present [104]. These analogs were first demonstrated to be useful in improving PCR efficiency [105]. isoG can take on the enol form, which base pairs with T, but it can also adopt the keto form, which base pairs readily with the thymine analog 5-methylisocytosine (MiC) [106]. The recombination protein RecA can mediate strand exchange with DNA containing iG and MiC base pairs at rates comparable to those of the natural bases, which expands the range of recombination-competent genetic material [104].
Figure 8.

isoC, isoG, and their base pairs (a) isocytosine : isoguanosine and (b) (enol) isoG : thymine [101–106].
7. Thymidine Analogs
Thymidine analogs have been particularly useful for probing DNA replication. Difluorotoluene, for example, is a synthetic dT analog, in which the hydrogen bonding capabilities seen in dA : dT base pairing are reduced or eliminated (Figure 9) [107–109]. Nevertheless, this analog can serve as a very good templating base for KF [107]. Difluorotoluene promotes efficiency of insertion as the incoming nucleotide only about fourfold less than that of natural dTTP [107]. When dA at the primer end is paired with difluorotoluene as the template base, dA is removed by KF exonucleolytic proofreading as efficiently as a natural base mismatch [97]. A similar effect was observed with human mitochondrial DNA polymerase gamma [110]. On the other hand, when difluorotoluene is at the primer terminus, the relative efficiency of removal is approximately 40-fold lower than that of natural base mismatches, again suggesting that specific interactions with natural bases govern removal by the exonuclease domain [97]. Difluorotoluene is an efficient template base for KF [97]. In contrast, difluorotoluene is poorly replicated by yeast pol eta and human pol kappa [112, 113], while S. solfataricus Dpo4 exhibits low activity but is able to carry out primer extension on templates containing difluorotoluene [114].
Figure 9.
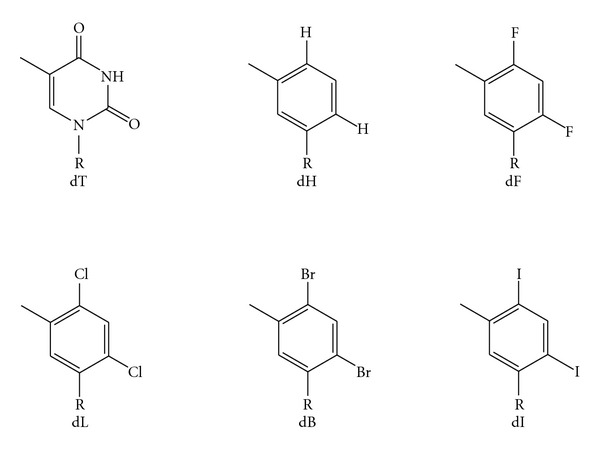
Small molecule thymidine (dT) analogs: 3-toluene-1-β-D-deoxyriboside (dH); 2,4-difluoro-5-toluene-1-β-D-deoxyriboside (dF); 2,4-dichloro-5-toluene-1-β-D-deoxyriboside (dL); 2,4-dibromo-5-toluene-1-β-D-deoxyriboside (dB); 2,4-diiodo-5-toluene-1-β-D-deoxyriboside (dI) [111]. The deoxyribose groups are indicated by R.
Hydrogen bonding capacities can be retained in a structure such as 2-thioTTP, in order to improve fidelity of PCR, which can be decreased by the tautomerization of dG to form the isoG minor tautomer [105, 115]. Use of 2-thioTTP increases fidelity of those PCR reactions that include isoC and isoG [92, 105, 116] by 5% using KlenTaq DNA polymerase [115]. This is due to introducing a specific steric interaction that prevents pairing between isoG and 2-thioTTP [115]. The yellow-colored 4-Se-T is also capable of hydrogen bonding with dA and is efficiently incorporated as 4-SeTTP into DNA by KF [117].
Thymidine analogs have also been used to study the steric interactions that govern nucleotide additions. Incrementally increasing the size of the substituent in place of the carbonyl oxygen on thymidine with a series of halide substitutions (F, Cl, Br, I) demonstrates that the replicative polymerase KF has a specific “tightness” that allows for only some substitutions to be incorporated. The highest efficiency of incorporation by KF was with base pairs that are larger than natural base pairs [111]. In contrast, T7 DNA polymerase is more stringent and has an optimum that is closer to the size of natural base pairs [111, 118]. Moving the substituents around the thymidine ring and probing the activity of KF led to the conclusion that KF is remarkably sensitive to the overall shape of the template base and incoming nucleotide [119]. KF achieves maximal fidelity of incorporation with the chlorosubstituted analog 2,4-dichloro-5-toluene-1-β-D-deoxyriboside (Figure 9) [111]. The catalytic efficiency of KF with these analogs showed that with the increase in size by 0.66 Å (H → Cl), KF was more efficient by a factor of ~180 [111]. This trend of increasing steric hindrance with these thymidine analogs utilized by KF is consistent with the steric hindrance seen with 4′ substituted dTTP analogs noted previously [120]. In contrast, the presence of 4′ substituted T analogs in the template are well tolerated by KF [121]. The model Y family DNA polymerase, S. solfataricus Dbh, incorporates 4′-modified dTTP analogs relatively efficiently and binds to the analogs nearly as well as binding to unmodified dTTP [122]. Similarly, Y family DNA polymerase Dpo4 exhibits much less size selectivity than KF, as determined with halogen-substituted thymine analogs [123]. Thus, although some Y family DNA polymerases require hydrogen bonding for efficient replication, these studies confirm their generally accommodating active sites.
8. Fluorescent Base Analogs
8.1. 2-Aminopurine
The most common fluorescent base analog in use today is 2-aminopurine (2AP), which can form hydrogen bonds and base pair with either of the pyrimidines thymine or cytosine (Figure 10) [124]. A recent crystal structure of DNA containing a 2AP : dC base pair in the active site of the Y567A variant of RB69 DNA polymerase suggests that the 2AP : dC pair may contain a bifurcated hydrogen bond between N 2-H of 2AP and N3 and O2 of dC [125]. In this example, the Y567A active site mutation in the nascent base-pair-binding pocket is both less discriminating in the formation of mismatched base pairs and is better able to extend mismatched primer termini [125]. The modified base 2AP is commercially available and has been used to study a number of DNA-binding protein interactions including KF [126], EcoRI, DNA methyltransferase [127], endonuclease [128, 129], and uracil DNA glycosylase [130]. The fluorescence of this analog is sequence-context dependent, with the most pronounced effect occurring when the base is surrounded by other purines; much like other fluorescent nucleobases, its fluorescence is quenched when it is within DNA [131]. KF has been shown to utilize 2AP, and the fluorescence has been used to give insights into the dynamics of this protein as it synthesizes DNA [126, 132, 133]. For example, in one FRET experiment with a labeled KF, the mechanism of the fingers closing conformational change was studied [133] and was found to be influenced by the added nucleotide. Specifically, mismatched nucleotides are detected before the polymerase “closes” on the DNA suggesting that the mismatched nucleotide itself may destabilize the “open” polymerase conformation [133]. The role in the conformational change of the divalent cation (usually Mg2+ or Ca2+ but, in this case, an “exchange inert” Rh(III)) was also probed using 2AP [134], and it was found that dNTP binding in the absence of the correct ion can induce the conformational shift [134]. The ability of the ion to diffuse to the proper position before the nucleophilic attack can occur may influence the reverse conformational shift observed in the presence of the incorrect nucleotide [134].
Figure 10.

2-Aminopurine (2AP) and its base pairs (a) 2AP base paired with thymine and (b) 2AP base paired with cytosine [124].
Fluorescence spectroscopy with 2AP can be used to study DNA polymerization on a millisecond time scale, and probe single events like nucleotide addition, base pairing interactions, and subsequent excision via nuclease activity [126, 132]. Insertion kinetics have been measured for the monophosphate version of 2AP (dAPMP versus dAMP); dAPMP is found to be misincorporated at similar rates to the incorporation of the natural triphosphate dATP opposite dT by KF [126]. This makes 2AP useful in studying polymerase activity as it is misincorporated about as frequently as dA is incorporated. However, this incorporation is influenced by the sequence surrounding the primer terminus, with double the rate of misincorporation of 2AP triphosphate if the nearest neighbor to the nascent base pair is dG, dC, or dA, as compared to dT [126].
Y family polymerases also have been studied using 2AP. Dbh adds dTTP correctly opposite 2AP in the template strand and binds various DNA substrates containing 2AP with K D values similar to those of natural DNA substrates [135]. Use of 2AP to monitor conformation changes during the base-skipping phenomenon, which can generate frameshift mutations as seen with Y family polymerases, provides evidence that the misincorporation pathway is distinct from the correct dNTP incorporation process [135]. Fluorescence from 2AP has been also used to probe the proofreading mechanism by which bases are excised via nuclease activity of phage T4 polymerase [136].
The analog 2AP has been used together with the base analog pyrrolo-dC as a FRET pair as the excitation and emission wavelengths of these two nucleotide probes are compatible [137], though this pair has not yet been utilized to study DNA polymerases. Pyrrolo-dC alone has been used to study DNA/RNA hybrids [138], single-stranded DNA hairpins [139], and base pair flipping [140]. Two potential drawbacks of using 2AP are the sequence dependence of its fluorescence and that it can perturb the DNA structure or be mutagenic if it forms a wobble pair with dT [124]. A 2AP : dT base pair destabilizes duplex DNA by ~8°C relative to a dA : dT base pair [141].
8.2. tC: 1,3-Diaza-2-oxophenothiazine
The synthetic cytosine analog tC was developed first by Lin et al. [142] but then used as a probe of DNA polymerases by Wilhelmsson and coworkers [143–145]. The fluorescence quantum yield of this nucleotide analog, unlike 2AP, is not sensitive to the surrounding environment [144, 146]. This base also is incorporated into DNA, shows canonical base pairing with guanosine (Figure 11), and does not perturb the B-form structure of DNA. In fact, a dG : tC base pair stabilized DNA by 3°C [124]. Different DNA polymerases have different efficiencies in utilizing tC in template DNA and in incorporating tC into the growing DNA primer strand. For example, KF utilizes template tC in preference to a template C, as KF apparently has a flexible enough active site to accommodate the extra cyclic ring system. Klenow also preferentially incorporates the tC nucleotide triphosphate in the growing DNA strand. E. coli DinB (pol IV), which is a Y family DNA polymerase [17], also utilizes the tC nucleotide triphosphate more efficiently than dCTP, similar to Klenow [147]. DinB also can extend from tC at the primer terminus [147]. However, DinB shows a 12-fold decrease in the catalytic efficiency of incorporation of dGTP opposite template tC as compared to the natural dC in the template strand and is unable to extend from the newly generated primer terminus [147]. Primer extension by DinB is inhibited unless the primer terminus is at least 3-4 nucleotides beyond the tC analog, which suggests that the “TLS patch” of nucleotides required beyond noncognate bases for DNA polymerases to resume efficient synthesis is shorter for a Y family DNA polymerase than for replicative polymerases. Moreover, the striking asymmetry of the DinB active site has also been observed in the case of B family DNA polymerases human polymerase alpha and herpes simplex virus I DNA polymerase when probed with nonnatural nucleotide analogs [148].
Figure 11.

Fluorescent cytosine analogs tC and tC° form canonical base pairs with dG, do not perturb B-form DNA structure, and can pair with a FRET donor to probe DNA polymerase dynamics [144–146, 149, 150].
8.3. tC°: 1,3-Diaza-2-oxophenoxazine
The oxo-analog of tC is tC°, 1,3-diaza-2-oxophenoxazine (Figure 11) [142], which has several similar properties to that of tC in that it stabilizes B-form DNA by 3°C and it base pairs with G in a standard Watson-Crick configuration [149]. It is exceptionally bright, on average 10–50 times brighter than 2AP, 3-MI, and 6-MAP [149]. The tC° analog, like tC, can be utilized by KF and by human DNA primase [146, 150, 151]. This analog has proven useful in high-density labeling of PCR products using a deep vent DNA polymerase and therefore should be useful in biotechnology applications [151].
9. Conclusions
Nonnatural nucleotides continue to provide an important tool for the study of DNA and its interacting protein partners. In particular, DNA polymerases that are responsible for the systematic replication of DNA, whether accurate or mutagenic, are required to specifically recognize and efficiently base pair with a large number of noncanonical DNA structures. An increasingly expanding genetic alphabet of nonnatural nucleobases provides the ability to obtain an unparalleled level of detail about how DNA polymerases discriminate among many different DNA structures. From the first introduction of artificial abasic sites [39] to the use of bright nonperturbing fluorescent analogs that are used to probe polymerase opening and closing dynamics on a nascent base pair [144, 146], nonnatural nucleotides are now fully integrated into DNA polymerase research. There remains however a need for novel DNA bases that have specific properties in order to better study the interactions of DNA polymerases with DNA. In particular, efficiently generating both phosphoramidite monomers and triphosphate versions of a given modified base can be a significant synthetic challenge. The understanding of DNA polymerase specificity for synthetic nucleobases, discussed in this paper and elsewhere [28, 38, 124, 152, 153], is increasing; in the future, synthetic bases will continue to be used for a variety of purposes including probing proteins and small molecules that bind to DNA, optimizing unnatural bases for coding as a synthetic genetic code [154], synthesizing unnatural biopolymers [155], and improving the prospects of DNA as a nanomaterial and a drug target [156].
Acknowledgments
This work was supported by the National Science Foundation CAREER Award, Grant no. MCB-0845033, and the Northeastern University Office of the Provost. P. J. Beuning is a Cottrell Scholar of the Research Corporation for Science Advancement. The authors thank Jana Sefcikova and Lisa Hawver for helpful suggestions on the paper.
References
- 1.Franklin RE, Gosling RG. Evidence for 2-chain helix in crystalline structure of sodium deoxyribonucleate. Nature. 1953;172(4369):156–157. doi: 10.1038/172156a0. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FH. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. Harvey Lectures. 1957;53:83–112. [PubMed] [Google Scholar]
- 4.Kleppe K, Ohtsuka E, Kleppe R, Molineux I, Khorana HG. Studies on polynucleotides. XCVI. Repair replication of short synthetic DNA’s as catalyzed by DNA polymerases. Journal of Molecular Biology. 1971;56(2):341–361. doi: 10.1016/0022-2836(71)90469-4. [DOI] [PubMed] [Google Scholar]
- 5.Cordonnier AM, Fuchs RP. Replication of damaged DNA: molecular defect in Xeroderma pigmentosum variant cells. Mutation Research. 1999;435(2):111–119. doi: 10.1016/s0921-8777(99)00047-6. [DOI] [PubMed] [Google Scholar]
- 6.Bresson A, Fuchs RP. Lesion bypass in yeast cells: pol eta participates in a multi-DNA polymerase process. EMBO Journal. 2002;21(14):3881–3887. doi: 10.1093/emboj/cdf363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pages V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21(58):8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 8.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nature Reviews Cancer. 2011;11(2):96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb LA, Monnat RJ., Jr. DNA polymerases and human disease. Nature Reviews Genetics. 2008;9(8):594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 10.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305(5690):1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 11.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (pol II, pol IV and pol V) are involved in induced mutagenesis. EMBO Journal. 2000;19(22):6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albertella MR, Green CM, Lehmann AR, O’Connor MJ. A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Research. 2005;65(21):9799–9806. doi: 10.1158/0008-5472.CAN-05-1095. [DOI] [PubMed] [Google Scholar]
- 13.Albertella MR, Lau A, O’Connor MJ. The overexpression of specialized DNA polymerases in cancer. DNA Repair. 2005;4(5):583–593. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Bassett E, King NM, Bryant MF, et al. The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Research. 2004;64(18):6469–6475. doi: 10.1158/0008-5472.CAN-04-1328. [DOI] [PubMed] [Google Scholar]
- 15.Chen YW, Cleaver JE, Hanaoka F, Chang CF, Chou KM. A novel role of DNA polymerase eta in modulating cellular sensitivity to chemotherapeutic agents. Molecular Cancer Research. 2006;4(4):257–265. doi: 10.1158/1541-7786.MCR-05-0118. [DOI] [PubMed] [Google Scholar]
- 16.Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Scharer OD. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Research. 2011 ;39:7455–7464. doi: 10.1093/nar/gkr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohmori H, Friedberg EC, Fuchs RP, et al. The Y-family of DNA polymerases. Molecular Cell. 2001;8(1):7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 18.Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. Journal of Molecular Biology. 2001;308(5):823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 19.Federley RG, Romano LJ. DNA polymerase: structural homology, conformational dynamics, and the effects of carcinogenic DNA adducts. Journal of Nucleic Acids. 2010;2010:15 pages. doi: 10.4061/2010/457176. Article ID 457176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgers PM, Eckstein F. A study of the mechanism of DNA polymerase I from Escherichia coli with diastereomeric phosphorothioate analogs of deoxyadenosine triphosphate. The Journal of Biological Chemistry. 1979;254(15):6889–6893. [PubMed] [Google Scholar]
- 21.Adler J, Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. IV. linkage of single deoxynucleotides to the deoxynucleoside ends of deoxyribonucleic acid. Proceedings of the National Academy of Sciences of the United States of America. 1958;44:641–647. doi: 10.1073/pnas.44.7.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bessman MJ, Lehman IR, Adler J, Zimmerman SB, Simms ES, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. III. The incorporation of pyrimidine and purine analogues into deoxyribonucleic acid. Proceedings of the National Academy of Sciences of the United States of America. 1958;44:633–640. doi: 10.1073/pnas.44.7.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bessman MJ, Lehman IR, Simms ES, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. II. General properties of the reaction. The Journal of Biological Chemistry. 1958;233(1):171–177. [PubMed] [Google Scholar]
- 24.Kornberg SR, Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic cleavage of deoxyguanosine triphosphate to deoxyguanosine and tripolyphosphate. The Journal of Biological Chemistry. 1958;233(1):159–162. [PubMed] [Google Scholar]
- 25.Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli . The Journal of Biological Chemistry. 1958;233(1):163–170. [PubMed] [Google Scholar]
- 26.Berdis AJ. Mechanisms of DNA polymerases. Chemical Reviews. 2009;109(7):2862–2879. doi: 10.1021/cr800530b. [DOI] [PubMed] [Google Scholar]
- 27.Reha-Krantz LJ. DNA polymerase proofreading: multiple roles maintain genome stability. Biochimica et Biophysica Acta. 2010;1804(5):1049–1063. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Lee I, Berdis AJ. Non-natural nucleotides as probes for the mechanism and fidelity of DNA polymerases. Biochimica et Biophysica Acta. 2010;1804(5):1064–1080. doi: 10.1016/j.bbapap.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedberg E, Walker GC, Siede WW, Wood RD, Schultz R, Ellenberger T. DNA Repair and Mutagenesis. 2nd edition. ASM Press; 2006. [Google Scholar]
- 30.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107(1):91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 32.Kiefer JR, Mao C, Hansen CJ, et al. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 Å resolution. Structure. 1997;5(1):95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 33.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Advances in Protein Chemistry. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 34.Fuchs RP, Fujii S. Translesion synthesis in Escherichia coli: lessons from the NarI mutation hot spot. DNA Repair. 2007;6(7):1032–1041. doi: 10.1016/j.dnarep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 35.Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO Journal. 2004;23(21):4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarosz DF, Cohen SE, Delaney JC, Essigmann JM, Walker GC. A DinB variant reveals diverse physiological consequences of incomplete TLS extension by a Y-family DNA polymerase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21137–21142. doi: 10.1073/pnas.0907257106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pages V, Janel-Bintz R, Fuchs RP. Pol III proofreading activity prevents lesion bypass as evidenced by its molecular signature within E. coli cells. Journal of Molecular Biology. 2005;352(3):501–509. doi: 10.1016/j.jmb.2005.07.063. [DOI] [PubMed] [Google Scholar]
- 38.Kool ET. Replacing the nucleobases in DNA with designer molecules. Accounts of Chemical Research. 2002;35(11):936–943. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- 39.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. The Journal of Biological Chemistry. 1987;262(21):10171–10179. [PubMed] [Google Scholar]
- 40.Strauss BS. The “A rule” of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? BioEssays. 1991;13(2):79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 41.Obeid S, Blatter N, Kranaster R, et al. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO Journal. 2010;29(10):1738–1747. doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruck I, Goodman MF, O’Donnell M. The essential C family DnaE polymerase is error-prone and efficient at lesion bypass. The Journal of Biological Chemistry. 2003;278(45):44361–44368. doi: 10.1074/jbc.M308307200. [DOI] [PubMed] [Google Scholar]
- 43.Le Chatelier E, Bécherel OJ, d’Alençon E, et al. Involvement of DnaE, the second replicative DNA polymerase from Bacillus subtilis, in DNA mutagenesis. The Journal of Biological Chemistry. 2004;279(3):1757–1767. doi: 10.1074/jbc.M310719200. [DOI] [PubMed] [Google Scholar]
- 44.McHenry CS. Breaking the rules: bacteria that use several DNA polymerase IIIs. EMBO Reports. 2011;12(5):408–414. doi: 10.1038/embor.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone JE, Kumar D, Binz SK, et al. Lesion bypass by S. cerevisiae Pol zeta alone. DNA Repair. 2011;10(8):826–834. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, Lamarche BJ, Tsai MD. Use of damaged DNA and dNTP substrates by the error-prone DNA polymerase X from African swine fever virus. Biochemistry. 2007;46(12):3814–3825. doi: 10.1021/bi061501l. [DOI] [PubMed] [Google Scholar]
- 47.Fiala KA, Hypes CD, Suo Z. Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. The Journal of Biological Chemistry. 2007;282(11):8188–8198. doi: 10.1074/jbc.M610718200. [DOI] [PubMed] [Google Scholar]
- 48.Maor-Shoshani A, Hayashi K, Ohmori H, Livneh Z. Analysis of translesion replication across an abasic site by DNA polymerase IV of Escherichia coli . DNA Repair. 2003;2(11):1227–1238. doi: 10.1016/s1568-7864(03)00142-3. [DOI] [PubMed] [Google Scholar]
- 49.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD’, RecA, and SSB and is specialized for translesion replication. The Journal of Biological Chemistry. 1999;274(45):31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 50.Tang M, Pham P, Shen X, et al. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404(6781):1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 51.Bebenek K, Tissier A, Frank EG, et al. 5′-deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science. 2001;291(5511):2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucleic Acids Research. 2001;29(4):928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO Journal. 2000;19(12):3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. The Journal of Biological Chemistry. 2003;278(50):50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 55.Sherrer SM, Fiala KA, Fowler JD, Newmister SA, Pryor JM, Suo Z. Quantitative analysis of the efficiency and mutagenic spectra of abasic lesion bypass catalyzed by human Y-family DNA polymerases. Nucleic Acids Research. 2011;39(2):609–622. doi: 10.1093/nar/gkq719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Yuan F, Wu X, et al. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro . Nucleic Acids Research. 2000;28(21):4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim N, Mudrak SV, Jinks-Robertson S. The dCMP transferase activity of yeast Rev1 is biologically relevant during the bypass of endogenously generated AP sites. DNA Repair. 2011;10:1262–1271. doi: 10.1016/j.dnarep.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pryor JM, Washington MT. Pre-steady state kinetic studies show that an abasic site is a cognate lesion for the yeast Rev1 protein. DNA Repair. 2011;10:1138–1144. doi: 10.1016/j.dnarep.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otsuka C, Loakes D, Negishi K. The role of deoxycytidyl transferase activity of yeast Rev1 protein in the bypass of abasic sites. Nucleic Acids Research. 2002;(2):87–88. doi: 10.1093/nass/2.1.87. [DOI] [PubMed] [Google Scholar]
- 60.Sabouri N, Johansson E. Translesion synthesis of abasic sites by yeast DNA polymerase epsilon. The Journal of Biological Chemistry. 2009;284(46):31555–31563. doi: 10.1074/jbc.M109.043927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao B, Xie Z, Shen H, Wang Z. Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Research. 2004;32(13):3984–3994. doi: 10.1093/nar/gkh710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matray TJ, Kool ET. A specific partner for abasic damage in DNA. Nature. 1999;399(6737):704–708. doi: 10.1038/21453. [DOI] [PubMed] [Google Scholar]
- 63.Sun L, Zhang K, Zhou L, et al. Yeast pol eta holds a Cis-Syn thymine dimer loosely in the active site during elongation opposite the 3′-T of the dimer, but tightly opposite the 5′-T. Biochemistry. 2003;42(31):9431–9437. doi: 10.1021/bi0345687. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, Lee I, Zhou X, Berdis AJ. Hydrophobicity, shape, and pi-electron contributions during translesion DNA synthesis. Journal of the American Chemical Society. 2006;128(1):143–149. doi: 10.1021/ja0546830. [DOI] [PubMed] [Google Scholar]
- 65.Reineks EZ, Berdis AJ. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43(2):393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 66.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406(6799):1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 67.Shachar S, Ziv O, Avkin S, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO Journal. 2009;28(4):383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jarosz D, Beuning P, Cohen S, Walker GC. Y-family DNA polymerases in Escherichia coli . Trends in Microbiology. 2007;15(2):70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 69.Walsh JM, Hawver LA, Beuning PJ. Escherichia coli Y family DNA polymerases. Frontiers in Bioscience. 2011;16(7):3164–3182. doi: 10.2741/3904. [DOI] [PubMed] [Google Scholar]
- 70.Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiology and Molecular Biology Reviews. 2009;73(1):134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Washington MT, Carlson KD, Freudenthal BD, Pryor JM. Variations on a theme: Eukaryotic Y-family DNA polymerases. Biochimica et Biophysica Acta. 2010;1804(5):1113–1123. doi: 10.1016/j.bbapap.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang W. Damage repair DNA polymerases Y. Current Opinion in Structural Biology. 2003;13(1):23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 73.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(40):15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maor-Shoshani A, Ben-Ari V, Livneh Z. Lesion bypass DNA polymerases replicate across non-DNA segments. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):14760–14765. doi: 10.1073/pnas.2433503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adar S, Livneh Z. Translesion DNA synthesis across non-DNA segments in cultured human cells. DNA Repair. 2006;5(4):479–490. doi: 10.1016/j.dnarep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 76.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N 2-alkyl guanine DNA adducts by human DNA polymerase kappa. The Journal of Biological Chemistry. 2006;281(30):21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 77.Choi JY, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N 2-guanine DNA lesions by human DNA polymerase eta. Journal of Molecular Biology. 2005;352(1):72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 78.Choi JY, Guengerich FP. Kinetic evidence for inefficient and error-prone bypass across bulky N 2-guanine DNA adducts by human DNA polymerase. The Journal of Biological Chemistry. 2006;281(18):12315–12324. doi: 10.1074/jbc.M600112200. [DOI] [PubMed] [Google Scholar]
- 79.Choi JY, Guengerich FP. Kinetic analysis of translesion synthesis opposite bulky N 2- and O 6-alkylguanine DNA adducts by human DNA polymerase REV1. The Journal of Biological Chemistry. 2008;283(35):23645–23655. doi: 10.1074/jbc.M801686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choi JY, Guengerich FP. Analysis of the effect of bulk at N 2-alkylguanine DNA adducts on catalytic efficiency and fidelity of the Processive DNA polymerases bacteriophage T7 exonuclease- and HIV-1 reverse transcriptase. The Journal of Biological Chemistry. 2004;279(18):19217–19229. doi: 10.1074/jbc.M313759200. [DOI] [PubMed] [Google Scholar]
- 81.Choi JY, Chowdhury G, Zang H, et al. Translesion synthesis across O 6-alkylguanine DNA adducts by recombinant human DNA polymerases. The Journal of Biological Chemistry. 2006;281(50):38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 82.Matsuda S, Henry AA, Romesberg FE. Optimization of unnatural base pair packing for polymerase recognition. Journal of the American Chemical Society. 2006;128(19):6369–6375. doi: 10.1021/ja057575m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsuda S, Leconte AM, Romesberg FE. Minor groove hydrogen bonds and the replication of unnatural base pairs. Journal of the American Chemical Society. 2007;129(17):5551–5557. doi: 10.1021/ja068282b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McMinn DL, Ogawa AK, Wu Y, Liu J, Schultz PG, Romesberg FE. Efforts toward expansion of the genetic alphabet: DNA polymerase recognition of a highly stable, self-pairing hydrophobic base. Journal of the American Chemical Society. 1999;121(49):11585–11586. [Google Scholar]
- 85.Hwang GT, Romesberg FE. Substituent effects on the pairing and polymerase recognition of simple unnatural base pairs. Nucleic Acids Research. 2006;34(7):2037–2045. doi: 10.1093/nar/gkl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Henry AA, Yu C, Romesberg FE. Determinants of unnatural nucleobase stability and polymerase recognition. Journal of the American Chemical Society. 2003;125(32):9638–9646. doi: 10.1021/ja035398o. [DOI] [PubMed] [Google Scholar]
- 87.Guo MJ, Hildbrand S, Leumann CJ, McLaughlin LW, Waring MJ. Inhibition of DNA polymerase reactions by pyrimidine nucleotide analogues lacking the 2-keto group. Nucleic Acids Research. 1998;26(8):1863–1869. doi: 10.1093/nar/26.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Piccirilli JA, Moroney SE, Benner SA. A C-nucleotide base pair: methylpseudouridine-directed incorporation of formycin triphosphate into RNA catalyzed by T7 RNA polymerase. Biochemistry. 1991;30(42):10350–10356. doi: 10.1021/bi00106a037. [DOI] [PubMed] [Google Scholar]
- 89.Trostler M, Delier A, Beckman J, et al. Discrimination between right and wrong purine dNTPs by DNA polymerase I from Bacillus stearothermophilus . Biochemistry. 2009;48(21):4633–4641. doi: 10.1021/bi900104n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang H, Bren U, Kozekov ID, Rizzo CJ, Stec DF, Guengerich FP. Steric and electrostatic effects at the C2 atom substituent influence replication and miscoding of the DNA deamination product deoxyxanthosine and analogs by DNA polymerases. Journal of Molecular Biology. 2009;392(2):251–269. doi: 10.1016/j.jmb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paul N, Nashine VC, Hoops G, et al. DNA polymerase template interactions probed by degenerate isosteric nucleobase analogs. Chemistry and Biology. 2003;10(9):815–825. doi: 10.1016/j.chembiol.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 92.Henry AA, Olsen AG, Matsuda S, Yu C, Geierstanger BH, Romesberg FE. Efforts to expand the genetic alphabet: identification of a replicable unnatural DNA self-pair. Journal of the American Chemical Society. 2004;126(22):6923–6931. doi: 10.1021/ja049961u. [DOI] [PubMed] [Google Scholar]
- 93.Eng K, Scouten-Ponticelli SK, Sutton M, Berdis A. Selective inhibition of DNA replicase assembly by a non-natural nucleotide: exploiting the structural diversity of ATP-binding sites. ACS Chemical Biology. 2010;5(2):183–194. doi: 10.1021/cb900218c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ghadessy FJ, Ramsay N, Boudsocq F, et al. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nature Biotechnology. 2004;22(6):755–759. doi: 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- 95.Zhang X, Lee I, Berdis AJ. The use of nonnatural nucleotides to probe the contributions of shape complementarity and pi-electron surface area during DNA polymerization. Biochemistry. 2005;44(39):13101–13110. doi: 10.1021/bi050585f. [DOI] [PubMed] [Google Scholar]
- 96.Zhang X, Lee I, Berdis AJ. A potential chemotherapeutic strategy for the selective inhibition of promutagenic DNA synthesis by nonnatural nucleotides. Biochemistry. 2005;44(39):13111–13121. doi: 10.1021/bi050584n. [DOI] [PubMed] [Google Scholar]
- 97.Morales JC, Kool ET. Importance of terminal base pair hydrogen-bonding in 3’-end proofreading by the Klenow fragment of DNA polymerase I. Biochemistry. 2000;39(10):2626–2632. doi: 10.1021/bi992173a. [DOI] [PubMed] [Google Scholar]
- 98.Hill F, Williams DM, Loakes D, Brown DM. Comparative mutagenicities of N 6-methoxy-2,6-diaminopurine and N 6-methoxyaminopurine 2′-deoxyribonucleosides and their 5′-triphosphates. Nucleic Acids Research. 1998;26(5):1144–1149. doi: 10.1093/nar/26.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hill F, Loakes D, Brown DM. Polymerase recognition of synthetic oligodeoxyribonucleotides incorporating degenerate pyrimidine and purine bases. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(8):4258–4263. doi: 10.1073/pnas.95.8.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. Journal of Molecular Biology. 1996;255(4):589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 101.Switzer CY, Moroney SE, Benner SA. Enzymatic recognition of the base pair between isocytidine and isoguanosine. Biochemistry. 1993;32(39):10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- 102.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343(6253):33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 103.Moser MJ, Marshall DJ, Grenier JK, et al. Exploiting the enzymatic recognition of an unnatural base pair to develop a universal genetic analysis system. Clinical Chemistry. 2003;49(3):407–414. doi: 10.1373/49.3.407. [DOI] [PubMed] [Google Scholar]
- 104.Rice KP, Chaput JC, Cox MM, Switzer C. RecA protein promotes strand exchange with DNA substrates containing isoguanine and 5-methyl isocytosine. Biochemistry. 2000;39(33):10177–10188. doi: 10.1021/bi0003339. [DOI] [PubMed] [Google Scholar]
- 105.Johnson SC, Sherrill CB, Marshall DJ, Moser MJ, Prudent JR. A third base pair for the polymerase chain reaction: inserting isoC and isoG. Nucleic Acids Research. 2004;32(6):1937–1941. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maciejewska AM, Lichota KD, Kuśmierek JT. Neighbouring bases in template influence base-pairing of isoguanine. The Biochemical Journal. 2003;369(3):611–618. doi: 10.1042/BJ20020922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moran S, Ren RX, Rumney S, Kool ET. Difluorotoluene, a nonpolar isostere for thymine, codes specifically and efficiently for adenine in DNA replication. Journal of the American Chemical Society. 1997;119(8):2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xia S, Eom SH, Konigsberg WH, Wang J. Structural basis for differential insertion kinetics of dNMPs opposite a difluorotoluene nucleotide residue. Biochemistry. 2012;51:1476–1485. doi: 10.1021/bi2016487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xia S, Konigsberg WH, Wang J. Hydrogen-bonding capability of a templating difluorotoluene nucleotide residue in an RB69 DNA polymerase ternary complex. Journal of the American Chemical Society. 2011;133(26):10003–10005. doi: 10.1021/ja2021735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee HR, Helquist SA, Kool ET, Johnson KA. Base pair hydrogen bonds are essential for proofreading selectivity by the human mitochondrial DNA polymerase. The Journal of Biological Chemistry. 2008;283(21):14411–14416. doi: 10.1074/jbc.M705006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim TW, Delaney JC, Essigmann JM, Kool ET. Probing the active site tightness of DNA polymerase in subangstrom increments. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(44):15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wolfle WT, Washington MT, Kool ET, et al. Evidence for a Watson-Crick hydrogen bonding requirement in DNA synthesis by human DNA polymerase kappa. Molecular and Cellular Biology. 2005;25(16):7137–7143. doi: 10.1128/MCB.25.16.7137-7143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Washington MT, Helquist SA, Kool ET, Prakash L, Prakash S. Requirement of Watson-Crick hydrogen bonding for DNA synthesis by yeast DNA polymerase eta. Molecular and Cellular Biology. 2003;23(14):5107–5112. doi: 10.1128/MCB.23.14.5107-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Irimia A, Eoff RL, Pallan PS, Guengerich FP, Egli M. Structure and activity of Y-class DNA polymerase DPO4 from Sulfolobus solfataricus with templates containing the hydrophobic thymine analog 2,4-difluorotoluene. The Journal of Biological Chemistry. 2007;282(50):36421–36433. doi: 10.1074/jbc.M707267200. [DOI] [PubMed] [Google Scholar]
- 115.Sismour AM, Benner SA. The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system. Nucleic Acids Research. 2005;33(17):5640–5646. doi: 10.1093/nar/gki873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sismour AM, Lutz S, Park JH, et al. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from human immunodeficiency virus-1. Nucleic Acids Research. 2004;32(2):728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Caton-Williams J, Huang Z. Synthesis and DNA-polymerase incorporation of colored 4-selenothymidine triphosphate for polymerase recognition and DNA visualization. Angewandte Chemie. 2008;47(9):1723–1725. doi: 10.1002/anie.200705213. [DOI] [PubMed] [Google Scholar]
- 118.Kim TW, Brieba LG, Ellenberger T, Kool ET. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase: probing T7 DNA polymerase with variably sized base pairs. The Journal of Biological Chemistry. 2006;281(4):2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 119.Sintim HO, Kool ET. Remarkable sensitivity to DNA base shape in the DNA polymerase active site. Angewandte Chemie. 2006;45(12):1974–1979. doi: 10.1002/anie.200504296. [DOI] [PubMed] [Google Scholar]
- 120.Strerath M, Cramer J, Restle T, Marx A. Implications of active site constraints on varied DNA polymerase selectivity. Journal of the American Chemical Society. 2002;124(38):11230–11231. doi: 10.1021/ja027060k. [DOI] [PubMed] [Google Scholar]
- 121.Strerath M, Summerer D, Marx A. Varied DNA polymerase-substrate interactions in the nucleotide binding pocket. ChemBioChem. 2002;3:578–580. doi: 10.1002/1439-7633(20020603)3:6<578::AID-CBIC578>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 122.Cramer J, Rangam G, Marx A, Restle T. Varied active-site constraints in the Klenow fragment of E. coli DNA polymerase I and the lesion-bypass Dbh DNA polymerase. ChemBioChem. 2008;9(8):1243–1250. doi: 10.1002/cbic.200700634. [DOI] [PubMed] [Google Scholar]
- 123.Mizukami S, Kim TW, Helquist SA, Kool ET. Varying DNA base-pair size in subangstrom increments: evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry. 2006;45(9):2772–2778. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]
- 124.Wilhelmsson LM. Fluorescent nucleic acid base analogues. Quarterly Reviews of Biophysics. 2010;43(2):159–183. doi: 10.1017/S0033583510000090. [DOI] [PubMed] [Google Scholar]
- 125.Reha-Krantz LJ, Hariharan C, Subuddhi U, et al. Structure of the 2-aminopurine-cytosine base pair formed in the polymerase active site of the RB69 Y567A-DNA polymerase. Biochemistry. 2011;50:10136–10149. doi: 10.1021/bi2014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bloom LB, Otto MR, Beechem JM, Goodman MF. Influence of 5’-nearest neighbors on the insertion kinetics of the fluorescent nucleotide analog 2-aminopurine by Klenow fragment. Biochemistry. 1993;32(41):11247–11258. doi: 10.1021/bi00092a039. [DOI] [PubMed] [Google Scholar]
- 127.Allan BW, Reich NO. Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry. 1996;35(47):14757–14762. doi: 10.1021/bi9615708. [DOI] [PubMed] [Google Scholar]
- 128.Lycksell PO, Graslund A, Claesens F, McLaughlin LW, Larsson U, Rigler R. Base pair opening dynamics of a 2-aminopurine substituted EcoRI restriction sequence and its unsubstituted counterpart in oligonucleotides. Nucleic Acids Research. 1987;15:9011–9025. doi: 10.1093/nar/15.21.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nordlund TM, Andersson S, Nilsson L, Rigler R, Graslund A, McLaughlin LW. Structure and dynamics of a fluorescent DNA oligomer containing the EcoRI recognition sequence: fluorescence, molecular dynamics, and NMR studies. Biochemistry. 1989;28(23):9095–9103. doi: 10.1021/bi00449a021. [DOI] [PubMed] [Google Scholar]
- 130.Stivers JT, Pankiewicz KW, Watanabe KA. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38(3):952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 131.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. The Journal of Biological Chemistry. 1969;244(5):1228–1237. [PubMed] [Google Scholar]
- 132.Hochstrasser RA, Carver TE, Sowers LC, Millar DP. Melting of a DNA helix terminus within the active site of a DNA polymerase. Biochemistry. 1994;33:11971–11979. doi: 10.1021/bi00205a036. [DOI] [PubMed] [Google Scholar]
- 133.Joyce CM, Potapova O, DeLucia AM, Huang X, Basu VP, Grindley ND. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47(23):6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 134.Lee HR, Wang M, Konigsberg W. The reopening rate of the fingers domain is a determinant of base selectivity for RB69 DNA polymerase. Biochemistry. 2009;48(10):2087–2098. doi: 10.1021/bi8016284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.DeLucia AM, Grindley ND, Joyce CM. Conformational changes during normal and error-prone incorporation of nucleotides by a Y-family DNA polymerase detected by 2-aminopurine fluorescence. Biochemistry. 2007;46(38):10790–10803. doi: 10.1021/bi7006756. [DOI] [PubMed] [Google Scholar]
- 136.Subuddhi U, Hogg M, Reha-Krantz LJ. Use of 2-aminopurine fluorescence to study the role of the beta hairpin in the proofreading pathway catalyzed by the phage T4 and RB69 DNA polymerases. Biochemistry. 2008;47(23):6130–6137. doi: 10.1021/bi800211f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marti AA, Jockusch S, Li Z, Ju J, Turro NJ. Molecular beacons with intrinsically fluorescent nucleotides. Nucleic Acids Research. 2006;34(6, article e50) doi: 10.1093/nar/gkl134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dash C, Rausch JW, Le Grice SFJ. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Research. 2004;32(4):1539–1547. doi: 10.1093/nar/gkh307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang X, Wadkins RM. DNA hairpins containing the cytidine analog pyrrolo-dC: structural, thermodynamic, and spectroscopic studies. Biophysical Journal. 2009;96(5):1884–1891. doi: 10.1016/j.bpj.2008.12.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang K, Stanley RJ. The extent of DNA deformation in DNA photolyase-substrate complexes: a solution state fluorescence study. Photochemistry and Photobiology. 2008;84(3):741–749. doi: 10.1111/j.1751-1097.2007.00251.x. [DOI] [PubMed] [Google Scholar]
- 141.Petruska J, Goodman MF. Influence of neighboring bases on DNA polymerase insertion and proofreading fidelity. The Journal of Biological Chemistry. 1985;260(12):7533–7539. [PubMed] [Google Scholar]
- 142.Lin KY, Jones RJ, Matteucci M. Tricyclic 2′-deoxycytidine analogs: syntheses and incorporation into oligodeoxynucleotides which have enhanced binding to complementary RNA. Journal of the American Chemical Society. 1995;117(13):3873–3874. [Google Scholar]
- 143.Wilhelmsson LM, Holmen A, Lincoln P, Nielsen PE, Norden B. A highly fluorescent DNA base analogue that forms Watson-Crick base pairs with guanine. Journal of the American Chemical Society. 2001;123:2434–2435. doi: 10.1021/ja0025797. [DOI] [PubMed] [Google Scholar]
- 144.Stengel G, Gill JP, Sandin P, et al. Conformational dynamics of DNA polymerase probed with a novel fluorescent DNA base analogue. Biochemistry. 2007;46(43):12289–12297. doi: 10.1021/bi700755m. [DOI] [PubMed] [Google Scholar]
- 145.Sandin P, Lincoln P, Brown T, Wilhelmsson LM. Synthesis and oligonucleotide incorporation of fluorescent cytosine analogue tC: a promising nucleic acid probe. Nature Protocols. 2007;2(3):615–623. doi: 10.1038/nprot.2007.80. [DOI] [PubMed] [Google Scholar]
- 146.Sandin P, Stengel G, Ljungdahl T, Borjesson K, Macao B, Wilhelmsson LM. Highly efficient incorporation of the fluorescent nucleotide analogs tC and tCo by Klenow fragment. Nucleic Acids Research. 2009;37(12):3924–3933. doi: 10.1093/nar/gkp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Walsh JM, Bouamaied I, Brown T, Wilhelmsson LM, Beuning PJ. Discrimination against the cytosine analog tC by Escherichia coli DNA polymerase IV DinB. Journal of Molecular Biology. 2011;409(2):89–100. doi: 10.1016/j.jmb.2011.03.069. [DOI] [PubMed] [Google Scholar]
- 148.Lund TJ, Cavanaugh NA, Joubert N, et al. B family DNA polymerases asymmetrically recognize pyrimidines and purines. Biochemistry. 2011;50:7243–7250. doi: 10.1021/bi2006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sandin P, Borjesson K, Li H, et al. Characterization and use of an unprecedentedly bright and structurally non-perturbing fluorescent DNA base analogue. Nucleic Acids Research. 2008;36:157–167. doi: 10.1093/nar/gkm1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stengel G, Purse BW, Wilhelmsson LM, Urban M, Kuchta RD. Ambivalent incorporation of the fluorescent cytosine analogues tC and tCo by human DNA polymerase alpha and Klenow fragment. Biochemistry. 2009;48(31):7547–7555. doi: 10.1021/bi9006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stengel G, Urban M, Purse BW, Kuchta RD. High density labeling of polymerase chain reaction products with the fluorescent base analogue tCo. Analytical Chemistry. 2009;81(21):9079–9085. doi: 10.1021/ac9017555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kool ET, Morales JC, Guckian KM. Mimicking the structure and function of DNA: insights into DNA stability and replication. Angewandte Chemie. 2000;39:990–1009. doi: 10.1002/(sici)1521-3773(20000317)39:6<990::aid-anie990>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 153.Henry AA, Romesberg FE. Beyond A, C, G and T: augmenting nature’s alphabet. Current Opinion in Chemical Biology. 2003;7(6):727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 154.Seo K, Yin J, Donthamsetti P, Chandani S, Lee C, Loechler E. Amino acid architecture that influences dNTP insertion efficiency in Y-family DNA polymerase V of E. coli . Journal of Molecular Biology. 2009;392(2):270–282. doi: 10.1016/j.jmb.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Loakes D, Holliger P. Polymerase engineering: towards the encoded synthesis of unnatural biopolymers. Chemical Communications. 2009;(31):4619–4631. doi: 10.1039/b903307f. [DOI] [PubMed] [Google Scholar]
- 156.Bailly C, Waring MJ. Use of DNA molecules substituted with unnatural nucleotides to probe specific drug-DNA interactions. Methods in Enzymology. 2001;340:485–502. doi: 10.1016/s0076-6879(01)40438-1. [DOI] [PubMed] [Google Scholar]


