Abstract
Acyl-CoA synthetase enzymes are essential for de novo lipid synthesis, fatty acid catabolism, and remodeling of membranes. Activation of fatty acids requires a two-step reaction catalyzed by these enzymes. In the first step, an acyl-AMP intermediate is formed from ATP. AMP is then exchanged with CoA to produce the activated acyl-CoA. The release of AMP in this reaction defines the superfamily of AMP-forming enzymes. The length of the carbon chain of the fatty acid species defines the substrate specificity for the different acyl-CoA synthetases (ACS). On this basis, five sub-families of ACS have been characterized. The purpose of this review is to report on the large family of mammalian long-chain acyl-CoA synthetases (ACSL), which activate fatty acids with chain lengths of 12 to 20 carbon atoms. Five genes and several isoforms generated by alternative splicing have been identified and limited information is available on their localization. The structure of these membrane proteins has not been solved for the mammalian ACSLs but homology to a bacterial form, whose structure has been determined, points at specific structural features that are important for these enzymes across species. The bacterial form acts as a dimer and has a conserved short motif, called the fatty acid Gate domain, that seems to determine substrate specificity. We will discuss the characterization and identification of the different spliced isoforms, draw attention to the inconsistencies and errors in their annotations, and their cellular localizations. These membrane proteins act on membrane-bound substrates probably as homo- and as heterodimer complexes but have often been expressed as single recombinant isoforms, apparently purified as monomers and tested in Triton X-100 micelles. We will argue that such studies have failed to provide an accurate assessment of the activity and of the distinct function of these enzymes in mammalian cells.
Introduction
In mammals, long-chain acyl-CoA synthetases (ACSL) are necessary for fatty acid degradation, phospho-lipid remodeling, and the production of long acyl-CoA esters that regulate various physiological processes. Acyl-CoA synthetases can be divided into five sub-families based on the chain length of their preferred acyl groups: acyl-CoA synthetase short-chain (ACSS), C2 to C4; medium-chain (ACSM), C4 to C12; long-chain (ACSL), C12 to C20; bubblegum (ACSBG), C14 to C24; very long-chain, annotated as solute carrier family 27A (SLC27A), C18 to C26. The five long-chain acyl-CoA synthetase (ACSL) genes are annotated as members 1, 3, 4, 5, and 6 (ACSL1 to ACSL6) and all are represented by many spliced transcript variants. The cDNAs of ACSL2 were found to correspond to the same gene as ACSL1; consequently ACSL2 was deleted from the databases. As many as five different isoforms per gene have been isolated in this family. This review will focus on their identification and will present the current information on the cellular and organelle distribution of these membranes proteins. In addition, we will also discuss the difficulty of defining their functions and to what extent their in vitro enzymatic characterization in a detergent environment has contributed to a better understanding of their function. These membrane-bound enzymes act on non-polar hydrophobic substrates (fatty acids) and their products (acyl-CoAs) are water-soluble and are powerful detergents.
The ACSL Genes and Their Isoforms
Alternative Spliced Elements: Overview
In mammals, five ACSL genes have been identified, and to date, each gene has as many as five different spliced variants. Most variations are defined by differences in the 5′-UTR regions, the first encoding exon, alternative coding-exons, and two exchangeable motifs located in the proximity to the ATP-binding site: the so-called fatty acid Gate-domain (1) (Table 1).
Table 1.
The Mammalian ACSLsa
| Characteristics | Spliced transcript variant | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Alternative elements |
Isoforms | Length (aa) | Human | Mouse | Rat | |||
| ACSL1 | F,Y-exon | Y-exon | 698 | v1 | DQ083029 | ||||
| F-exon | 698/699b | v2 | NM_001995.2 | NM_007981.3 | NM_012820.1 | ||||
| no F,Y-exon | 672 | v3 | DQ083028 | ||||||
| ACSL3 | 5′-UTRs | 720 | v1 | NM_004457.3 | v1 | NM_028817.2 | NM_057107.1 | ||
| 720 | v2 | NM_203372.1 | v2 | NM_001033606.1 | |||||
| ACSL4 | 5′-UTRs / AUGs | AUG2 | 670 | v1 | NM_004458.1 | v3 | NM_001033600.1 | ||
| 670 | v2 | NM_019477.3 | NM_053623.1 | ||||||
| AUG1 | 711 | v2 | NM_022977.1 | v1 | NM_207625.2 | ||||
| ACSL5 | 5′-UTRs / AUGs | AUG1 | 739 | v1 | NM_016234.3 | ||||
| AUG2 | 683 | v2 | NM_203379.1 | NM_027976.2 | NM_053607.1 | ||||
| 683 | v3 | NM_203380.1 | |||||||
| ACSL6 | 5′-UTRs / AUGs / exon8 / F,Y-exon |
AUG1 / Y-exon | 722 | v1 | NM_015256.2 | v2 | NM_001033597.1 | EF490998c | |
| AUG1 / F-exon | 722 | v2 | NM_001009185.1 | v1 | NM_144823.4 | ||||
| AUG2 / Y-exon | 697 | v4 | NM_001033599.1 | v1 | NM_130739.1 | ||||
| AUG2 / F-exon | 697 | v3 | NM_001033598.1 | v2 | AY625254.1 | ||||
| AUG2 / no F,Y-exon | 622 | v3 | BC047453.1 | ||||||
| AUG? / ΔF-exon | d | v4 | DQ083030 | ||||||
| AUG2 /Ex.8 / F-exon | 712 | v5 | DQ083031 | ||||||
GenBank accession numbers of the various spliced variants of the five ACSL members. Alternative elements are 5′-UTRs, translational codon initiators (AUG1, AUG2), alternative exon (exon 8 of ACSL6), exclusive exon pair encoding the Y-Gate and the F-Gate domain (Y-exon, F-exon). Isoforms without Gate-domain are indicated as no F,Y-exon and isoform with truncated F-Gate domain as ΔF-exon. Length of the product of each spliced variant is indicated (aa, amino acid).
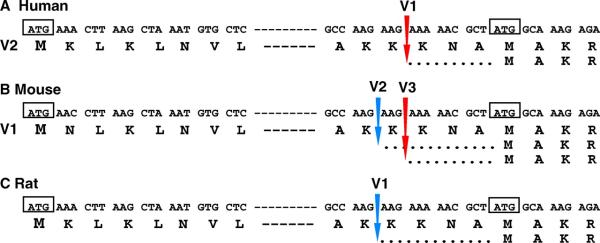
ACSL1 form of rodent is one residue longer than the form of other mammals (see text and Fig. 4).
Partial cDNA sequence (see text and Fig. 7).
The full-length sequence of variant 4 of ACSL6 is not known.
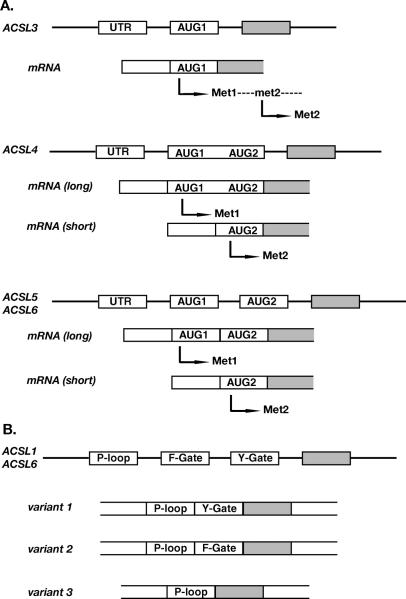
ACSL3, ACSL4, ACSL5, and ACSL6 have two in-frame AUG-translational initiators that produce isoforms of different lengths. A longer product is initiated at the first AUG codon, AUG1, and a shorter product is initiated at the downstream AUG2. Such isoforms are reported for ACSL3 in rat, ACSL4 in human and mouse, ACSL5 in human and ACSL6 in human, mouse and rat (Figs. 1A and 2A). As predicted by signal peptide analysis, these variations could result in different cellular localization (Fig. 3). ACSL1 is the only member of this family not known to produce isoforms with a different N-terminus.
Figure 1.
Representation of the spliced events affecting initiation of translation (A) and the fatty acid Gate domains (B). (A) Cartoon showing the exon-intron organization of the 5′-end of the ACSL genes, corresponding spliced variants (mRNA) and their products. The long isoforms start at the first AUG codon (AUG1, Met1) and the short isoform at the second AUG codon (AUG2, Met2). Note that two isoforms of ACSL3 are generated from the same mRNA. No variation has been reported for ACSL1. (B) Representation of the exclusive-exon pair encoding the two versions of the fatty acid Gate-domains (F- and Y-Gate) downstream of the exon encoding the ATP-binding site (P-loop). ACSL3, ACSL4, and ACSL5 genes lack the F-exon and encoded only the Y-Gate-domain version.
Figure 2.
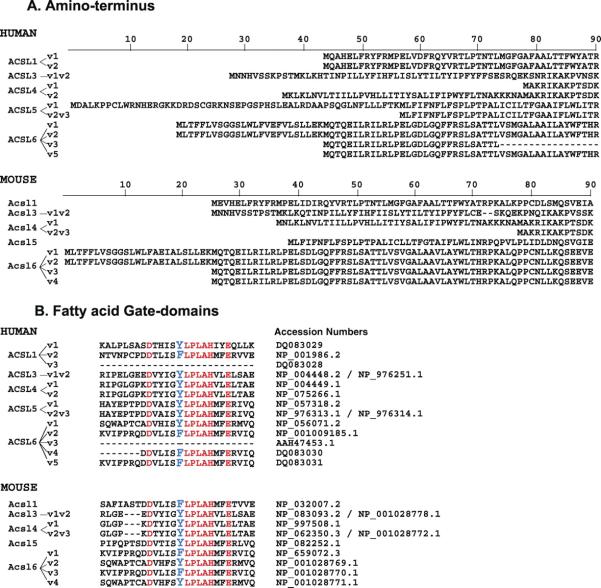
Residue alignments of the amino-terminus (A) and fatty acid Gate-domains (B) of all known human and mouse ACSL isoforms. The spliced variant annotation is shown on the left and the GenBank accession numbers are indicated in panel B. A color version of this figure is available in the online journal.
Figure 3.
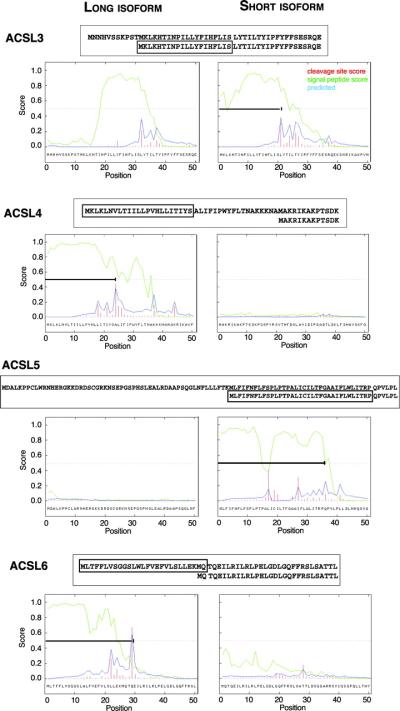
In silico analysis of the amino-terminus of the human ACSL isoforms. Prediction analysis of signal sequence was performed with SignalP on the long (left) and short (right) isoforms. The plots represent the probability of each residue to not be a part of the mature product (display as the signal peptide score, green curve), the probably of the presence of a site for signal peptidase (cleavage site score, red bar) and the predicted start of the mature product based on these two parameters (predicted, blue curve). ACSL1 encoded three isoforms that are identical in their N-terminus and are not predicted to carry a signal peptide. The residue alignments of the two isoforms are shown above the graphs with the putative signal peptide indicated in a box. A bar indicates the signal peptide position on the graphs.
ACSL4, ACSL5, and ACSL6 generate isoforms of different length by alternative splicing affecting the first encoding exon, whereas rat ACSL3 generates two products by alternative usage of two in-frame AUGs present in the same mRNA (2) (Fig. 1A). ACSL4 was proposed to generate a brain-specific spliced variant by splicing the first of the two putative AUG-encoding exons, but the two AUGs are in fact encoded by the same exon (Fig. 1A). Thus, an alternative 3′ acceptor splice site must be present in between the two AUG-initiators to account for the production of the two isoforms. ACSL5 and ACSL6 are to date the only two members that are represented by variants produced by alternative splicing of two AUG-encoding exons (Fig. 1A). Unlike human and mouse ACSL6, rat ACSL6 appeared to be unique in producing only the shorter isoform and having only one AUG-encoding exon in its gene. Recently though, we identified the missing exon, AUG1, and cloned a cDNA containing the two AUG-exons representing the full-length isoform (see ACSL6 section). Thus, the significant number of biochemical studies reported previously for rat ACSL6 were performed with an isoform lacking 22 residues and a predicted signal peptide.
ACSL1 and ACSL6 are the only members represented by isoforms with different Gate-domains. For these two members, the two motifs are encoded by two mutually-exclusive exons. Only one of the two exons is present in members 3, 4 and 5. All three of the predicted spliced variants, representing the three possible spliced events, have been identified in human: the two isoforms with one of the two exons and the isoform lacking both exons (Figs. 1B and 2B).
Distinctive Attributes of the Five ACSL Isoforms
The nomenclature of the mammalian long-chain acyl-CoA synthetase genes has recently been revised but not all variants were included in that report, such as the spliced variant 1 of mouse Acsl3 and Acsl4, and variant 3 of human ACSL6 (3). Moreover, some mistakes were introduced, i.e., the accession number for the protein of rat ACSL6 was mistaken for its cDNA. This erroneous accession number has been used in at least one study done subsequently to the revision of the nomenclature (4). Unfortunately, after successive annotation updates and the exclusion of spliced variants of mouse, the nomenclature is no longer uniform between human and mouse, and should be revised. A number of ESTs have been identified for rat ACSL but many forms are represented by only one spliced variant. An updated list is shown in Table 1.
ACSL1
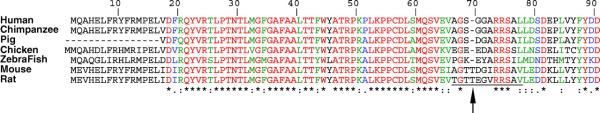
Two features are of interest. First, the rodent protein is one residue longer (699 amino acids) than the human protein (698 amino acids) (Fig. 4). This extra residue is a threonine, T70, downstream from another threonine residue (a serine in human), and encoded by an identical codon, ACC. This duplication event does not occur at a spliced site and does not appear to be a sequencing error since it is supported by several cDNAs from various sources. Other reported mammalian ACSL1 orthologs lack this codon and are similar to the human forms. Worthy of note, an antibody against rat ACSL1 was obtained with a peptide overlapping this region.
Figure 4.
Residue alignment of the amino-terminus of the ACSL1 forms of various species. The alignment was performed with CLUSTALW and shows the presence of an extra residue (Threonine), indicated with an arrow, in the rodent forms. The peptide used to raise a rat ACSL1 antibody is shown underlined (21). GenBank accession numbers were: human, NP_001986.2; Pan troglodytes, XP_517555.1; Sus scrofa, AAT79534; Gallus gallus, NP_001012596; Danio rerio, NP_001027007.1; mouse, NP_032007.2; rat, NP_036952.1. A color version of this figure is available in the online journal.
Second, alternative splicing of an exon pair in the proximity to the ATP-binding site results in the production of isoforms with a different so-called ‘fatty acid Gate-domain’ (Figs. 1B and 2B). Based on the structure of the ACSL bacterial homologue (5), this domain is probably involved in controlling access of the fatty acid substrate to the catalytic site of the enzyme (1). In the bacterial form, it also defines the specificity of the enzyme toward the fatty acid substrate. These domains are coded by the F-exon and the Y-exon (see discussion of ACSL6 below). These two exons are mutually exclusive and cannot co-exist in the same mRNA, but both can be removed. In human, three spliced isoforms have been isolated representing the two types of Gate-domains plus the isoform in which both domains were removed (Table 1).
ACSL3
The only known spliced variants differ in the un-translated region at the 5′ end of the mRNA (Table 1). Two products of slightly different lengths have been detected for rat ACSL3 cDNA (2). No such forms have been reported in other mammals. The shorter product of rat ACSL3 lacks the first 11 residues due to alternative translation initiation of two in-frame AUGs. The two putative translational codon initiators are encoded by the same exon and the same mRNA produced both isoforms (Fig. 1A). Thus, these two products are likely not unique to rat ACSL3 and can be expected to be present in other mammals.
ACSL4
One, two and three spliced isoforms have been identified in rat, human, and mouse, respectively (Table 1). Mouse variant 1 corresponds to human variant 2 and mouse variant 3 corresponds to human variant 1 (Fig. 2A and Fig. 5). Mouse variant 2 corresponds to the single variant identified in rat. Mouse variant 2 and variant 3 encode the same short isoform and differ by only 3 three bases at a spliced site in the first encoding exon (Fig. 5). As stated previously, this exon contains two in-frame AUGs. AUG1 is the initiator codon for the long isoform 1 and can be removed by the usage of an alternatively spliced acceptor site present in this exon between the two AUGs (Fig. 1A). The downstream in-frame AUG2 is used for the shorter isoform. The three nucleotides, AAG, missing in variant 3, indicate the presence of a second alternatively spliced acceptor site, AAGAAG/AAA, which is located three nucleotides downstream of the site used in variant 2, AAG/AAGAAA (Fig. 5). The alternative splicing event of mouse variant 3 is identical to the one occurring for human variant 1, whereas the splicing site of mouse variant 2 is identical to the one producing the rat variant. This suggests that the equivalent of the three mouse variants might also exist in human and rat, and have yet to be identified. The amino-terminal extension of the longer isoform has been proposed to represent a brain-specific isoform and is speculated to carry a leader transit peptide that directs the specific localization of this isoform (6). Mutations and deletions in ACSL4 have been established in nonspecific X-linked mental retardation syndrome (6–8) but another study reported this linkage to its neighboring gene, PAK3 (9). Similarly, a chromosomal translocation [t(5;12)(q31;p13)] resulting in the fusion of the ETV6 gene to ACSL6 has been identified in myelodysplastic syndrome and acute myelogenous leukemia (10) but a positional effect on the expression of closely located genes (perhaps IL-3) may be responsible for the development of those hematologic malignancies (11).
Figure 5.
Spliced variants of human (A), mouse (B), and rat (C) ACSL4. Spliced sites occurring between the two codon initiators of translation, boxed ATG, are indicated by an arrow. Note that only a partial sequence of the exon is shown, indicated with a dashed line. Genbank accession numbers are given in Table 1. A color version of this figure is available in the online journal.
ACSL5
Different spliced isoforms are generated by the alternative use of two AUG-encoding exons and the two shorter forms differ by 5′-UTR sequences (Table 1, Figs. 1A and 2A). The longer isoform of human ACSL5 (variant 1), which has not been identified in rodent, is the longest ACSL isoform known to date.
ACSL6
Five spliced variants representing five different isoforms have been identified in human (1), four in mouse (12), and two in rat (4) (Table 1). Human variant 5 contains an exon not found in any of the other isoforms (1). All other products only differ in two regions: the amino-terminus and a short motif in proximity to the ATP-binding site (Figs. 1B and 2B). The 25 residues of the extended N-terminus in the long isoforms are removed by alternative splicing of the AUG1-encoding exon. The translation initiation of the shorter forms occurs at the downstream AUG2 of the next exon (Figs. 1A and 2A). A mutually exclusive exon pair encoding two versions of the fatty acid Gate-domain, Y-exon and F-exon, can be alternatively spliced or skipped completely (no F- nor Y-exon) (Figs. 1B and 2B). Similar to ACSL1, the ACSL6 isoform lacking the F- and Y-exon (variant 3) has only been isolated in human. Variant 3 also lacks the AUG1-encoding exon and contains a unique 5′-UTR not found in other forms. Only in mouse, variants representing the 4 possible combinations of these exons, AUG1 with Y- or F-exon and AUG2 with Y- or F-exon, have been identified (12) (Table 1 and Fig. 2).
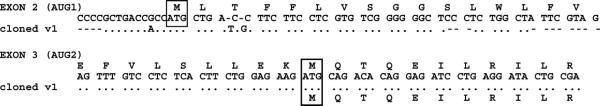
In human, the first cDNA representing ACSL6 was isolated from red blood cell precursors, NM_015256.1. It was originally reported to encode a short isoform that lacked the AUG1-encoding exon (13). However, sequence errors, short deletions and insertions of bases seemed to have occurred during the isolation of this clone, particularly at the 5′-end. As no product could be translated, it was concluded that this region was a 5′-UTR sequence and that the second AUG, AUG2, was the codon initiator. However, this predicted 5′-UTR does not match the genomic sequence. Rather, it matches the first coding exon (Fig. 6). The corrected ACSL6 variant 1, NM_015256.2, now includes AUG1. It appears that for similar reasons, the reported ortholog from Bos taurus is also probably erroneously translated from AUG2. The product (AAI11156.1) starts at AUG2 because the cDNA containing the AUG1-encoding exon also contains an in-frame stop codon TAA (BC111155). It is likely that a sequence error accounts for the predicted initiation at the second AUG. As already mentioned, the only two known spliced variants (Y- or F-exon) of rat represent the shorter version (Table 1, Figs. 2A and 7A).
Figure 6.
Sequence alignment of human ACSL6 gene and cloned variant 1. Alignment showing the missing bases and mismatches in the first encoding exon (exon 2) of the original cDNA of ACSL6, isolated from K562 cells (13). Missing bases are indicated with dashes (–) and matching bases with dots (.). This exon was interpreted as a UTR and the cloned cDNA was annotated as encoding a short isoform initiated at the ATG present in exon 3. Note that only a partial sequence of exons is shown. The predicted translated product of the full-length isoform is shown above the exon sequence with the two in-frame codon initiators boxed (ATG) and the product of the cloned variant is indicated below the sequence of the isolated cDNA.
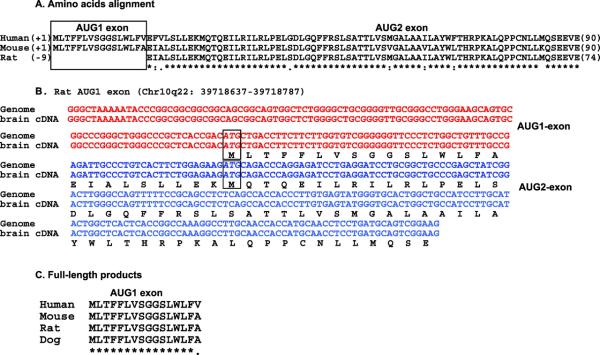
Figure 7.
Identification of the full-length rat ACSL6 isoform. (A) Residue alignment of the long products of human and rodent ACSL6 isoforms. Rat ACSL6 lacks the first encoding exon (AUG1 exon). (B) Identification of the first encoding exon of rat ACSL6. The alignment shows the nucleotide sequence of rat chromosome 10, position q22 39718637 to 39718787, compared with the sequence of an isolated cDNA obtained by reverse transcription-PCR of total RNA isolated from rat brain. The sequence was deposited at GenBank under GenBank accession number EF490998. (C) Residue alignment showing conservation of the first encoding exon in mammalian ACSL6 orthologues. A color version of this figure is available in the online journal.
In rat, the AUG1-encoding exon is not annotated in the gene. However, both a BLAT search of the rat genome with the mouse peptide sequence and a BLAST2seq search on the contig containing the rat ACSL6 gene with the human AUG1-exon identified the missing AUG1-exon and the missing residues (Fig. 7B). This sequence is located ≈16-kbp upstream of the AUG2 exon and contains an in-frame AUG. This organization is similar to the human and mouse genes, in which the AUG1-exon is ≈18 and ≈16-kbp upstream of the AUG2-exon, respectively. To confirm this, we isolated and cloned a cDNA from rat brain total RNA containing the two correctly spliced AUG1-AUG2 exons, which represents the longer isoform (Fig. 7B and C). Thus, the two rat ACSL6 isoforms cloned, expressed, partially purified, and studied in vitro represent isoforms truncated of their amino-terminus (4). Similar to rat ACSL3 described above, it appears that rat ACSL6 is not unique and does not differ from other orthologues. It is possible that the limited number of ESTs, an unfinished genome sequence, and a limited analysis contributed to the premature conclusions that rat ACSL6 was unique (4).
Challenges in the Current Annotation System
The current annotation system for the spliced variants of the five members of the long chain ACSL family poses challenges for researchers. The current system is nonuniform across species and the annotation information available in the Reference Sequence (RefSeq) collection, maintained by the National Center for Biological Information, can be confusing. When a new spliced variant of a gene is accepted for inclusion in the database, RefSeq must decide whether to create a new entry or update an existing entry. Often the decision is made to upgrade the version of an existing entry rather than create a new unique accession number. Of particular concern is the incessant modification of the 5′ UTR and putative alternative AUG-encoding exons predicted by homology to the 5′ end of cDNA sequences found in databases. These modifications disregard the fact that those cDNAs may contain differences elsewhere, and would thus represent different spliced variants. Another additional source of confusion in using the public domain databases is that the defined amino acid sequence of a protein (NP_), annotated as the product of a transcript (NM_), is not necessarily the translated product of that cDNA (see as an example mouse ACSL6). As indicated below, this shuffling of sequences of the acyl-CoA synthetase family has resulted in a non-uniform annotation of the various spliced variants of each gene across different animal species.
Particularly the annotation of mouse Acsl6 variants is problematic. The RefSeq annotation currently identifies the product of the original spliced variant 1, isoform 1 (AUG1, Y-exon), as the product of a different variant encoding isoform 2 (AUG1, F-exon) (Fig. 8). In other words, during an update of the sequence of variant 1, NM_144823, from version 0.1 to version 0.4, RefSeq mistakenly substituted isoform 1 for isoform 2 as being the product of variant 1 (Fig. 8C) (see legend of Fig. 8 for further details). Because different ESTs corresponding to different isoforms were successively and inappropriately used as reference cDNA to update the entry NM_144823 from version 0.1 to 0.4, the same accession number, NM_144823, actually represents 3 different isoforms. Each isoform is currently annotated with its own accession number but they are also the product of NM_144823 version 0.1, 0.2 and 0.4. Perhaps not surprisingly, RefSeq is still annotating only one of the two known spliced variants of rat ACSL6. In summary, isoform 1 of mouse corresponds to isoform 2 of human, and mouse isoform 2 correspond to isoform 1 of human, and only one isoform is annotated for rat. It is conceivable that other errors exist in the annotations for other variants in the database. If other errors do indeed exist, results of the update strategy of RefSeq may need to be critically evaluated for accuracy, and a systematic check to evaluate if a translated product of manipulated nucleotide sequences corresponds to the intended correction may be warranted.
Figure 8.
Erroneous translation (A), update (B), and annotation (C) of mouse Acsl6 isoform 1. (A) Sequence of cDNA and product of version 0.1. The first 25 codons present in the annotated transcript (NM_144823.1) were mistakenly not translated and the annotated product (NP_659072.1) was initiated at the second ATG. (B) Residue alignment of the fatty acid Gate-domain of version 0.1 and the updated version, 0.3, of the Acsl6 product, NP_659072. Version 0.1 (NP_659072.1) lacked the first 25 residues (697 aa in length, residues shown +295 to +335) and contained the Y-Gate domain, whereas version 0.3 (NP_659072.3) was initiated at the first ATG (722 aa in length, residues shown +320 to +360) but contained the F-Gate domain version. (C) Residue alignment of the fatty acid Gate-domains of the two isoforms before and after the update. The original isoform 1 (NP_659072.1) and isoform 2 (AAO38689.1) represent the two versions of the Gate-domain, Y-Gate and F-Gate, respectively, which was mistakenly switched to the F-Gate (NP_659072.3) and Y-Gate (NP_001028769.1) during the update. RefSeq mistakes occurred during the correction of another mistake of the RefSeq annotated product of NM_144823.1. The first annotated product NP_659072.1 (AUG2, Y-exon) was erroneously translated from the second AUG and lacked the first 25 residues predicted by the nucleotide sequence of NM_144823 version 0.1 (AUG1, Y-exon) (panel A). Instead of correcting the wrongly translated product of NM_144823 version 0.1, NP_659072.1, and updating the product to version 0.2 (AUG1, Y-exon), RefSeq chose to update the cDNA sequence to version 0.2. It did so by truncating the AUG1-exon present in NM_144823.1, using an EST lacking the first AUG, and whose translated product would match the wrong product NP_659072.1. Then, version 0.2 of the cDNA and 0.1 of the product were both updated to version 0.3 and 0.2, respectively, using a third different EST. This EST contains AUG1, as in version 0.1, but also contains the F-exon instead of the Y-exon. Hence, version 0.2 of the product represented a different isoform (AUG1, F-exon). Moreover, the third cDNA also contained 5 base mismatches (two residues), hence a fourth EST was used to perform another update of both the cDNA and the product sequence, to version 0.4 and 0.3 respectively (panel B). Genbank accession numbers are given in Table 1. A color version of this figure is available in the online journal.
Topology and Localization
ACSL proteins are predicted to be membrane proteins but only limited information is available on their topology and actual localization in the cell. Although they are usually identified to be associated with membranes, some have been detected in the cytosol of cells. The specific functions of most of the isoforms, and how their structures determine their locations and activities are largely unknown. Studying their roles associated with specific cell organelle function may shed light on their localization and provide a better understanding how different isoforms of these proteins contribute specifically to the diverse metabolic pathways. Free fatty acid, the substrate of these enzymes, can cross (plasma) membranes, but its product, acyl-CoA, is unable to do so. Hence, esterification of free fatty acids by ACSL results in trapping the non-membrane permeant acyl-CoA inside the cells. ACSL1 and ACSL6 are associated with the plasma membrane; they are the only two members that can produce spliced isoforms with two different fatty acid-Gate domains. Although, the specific functions of these two fatty acid-Gate domain versions are not known, they might be needed in the plasma membrane of cells where fatty acid uptake takes place. ACSL1 has been found to interact with the fatty acid transporter FATP1 in adipocytes, and they constitute the first described enzyme pair involved in a vectorial acylation system in mammals (14).
Topology Prediction
ACSL members 3, 4, 5, and 6 are expressed as isoforms of different lengths. The long and short isoforms are predicted to differ by leader sequences that may act as signal peptides cleaved in the mature form (Fig. 3). In other words, the amino-terminus of the different proteins might target the various isoforms to different membranes. The long isoforms of ACSL4 and ACSL6 are predicted to carry a leader peptide whereas it is predicted for the shorter isoform of ACSL3 and ACSL5.
For ACSL3, the 11 residues missing in the short isoform, upstream of the second AUG, are strongly predicted not to be part of a signal peptide whereas the following residues do appear to be.
The long isoform of ACSL4 has been proposed to be a brain-specific form and the leader sequence has been speculated to target this form specifically to this tissue (6).
ACSL5 is present on the cytosolic side of the mitochondrial membrane, thus a secretory peptide is probably needed to cross the mitochondrial membrane. Removal of the first encoding exon in the shorter form of ACSL5 results in exposure of an otherwise buried signal peptide.
For ACSL6, the shorter isoform lacks the predicted leader signal but is still predicted to carry one transmembrane spanning segment at its amino-terminus. However, a recombinant form truncated of this motif was still found to be associated with the membrane fraction when expressed in Escherichia coli (unpublished data). Thus, it is still unclear if ACSL6 is inserted into the membrane or if, as in the case of ACSL4 (15), it is only associated to membranes. The E. coli ACSL enzyme, FadD (16), is loosely bound to the membrane (17–19), and a complex topology has been described for a mouse very-long chain ACS member (20). This enzyme is inserted into the membrane by two transmembrane spanning segments present at the N-terminus but is also associated and bound to the cytosolic side of the lipid layer through hydrophobic regions of the protein, not requiring the N-terminus (20).
Evidence for Subcellular Localization
Detection by Antibody and by Mass Spectrometry
A summary of the reported localization of the different ACSL proteins and the techniques used is presented in Table 2. The five ACSL members can be divided into two groups based on sequence homology and gene organization: ACSL1/ACSL5/ACSL6 and ACSL3/ACSL4 (1). Interestingly, only members 1, 5, and 6 have been reported to be present in the plasma membrane while such is not the case for members 3 and 4.
Table 2.
Localization of the ACSL Forms
| Detection | ACSL1 | ACSL3 | ACSL4 | ACSL5 | ACSL6 | ||
|---|---|---|---|---|---|---|---|
| Antibody | |||||||
| Production | Protein | Peptide | Peptide | None | Peptide | Peptide | Peptide |
| Form detected |
ACSL1-4-5 | ACSL1 | unknown (68kDa)a,b |
ACSL4 (80–74 kDa)b |
ACSL5 (76–74.5–73 kDa)b |
ACSL6 | |
| Tissuesc | adipocytes (rat) | adipocytes (mm) | Liver (rat) | Liver (rat) | Liver (rat) | RBC (hs) | |
| Fraction | |||||||
| Plasma | + | + | n.t.d | n.t. | + | + | |
| Internal | Low density Mce | High density Mcf Mitochondria nucleus |
Microsome ER (MAM) |
Peroxisomes ER (MAM) |
ER (MAM) Mitochondria |
||
| Cytosol | n.t. | − | + | − | − | + (K562cells) | |
| References | (49) | (21) | (24) | (15, 24) | (24) | (13) | |
| Mass spectrometry | |||||||
| Tissues | |||||||
| Adipocytes (mm) (22) | Lipid droplet | Lipid dropletg | Lipid dropletg | ||||
| Adipocytes (rat) (23) | GLUT4-vesicles | ||||||
| Braini (rat) (25) | Mitochondria | ||||||
| Liver (rat) (30) | Lipid raft | Lipid raft | |||||
| RBC (hs) (29) | Plasma membraneh | ||||||
| Hela cells (50) | Lipid raft | ||||||
| Braini | n.d.j | n.d. | n.d. | n.d. | Microsome- mitochondria |
||
| Heartk | Microsome-mitochondria**-nucleus | n.d. | n.d. | n.d. | n.d. | ||
| Kidney | Microsome-mitochondria**-nucleus | n.d. | n.d. | Mitochondria | n.d. | ||
| Liver | Plasma-microsome (ER)**-Mitochondria***-cytosol | n.d. | n.d. | Microsome (ER)**- Mitochondria |
Mitochondria** | ||
| Lung | Microsome-mitochondria | n.d. | n.d. | Microsome- mitochondria-nucleus |
n.d. | ||
| Model | Plasma membrane | Plasma membrane | Plasma membrane | ||||
| ER and mitochondria cytosol | ER and eroxisomes |
ER and mitochondria | Mitochondria cytosol | ||||
Calculated molecular mass of ACSL1 is 78kDa and was detected at the expected position by other antibody (21).
Molecular masses of detected bands were assessed on SDS-PAGE 7.5% gel with Molecular Mass standard 97.4 kDa and 66.3 kDa (24).
Detection in cytosol, plasma membrane, and membrane of organelles in various tissues of human (hs), mouse (mm), and rat. When tested, presence or absence in the plasma membrane and cytosol is indicated by a+or −, respectively. Mc, microsome; ER, endoplasmic reticulum; MAM, mitochondria-associated membrane, presumably from the ER; RBC, red blood cell; plasma, plasma membrane.
Not tested.
Low density microsomes contain Golgi apparatus, endosomes, and secretory vesicles.
High density microsomes contain ER, trans-Golgi network, and endosomes.
Detected only after stimulation of lipolysis.
ACSL1 and ACSL3 were detected in the soluble and membrane fraction, respectively. Identification was indexed as suspicious for ACSL1 and uncertain for ACSL3. Only ACSL6 meet all criteria.
Compilation of three different organelles proteomic survey for ACSL members. Mootha et al, 2003 (28), surveyed mouse mitochondria samples free of markers for ER (calreticulin-calnexin) and mitochondria-associated membrane (phosphatidylethanolamine N-methyltransferase) in 4 tissues: liver, brain, heart, and kidney. Kislinger et al, 2006 (27), surveyed the Cytosol-Microsome (ER-Golgi-endosomes)-Mitochondria-Nuclei fractions of 5 mouse tissues: brain, heart, kidney, liver, lung, and placenta (not shown). Foster et al, 2006 (26), surveyed mouse liver fractionated on sucrose-density gradients into seven distinct organelles preparation identified with unique markers for Golgi, PM, early endosome, recycling endosome, ER, ER/Golgi-derived vesicles, mitochondria. Detection of an isoform in the same organelle and tissue in 2 and 3 studies is indicated with 2 and 3 asterisks (*), respectively. Note that Mootha et al surveyed only mitochondria and that Forster et al surveyed only liver tissue.
Not detected.
ACSL1 was also found in human heart mitochondria samples free of the marker for peroxisome (catalase), ER (calnexin), and mitochondria-associated membrane (phosphatidylethanolamine N-methyltransferase) (51).
ACSL1 was proposed to be an integral membrane protein based on the failure to remove it from membrane by extensive washing (21). However, ACSL1, as evidenced by immunodetection and mass spectrometry, was found in the cytosol of liver tissues (Table 2). ACSL1 seems to be present in both the plasma membrane and mitochondria of specialized cells such as adipocytes (21). In these cells the protein is also found in intracellular structures formed by accumulation of triacylglycerol (lipid droplets), and in GLUT4-loaded transit vesicles (22, 23). The localization of ACSL1 in the lipid bilayer of mitochondria seems a controversial issue, as ACSL1 was detected in mitochondria membranes with one antibody (21) but not with another (24). The antibody raised against a peptide unique to ACSL1 identified a protein of the correct predicted mass of 78 kDa in the mitochondrial membrane (21). The binding of this antibody was competed by the peptide used to generate it, which provides evidence of the specificity of the detection. ACSL1 was detected in mitochondria fractions of different tissues, shown to be free of other cellular contaminants by a battery of selective markers, by several proteomic studies (25–28). The antibody that failed to detect ACSL1 in mitochondria (24), also failed to detect a purified recombinant ACSL1 form (24). This antibody was raised against a peptide spanning residues 1 to 19 and the recombinant protein was made with a FLAG peptide fused to residue 3 of ACSL1. Although removal of 2 of the 19 residues might account for the failure of detection, this antibody has not been established to recognize the full-length ACSL1 protein. It is unclear which protein(s) cross-reacted with this antibody, as the apparent molecular mass was significantly different from its predicted mass, as identified by the specific ACSL1 antibody (21). Thus, failure of detection of ACSL1 in mitochondria by this antibody seems inconclusive (see below).
ACSL3 and ACSL4 were detected in lipase-activated lipid droplets but not in un-induced droplets (22). ACSL4 might be the only form of ACSL in peroxisomes and the ability to extract rat ACSL4 from the microsomal membranes by extensive washing suggests that this protein is not an integral membrane protein (15).
ACSL5 and ACSL6 were detected in the plasma membrane (13, 24, 29) and were reported to be enriched in cholesterol rich domains also identified as “lipid rafts”. (30).
Detection by Enzyme Activity
It was postulated that the reported detection of ACSL1 in mitochondria (21) was the result of improper purification of the mitochondria fraction (24), and that ACSL5 is the only acyl-CoA synthetase for long-chain fatty acids present in the mitochondria (24). However, inhibition of the long-chain acyl-CoA synthetase activity in the mitochondrial membrane by triacsin C indicated that ACSL5 cannot be the only member present in this organelle. Triacsin C inhibits the activity of ACSL1, ACSL3, and ACSL4 in a concentration-dependent manner (31). ACSL5 and 6 are resistant to this compound (4, 24). The ACSL activity of mitochondria is inhibited by triacsin C, and therefore, a significant amount of the mitochondrial ACSL activity must be a form that is sensitive to this compound: ACSL1, ACSL3 and ACSL4. The presence of ACSL4 was ruled out by the use of a different drug (thiazolidinedione), specific for this enzyme (24). As only five forms of ACSL are present in mammals and much evidence supports the presence of ACSL1 in the mitochondrial membrane, it is more likely that ACSL1 activity was the one inhibited in mitochondria rather than the activity of “a hitherto unidentified” sixth form as suggested (24). Therefore, a revision of the model hypothesizing the specific physiological function of the five ACSL members in the different membranes of a living cell that assumes the absence of ACSL1 from the mitochondrial membrane may be warranted (32).
Structure and Activity
No conclusive structural information is available for any of the mammalian acyl-CoA synthetases. The first and currently only known structure of a long-chain acyl-CoA synthetase is that of a homologue of a hyperthermophilic bacterium (5). Using this as a template to predict the structure of the mammalian homologue provides a wealth of information.
The bacterial form of the enzyme acts as a dimeric complex of two identical subunits, each forming a catalytic site (5). The E. coli FadD and human ACSL6 homologues have also been shown to form homodimers (1, 17). The bacterial protein contains a domain implicated in substrate specificity: the fatty acid Gate domain. The sequence of this domain is conserved in mammalian ACSL forms, although with some difference, and as mentioned before, member 1 and 6 generate different spliced isoforms carrying two slightly different domains and one truncated isoform (1). We have proposed that the difference between the two Gate versions lies in a single aromatic residue, which defines the “gating residue” of the Gate domain (1). In the bacterial homologue, a Tryptophan is positioned between the fatty acid channel and the ATP-binding site, in the catalytic region of the enzyme. This aromatic residue blocks entry of the fatty acid until ATP binds to the nearby P-loop. Upon binding, the Tryptophan rotates and gives access to the hydrophilic carboxyl moiety of the fatty acid to the active site. The fatty acid is first activated by esterification to AMP and release of pyrophosphate, followed by exchange with Co-enzyme A, and liberation of the acyl-CoA ester on the cytosolic side of the enzyme. In mammals, this tryptophan is absent and the gating residue appears to be a tyrosine for all isoforms of ACSL3, ACSL4, and ACSL5 and for spliced isoform 1 of ACSL1 and ACSL6. The gating residue is a phenylalanine for variant 2 of ACSL1 and ACSL6 (Table 1, Figs. 1B and 2B). The effects of a switch of these two Gate domains on the activity, selectivity and function of the enzyme are currently unknown.
Conversion of Non-Polar to Powerful Detergent Molecules at the Water-Lipid Interface
Regardless of their location, all ACSL isoforms act on a lipophilic substrate (fatty acid) at the water membrane interface and generate a product (acyl-CoA) that is used by other membrane bound enzymes such as the acyl-CoA:lysophos-pholipid-acyltransferases (LPLAT). Therefore, the activity of these enzymes will be affected by both lipid-lipid, lipid-protein and protein-protein interactions at the membrane water interface (19, 33). A number of studies of the activity of the different forms of this family of enzymes have been reported, but we argue that the majority of such data should be evaluated very carefully. At best, these reports indicate the activity of such proteins in an artificial environment of lipids and detergents with a composition and organization very different from their native environment. Enzyme kinetics collected under these conditions will not necessarily report on the kinetics of these enzymes in vivo. This challenge is not unique for this set of proteins and will apply to many studies of membrane proteins. Studies of ACSL activity are particularly complicated because their substrate is a component of the lipid layer and their product is a lipid detergent by nature.
Challenges of Sample Preparation
Measurements and detection of long-chain acyl-CoA synthetase activity in vivo have been obtained in many cells and tissues but definition of enzyme kinetics of a single form requires its isolation, purification, and reconstitution. In addition to technical concerns, the non-uniform and changing annotation of the many isoforms makes it often unclear which protein was analyzed. For example, the first published sequence of human ACSL6 represented the spliced transcript of variant 1 but the cDNA that was cloned and expressed in E. coli represented spliced variant 2 (13). The activity assays were in fact performed with isoform 2 and not with isoform 1. A general concern with these proteins when over-expressed in E. coli is their tendency for aggregation, formation of inclusion bodies, a high degree of proteolysis, and destabilization of the dimeric complexes during purification. Despite the indications that the mammalian enzymes may act as a dimer, none of the activity reports have taken this into account. The detergent Triton X-100 has been a compound of choice to dissolve these proteins. This detergent has been shown to have significant effects on the activity of many membrane proteins, and has been shown to be a poor surrogate for the natural environment of membrane proteins. The ACSL enzymes are no exception and Triton X-100 has been shown to differentially affect the activity of all forms (4, 34). Even at low concentrations (3 mM) ACSL4 lost its activity, whereas the activity of ACSL5 was increased 600% (34). Unexpectedly, the most sensitive form, ACSL4, was solubilized from the membrane in the presence of 1% Triton X-100 (≈16 mM; 5 times higher than the inhibitory concentration) during its purification for enzymatic characterization (34). These data indicate that the lipid environment generated by the detergents affects the enzyme kinetics. Despite these concerns, which seem to preclude a direct comparison of the activity of various forms in the presence of various amounts of Triton X-100, kinetics have been reported under these variable conditions (4, 34).
Challenges in Measuring the Activity of the ACSL Enzymes
Three techniques have been used to determine the activity of these enzymes. Detection of the product acyl-CoA by HPLC, isotopic measurement using 14C/3H-labelled fatty acid (35) or 3H-labelled Co-enzyme A (36) in the formation of acyl-CoA, and spectrophotometric measurement of the utilization of Co-enzyme A (37–40). The later measurements are indirect and can be achieved by various coupled-assays: oxidation of NADH, of the formed acyl-CoA, or by the effect on added sulfhydryl-binding reagents. These activity measurements cannot be performed in complex unpurified preparations due to interference by other compounds, including CoA- and NADH-utilizing enzymes, and acyl-CoA hydrolases. The different approaches have their specific pitfalls and comparison of the kinetics of different forms of ACSL obtained under different conditions and with different techniques is unfortunately questionable. In all cases, the measured activities are only relative and are specific for a given experimental condition. Unless rigorous controls are in place and activity of the different isoforms are measured under identical conditions, their usefulness in the description of their enzymatic characteristics is limited.
The first mammalian long-chain acyl-CoA synthetase was isolated from rat liver in 1990, currently annotated as ACSL1 (41). The first form isolated from brain tissue was ACSL6 but was mistakenly identified as a PI 4-kinase (phosphatidylinositol 4-kinase) (42). ACSL6 was also the first to be found to ‘form insoluble aggregates’ and to require Triton X-100 for its extraction. Yamamoto and collaborators have identified and characterized all five ACSL members (43–46). They also purified them to homogeneity and established the substrate preference of each forms (Table 3).
Table 3.
In Vitro Substrate Preferences of Purified ACSL Membersa
| Fatty acids preference |
|
|---|---|
| ACSL1 | 16:0 • 16:1 • 18:1 • 18:2 > 14:0 • 18:0 • 18:3 • 20:4 > 20:5 >> 20:0 • 22:0 • 22:6 • 24:0 |
| ACSL3 | 14:0 • 20:4 • 20:5 > 16:1 • 18:1 • 18:2 • 18:3 > 16:0 • 18:0 >> 20:0 • 22:0 • 22:6 |
| ACSL4 | 20:4 • 20:5 >> 16:0 • 18:0 • 18:2 > 16:1 • 18:3 • 22:6 > 14:0 • 18:1 • 20:0 >> 22:2 |
| ACSL5 | 16:0 • 16:1 • 18:1 • 18:2 • 18:3 > 20:4 > 14:0 • 18:0 >> 20:0 • 22:0 • 24:0 |
| ACSL6 | 16:0 • 18:1 • 20:4 • 20:5 > 14:0 • 22:6 > 16:1 • 18:0 • 18:2 • 18:3 > 20:0 • 22:0 • 24:0 |
Since then, how much has been learned? The activity of these enzymes is strongly dependent on their lipid environment. The presence of lipids enhance their activity and the lipid composition of the membranes in which the enzymes are measured has been reported to affect their activity (19, 33). Rescue of acyl-CoA synthetase mutants by the mammalian forms were performed in both E. coli and S. cerevisiae strains. In E. coli, only ACSL5 rescued a fadD mutant. On the other hand, ACSL5 was the only form that failed to do so in the yeast mutant (33, 47). It is unclear what one can learn about the specific function of the five ACSL forms from those studies, besides the fact that those enzymes behave differently in the membrane(s) of E. coli and yeast.
Other studies have attempted to define specific functions for spliced isoforms. These studies assessed activity of partially purified enzymes expressed in E. coli and solubilized in Triton X-100, and present a number of inconsistencies. Different enzymes appeared to lose their activity during the purification process and the degree of activity loss was different for each isoform. While some show a moderate increase in specific activity after purification, 30-fold compared to a crude protein extract, others were reported to be increased only 3-fold (ACSL6 variant 1, see below) (4, 34). Human and rat ACSL6 forms show high degrees of proteolysis when expressed in E. coli, which can be detected using an antibody reacting against the tag used for their affinity purification (1). The effect of those truncated products on the activity of the full-length products is unknown. Moreover, the level of contamination of the various purified enzymes by those products as well as inactivated enzyme fractions precludes correct estimation of the specific activity, as these calculations are normalized to the amount of protein(s) present in the sample tested.
Such concerns can be illustrated by the reported kinetic data for these enzymes such as the apparent Km for ATP as measured for isoform 1 and 2 of rat ACSL6. These two spliced isoforms differ in a motif located near the ATP-binding site, the fatty acid Gate-domain. The isoform with the lowest affinity for ATP was also the enzyme preparation only enriched 3-fold compared to the more highly enriched isoforms (4). The level of contamination by E. coli ATPases of the partially purified enzymes was not taken into account, and the kinetic parameters of the enzymes for coenzyme A and various fatty acids substrates were assessed under limiting conditions for ATP (4). Of particular concern is the fact that, in contrast to what would be expected, the kinetic constants obtained in several studies were not constant. The values differ between experiments and sample preparations. As an example, purified ACSL4 strongly prefers long and poly-unsaturated fatty acids over palmitate with an apparent Km of 15 μM for arachidonic acid and 100 μM for palmitate (45). However, others have reported that partially purified ACSL4 showed a similar affinity to arachidonic acid (Km = 10 μM) and to palmitate (Km = 5.4 μM) (4, 34). This discrepancy strongly suggests that activity measurements do not necessarily reflect real enzymatic kinetics and apparently, not even the substrate preference of the different forms can be conclusively determined.
Whereas biochemical dissection on the role of specific residues and/or motifs can be assessed by testing their activity in Triton X-100 micelles little information is gained with respect to the physiological functions of the ACSL proteins. Importantly, it is not known how many spliced isoforms of each of the ACSL members are present in tissues and membranes in which one form has been detected, and how they may affect the activity of each other.
In red blood cells, there are 4 different spliced isoforms of member 6, v1-v2-v4, and v5 (1). ACSL6 appears to form dimeric complexes, as established for the bacterial homologues (5, 17). The assumption that both homo and hetero dimers can be formed leads to a population of 10 potentially active species (v1/v1; v1/v2; v1/v4; v1/v5; v2/v2; v2/v4; v2/ v5; v4/v4; v4/v5; v5/v5). To our knowledge, only one study has addressed this issue. Li and collaborators have isolated three spliced isoforms of Acsbg1, a member of the fifth family of mammalian acyl-CoA synthetase, with long/very-long chain ACS activity (48). One of the two shorter isoforms is truncated of the Gate-domain. This isoform has no detectable acyl-CoA synthetase activity, which confirms the role of this domain in the catalytic activity of ACS enzymes. This inactive short isoform, however, inhibited the activity of the long isoform when co-expressed in COS-1 cells. This result is the first evidence that in vivo long-chain ACS proteins can act as oligomeric complexes, probably as dimers. It also provides the first indication of the regulatory function of inactive spliced isoforms identified for many members of the long-chain and very-long chain family in mammals. Thus, there is a strong need for in vivo studies of the activity of the ACSL enzymes and extreme caution appears warranted when attempting to interpret results of the expression of a single isoform.
Conclusion
In the last 20 years, major progress has been made in the identification and characterization of members of the acyl-CoA synthetase family of enzymes. These proteins play an essential role in lipid synthesis, fatty acid catabolism, and remodeling of membranes. The current knowledge, as reviewed here, of the large family of mammalian long-chain acyl-CoA synthetases (ACSL), which activate fatty acids with chain lengths of 12 to 20 carbon atoms, indicates that many questions still remain to be answered to better understand their localization and specific modes of action in different sites of the cell. Given the many different (spliced) isoforms it is essential that a uniform nomenclature and description be available for researchers studying these proteins. Failure to do so will lead to confusion as is indicated. It can be anticipated that the structure of the mammalian forms of these proteins will be solved in the next few years. This in turn may shed light on their action as homo- or heterodimers at the membrane interface, and their interaction with other proteins as well as with the lipid bilayer. However, as is the case with other proteins that act on membranes at the water lipid interface, accurate assessment of their activity will be a major challenge. These proteins use fatty acids as substrate and generate a product that potentially can alter the membrane environment. Hence, enzyme kinetics will be difficult to assess properly. In addition, expression in model systems and purification/reconstitution will inevitably render an artificial environment, very different compared to their cellular environment. This will affect their activity and make comparison with their action in their natural environment difficult to interpret. Many questions remain with respect to the cellular localization and specific biological function of the different forms of these enzymes. Such functions will become more into focus as we extend our knowledge on the structure function relationship of these enzymes as well as their interaction with membrane lipids and other acyl-CoA requiring enzymes such as the acyl-CoA lysophospholipid acyltransferases. It is reasonable to assume that different forms of these enzymes exert tissue specific functions by generating specific fatty acyl-CoA thioesters that play a role in anabolic or catabolic reactions. Tissue and species specific modulation of cellular metabolism seems needed to regulate function of organs such as the adrenal glands, the brain, and reproductive organs (48). New approaches to evaluate the activity of these proteins in well-defined membranes that closely resemble their natural environment are needed.
Acknowledgments
This work was supported by the National Institute of Health, grant NIHIU54 HL070583.
References
- 1.Soupene E, Kuypers FA. Multiple erythroid isoforms of human long-chain acyl-CoA synthetases are produced by switch of the fatty acid gate domains. BMC Mol Biol. 2006;7:21. doi: 10.1186/1471-2199-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujino T, Man-Jong K, Minekura H, Suzuki H, Yamamoto TT. Alternative translation initiation generates acyl-CoA synthetase 3 isoforms with heterogeneous amino termini. J Biochem (Tokyo) 1997;122:212–216. doi: 10.1093/oxfordjournals.jbchem.a021731. [DOI] [PubMed] [Google Scholar]
- 3.Mashek DG, Bornfeldt KE, Coleman RA, Berger J, Bernlohr DA, Black P, DiRusso CC, Farber SA, Guo W, Hashimoto N, Khodiyar V, Kuypers FA, Maltais LJ, Nebert DW, Renieri A, Schaffer JE, Stahl A, Watkins PA, Vasiliou V, Yamamoto TT. Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. J Lipid Res. 2004;45:1958–1961. doi: 10.1194/jlr.E400002-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Van Horn CG, Caviglia JM, Li LO, Wang S, Granger DA, Coleman RA. Characterization of recombinant long-chain rat acyl-CoA synthetase isoforms 3 and 6: identification of a novel variant of isoform 6. Biochemistry. 2005;44:1635–1642. doi: 10.1021/bi047721l. [DOI] [PubMed] [Google Scholar]
- 5.Hisanaga Y, Ago H, Nakagawa N, Hamada K, Ida K, Yamamoto M, Hori T, Arii Y, Sugahara M, Kuramitsu S, Yokoyama S, Miyano M. Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. J Biol Chem. 2004;279:31717–31726. doi: 10.1074/jbc.M400100200. [DOI] [PubMed] [Google Scholar]
- 6.Piccini M, Vitelli F, Bruttini M, Pober BR, Jonsson JJ, Villanova M, Zollo M, Borsani G, Ballabio A, Renierei A. FACL4, a new gene encoding long-chain acyl-CoA synthetase 4, is deleted in a family with Alport syndrome, elliptocytosis, and mental retardation. Genomics. 1998;47:350–358. doi: 10.1006/geno.1997.5104. [DOI] [PubMed] [Google Scholar]
- 7.Longo I, Frints SG, Fryns JP, Meloni I, Pescucci C, Ariani F, Borghgraef M, Raynaud M, Marynen P, Schwartz C, Renieri A, Froyen G. A third MRX family (MRX68) is the result of mutation in the long chain fatty acid-CoA ligase 4 (FACL4) gene: proposal of a rapid enzymatic assay for screening mentally retarded patients. J Med Genet. 2003;40:11–17. doi: 10.1136/jmg.40.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meloni I, Muscettola M, Raynaud M, Longo I, Bruttini M, Moizard MP, Gomot M, Chelly J, des Portes V, Fryns JP, Ropers HH, Magi B, Bellan C, Volpi N, Yntema HG, Lewis SE, Schaffer JE, Renieri A. FACL4, encoding fatty acid-CoA ligase 4, is mutated in nonspecific X-linked mental retardation. Nat Genet. 2002;30:436–440. doi: 10.1038/ng857. [DOI] [PubMed] [Google Scholar]
- 9.Verot L, Alloisio N, Morle L, Bozon M, Touraine R, Plauchu H, Edery P. Localization of a non-syndromic X-linked mental retardation gene (MRX80) to Xq22-q24. Am J Med Genet A. 2003;122:37–41. doi: 10.1002/ajmg.a.20221. [DOI] [PubMed] [Google Scholar]
- 10.Yagasaki F, Jinnai I, Yoshida S, Yokoyama Y, Matsuda A, Kusumoto S, Kobayashi H, Terasaki H, Ohyashiki K, Asou N, Murohashi I, Bessho M, Hirashima K. Fusion of TEL/ETV6 to a novel ACS2 in myelodysplastic syndrome and acute myelogenous leukemia with t(5;12)(q31;p13) Genes Chromosomes Cancer. 1999;26:192–202. doi: 10.1002/(sici)1098-2264(199911)26:3<192::aid-gcc2>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 11.Cools J, Mentens N, Odero MD, Peeters P, Wlodarska I, Delforge M, Hagemeijer A, Marynen P. Evidence for position effects as a variant ETV6-mediated leukemogenic mechanism in myeloid leukemias with a t(4;12)(q11-q12;p13) or t(5;12)(q31;p13) Blood. 2002;99:1776–1784. doi: 10.1182/blood.v99.5.1776. [DOI] [PubMed] [Google Scholar]
- 12.Lee EJ, Kim HC, Cho YY, Byun SJ, Lim JM, Ryoo ZY. Alternative promotion of the mouse acyl-CoA synthetase 6 (mAcsl6) gene mediates the expression of multiple transcripts with 5′-end heterogeneity: genetic organization of mAcsl6 variants. Biochem Biophys Res Commun. 2005;327:84–93. doi: 10.1016/j.bbrc.2004.11.141. [DOI] [PubMed] [Google Scholar]
- 13.Malhotra KT, Malhotra K, Lubin BH, Kuypers FA. Identification and molecular characterization of acyl-CoA synthetase in human erythrocytes and erythroid precursors. Biochem J. 1999;344:135–143. [PMC free article] [PubMed] [Google Scholar]
- 14.Richards MR, Harp JD, Ory DS, Schaffer JE. Fatty acid transport protein 1 and long-chain acyl coenzyme A synthetase 1 interact in adipocytes. J Lipid Res. 2006;47:665–672. doi: 10.1194/jlr.M500514-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Lewin TM, Van Horn CG, Krisans SK, Coleman RA. Rat liver acyl-CoA synthetase 4 is a peripheral-membrane protein located in two distinct subcellular organelles, peroxisomes, and mitochondrial-associated membrane. Arch Biochem Biophys. 2002;404:263–270. doi: 10.1016/s0003-9861(02)00247-3. [DOI] [PubMed] [Google Scholar]
- 16.Black PN, DiRusso CC, Metzger AK, Heimert TL. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J Biol Chem. 1992;267:25513–25520. [PubMed] [Google Scholar]
- 17.Kameda K, Nunn WD. Purification and characterization of acyl coenzyme A synthetase from Escherichia coli. J Biol Chem. 1981;256:5702–5707. [PubMed] [Google Scholar]
- 18.Overath P, Pauli G, Schairer HU. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur J Biochem. 1969;7:559–574. [PubMed] [Google Scholar]
- 19.Weimar JD, DiRusso CC, Delio R, Black PN. Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids. J Biol Chem. 2002;277:29369–29376. doi: 10.1074/jbc.M107022200. [DOI] [PubMed] [Google Scholar]
- 20.Lewis SE, Listenberger LL, Ory DS, Schaffer JE. Membrane topology of the murine fatty acid transport protein 1. J Biol Chem. 2001;276:37042–37050. doi: 10.1074/jbc.M105556200. [DOI] [PubMed] [Google Scholar]
- 21.Gargiulo CE, Stuhlsatz-Krouper SM, Schaffer JE. Localization of adipocyte long-chain fatty acyl-CoA synthetase at the plasma membrane. J Lipid Res. 1999;40:881–892. [PubMed] [Google Scholar]
- 22.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 23.Sleeman MW, Donegan NP, Heller-Harrison R, Lane WS, Czech MP. Association of acyl-CoA synthetase-1 with GLUT4-containing vesicles. J Biol Chem. 1998;273:3132–3135. doi: 10.1074/jbc.273.6.3132. [DOI] [PubMed] [Google Scholar]
- 24.Lewin TM, Kim JH, Granger DA, Vance JE, Coleman RA. Acyl-CoA synthetase isoforms 1, 4, and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J Biol Chem. 2001;276:24674–24679. doi: 10.1074/jbc.M102036200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Fu J, Zhou Z. Changes in the brain mitochondrial proteome of male Sprague-Dawley rats treated with manganese chloride. Toxicol Appl Pharmacol. 2005;202:13–17. doi: 10.1016/j.taap.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 28.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 29.Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, Mann M. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- 30.Bae TJ, Kim MS, Kim JW, Kim BW, Choo HJ, Lee JW, Kim KB, Lee CS, Kim JH, Chang SY, Kang CY, Lee SW, Ko YG. Lipid raft proteome reveals ATP synthase complex in the cell surface. Proteomics. 2004;4:3536–3548. doi: 10.1002/pmic.200400952. [DOI] [PubMed] [Google Scholar]
- 31.Tomoda H, Igarashi K, Cyong JC, Omura S. Evidence for an essential role of long chain acyl-CoA synthetase in animal cell proliferation. Inhibition of long chain acyl-CoA synthetase by triacsins caused inhibition of Raji cell proliferation. J Biol Chem. 1991;266:4214–4219. [PubMed] [Google Scholar]
- 32.Coleman RA, Lewin TM, Van Horn CG, Gonzalez-Baro MR. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J Nutr. 2002;132:2123–2126. doi: 10.1093/jn/132.8.2123. [DOI] [PubMed] [Google Scholar]
- 33.Caviglia JM, Li LO, Wang S, DiRusso CC, Coleman RA, Lewin TM. Rat long chain acyl-CoA synthetase 5, but not 1, 2, 3, or 4, complements Escherichia coli fadD. J Biol Chem. 2004;279:11163–11169. doi: 10.1074/jbc.M311392200. [DOI] [PubMed] [Google Scholar]
- 34.Kim JH, Lewin TM, Coleman RA. Expression and characterization of recombinant rat Acyl-CoA synthetases 1, 4, and 5. J Biol Chem. 2001;276:24667–24673. doi: 10.1074/jbc.M010793200. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka T, Hosaka K, Hoshimaru M, Numa S. Purification and properties of long-chain acyl-coenzyme-A synthetase from rat liver. Eur J Biochem. 1979;98:165–172. doi: 10.1111/j.1432-1033.1979.tb13173.x. [DOI] [PubMed] [Google Scholar]
- 36.Polokoff MA, Bell RM. Millipore filter assay for long-chain fatty acid: CoASH ligase activity using 3H-labeled coenzyme A. J Lipid Res. 1975;16:397–402. [PubMed] [Google Scholar]
- 37.Hosaka K, Mishina M, Tanaka T, Kamiryo T, Numa S. Acyl-coenzyme-A synthetase I from Candida lipolytica. Purification, properties and immunochemical studies. Eur J Biochem. 1979;93:197–203. doi: 10.1111/j.1432-1033.1979.tb12811.x. [DOI] [PubMed] [Google Scholar]
- 38.Ichihara K, Shibasaki Y. An enzyme-coupled assay for acyl-CoA synthetase. J Lipid Res. 1991;32:1709–1712. [PubMed] [Google Scholar]
- 39.Lageweg W, Steen I, Tager JM, Wanders RJ. A fluorimetric assay for acyl-CoA synthetase activities. Anal Biochem. 1991;197:384–388. doi: 10.1016/0003-2697(91)90408-l. [DOI] [PubMed] [Google Scholar]
- 40.Wu P, Bremer J. Activation of alkylthioacrylic acids in subcellular fractions of rat tissues: a new spectrophotometric method for assay of acyl-CoA synthetase. Biochim Biophys Acta. 1994;1215:87–92. doi: 10.1016/0005-2760(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki H, Kawarabayasi Y, Kondo J, Abe T, Nishikawa K, Kimura S, Hashimoto T, Yamamoto T. Structure and regulation of rat long-chain acyl-CoA synthetase. J Biol Chem. 1990;265:8681–8685. [PubMed] [Google Scholar]
- 42.Yamakawa A, Nishizawa M, Fujiwara KT, Kawai S, Kawasaki H, Suzuki K, Takenawa T. Molecular cloning and sequencing of cDNA encoding the phosphatidylinositol kinase from rat brain. J Biol Chem. 1991;266:17580–17583. [PubMed] [Google Scholar]
- 43.Fujino T, Kang MJ, Suzuki H, Iijima H, Yamamoto T. Molecular characterization and expression of rat acyl-CoA synthetase 3. J Biol Chem. 1996;271:16748–16752. doi: 10.1074/jbc.271.28.16748. [DOI] [PubMed] [Google Scholar]
- 44.Iijima H, Fujino T, Minekura H, Suzuki H, Kang MJ, Yamamoto T. Biochemical studies of two rat acyl-CoA synthetases, ACS1 and ACS2. Eur J Biochem. 1996;242:186–190. doi: 10.1111/j.1432-1033.1996.0186r.x. [DOI] [PubMed] [Google Scholar]
- 45.Kang MJ, Fujino T, Sasano H, Minekura H, Yabuki N, Nagura H, Iijima H, Yamamoto TT. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc Natl Acad Sci U S A. 1997;94:2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oikawa E, Iijima H, Suzuki T, Sasano H, Sato H, Kamataki A, Nagura H, Kang MJ, Fujino T, Suzuki H, Yamamoto TT. A novel acyl-CoA synthetase, ACS5, expressed in intestinal epithelial cells and proliferating preadipocytes. J Biochem (Tokyo) 1998;124:679–685. doi: 10.1093/oxfordjournals.jbchem.a022165. [DOI] [PubMed] [Google Scholar]
- 47.Tong F, Black PN, Coleman RA, DiRusso CC. Fatty acid transport by vectorial acylation in mammals: roles played by different isoforms of rat long-chain acyl-CoA synthetases. Arch Biochem Biophys. 2006;447:46–52. doi: 10.1016/j.abb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Sheng Y, Tang PZ, Tsai-Morris CH, Dufau ML. Tissue-cell- and species-specific expression of gonadotropin-regulated long chain acyl-CoA synthetase (GR-LACS) in gonads, adrenal and brain identification of novel forms in the brain. J Steroid Biochem Mol Biol. 2006;98:207–217. doi: 10.1016/j.jsbmb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 49.Wang YL, Guo W, Zang Y, Yaney GC, Vallega G, Getty-Kaushik L, Pilch P, Kandror K, Corkey BE. Acyl coenzyme a synthetase regulation: putative role in long-chain acyl coenzyme a partitioning. Obes Res. 2004;12:1781–1788. doi: 10.1038/oby.2004.221. [DOI] [PubMed] [Google Scholar]
- 50.Foster LJ, De Hoog CL, Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc Natl Acad Sci U S A. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaucher SP, Taylor SW, Fahy E, Zhang B, Warnock DE, Ghosh SS, Gibson BW. Expanded coverage of the human heart mitochondrial proteome using multidimensional liquid chromatography coupled with tandem mass spectrometry. J Proteome Res. 2004;3:495–505. doi: 10.1021/pr034102a. [DOI] [PubMed] [Google Scholar]